A Computer Vision-Based Approach for Tick Identification Using Deep Learning Models
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
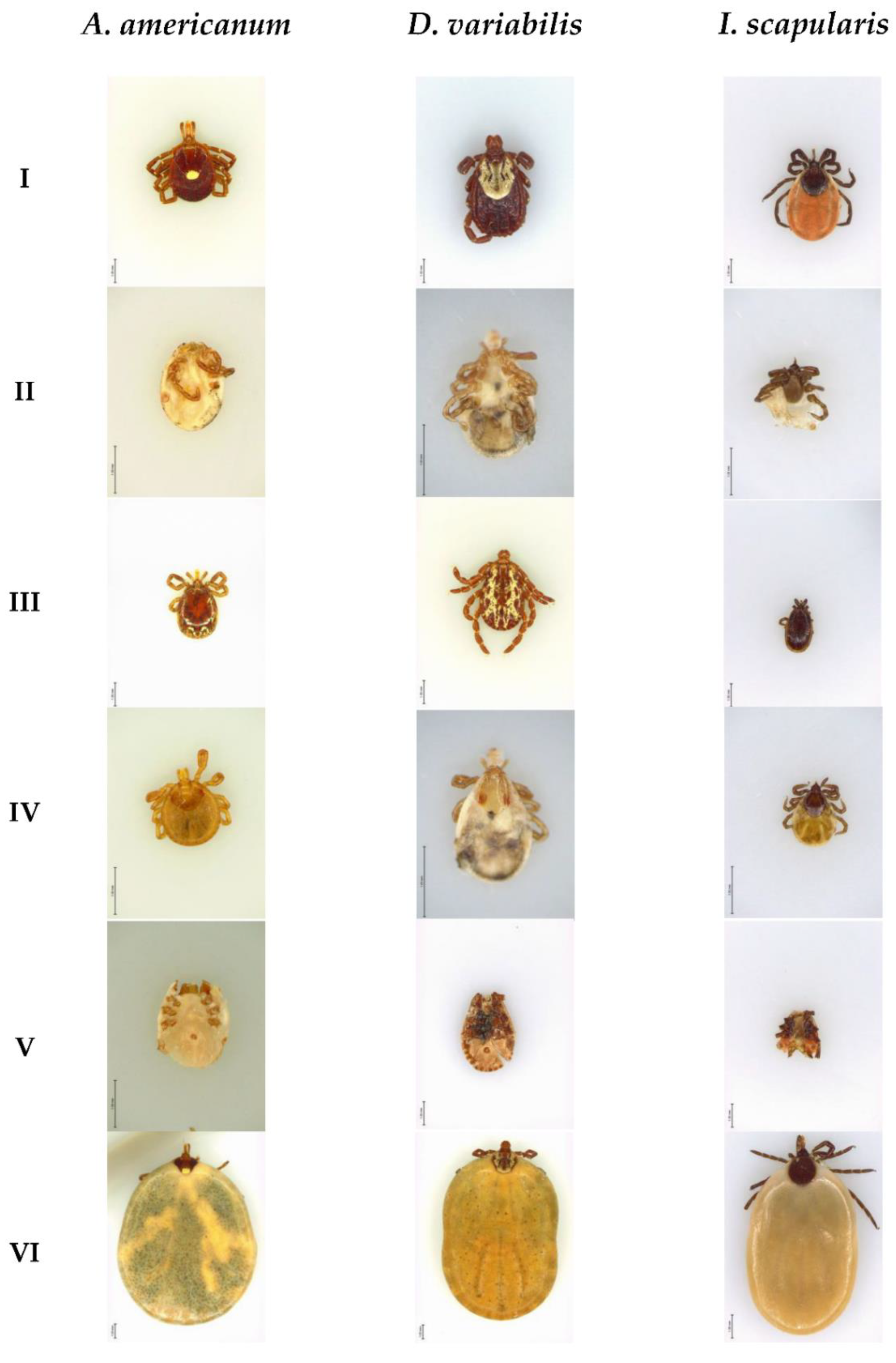
2.1. Data Sources
2.2. DCNN Model Architectures
2.3. The Hardware and Software Environment
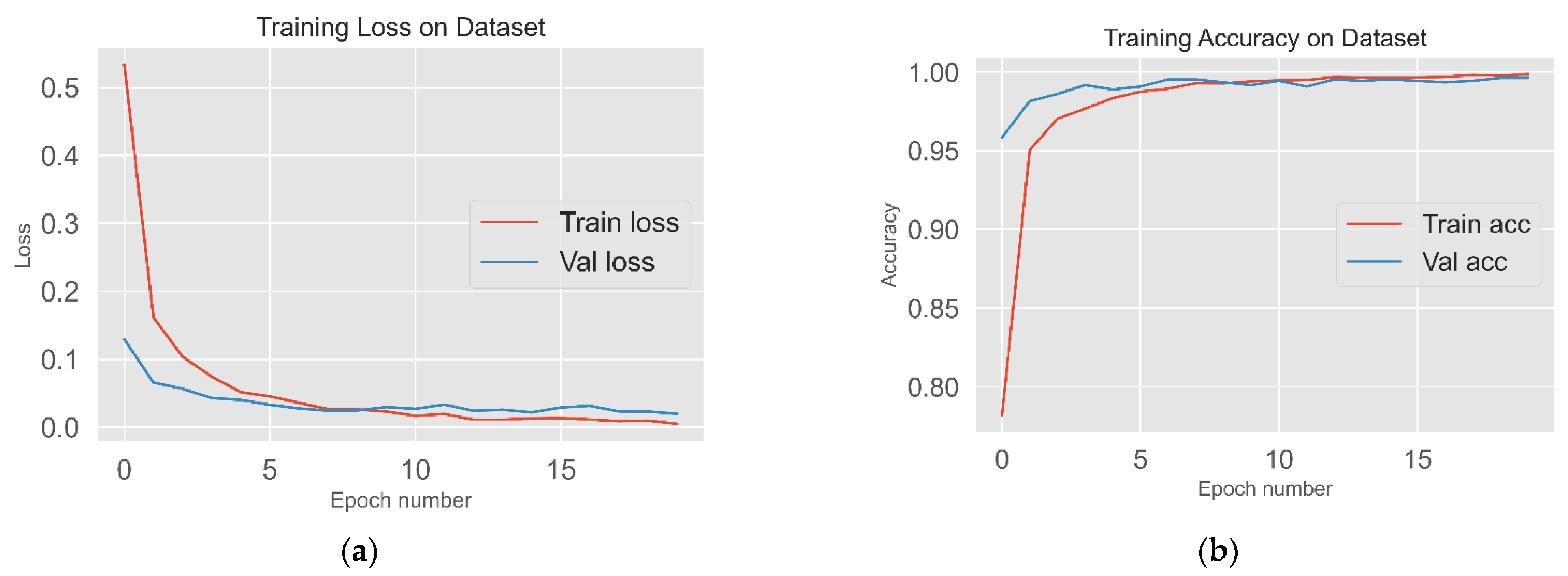
2.4. Training Process
3. Results
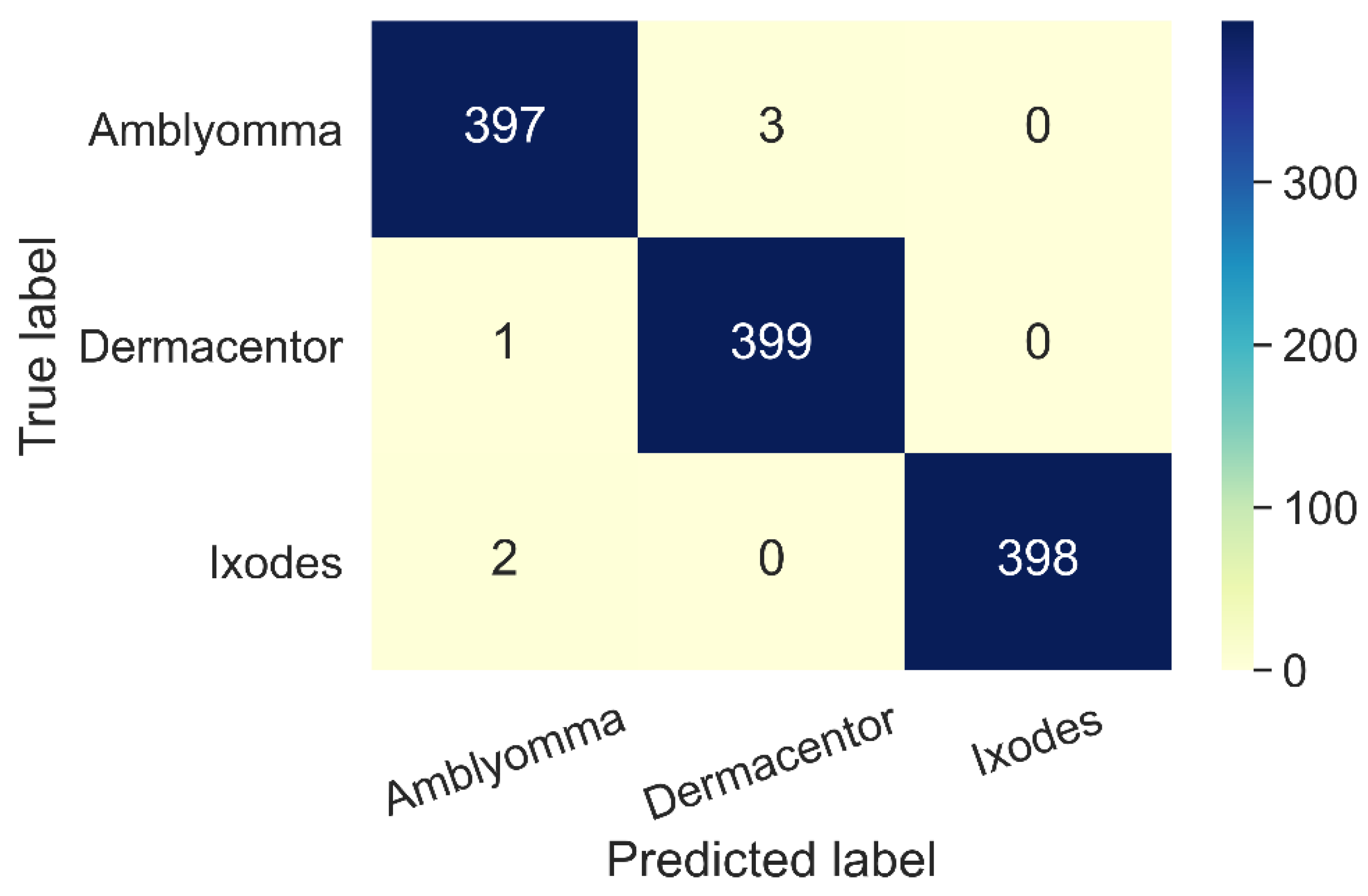
3.1. Performance Metrics
3.2. Classification Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Tickborne Disease Surveillance Data Summary | Ticks | CDC. Available online: https://www.cdc.gov/ticks/data-summary/index.html (accessed on 20 July 2021).
- Rosenberg, R.; Lindsey, N.P.; Fischer, M.; Gregory, C.J.; Hinckley, A.F.; Mead, P.S.; Paz-Bailey, G.; Waterman, S.H.; Drexler, N.A.; Kersh, G.J.; et al. Vital Signs: Trends in Reported Vectorborne Disease Cases—United States and Territories, 2004–2016. Morb. Mortal. Wkly. Rep. 2018, 67, 496–501. [Google Scholar] [CrossRef] [Green Version]
- Dantas-Torres, F.; Chomel, B.B.; Otranto, D. Ticks and tick-borne diseases: A One Health perspective. Trends Parasitol. 2012, 28, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Kopsco, H.L.; Duhaime, R.J.; Mather, T.N. An analysis of companion animal tick encounters as revealed by photograph-based crowdsourced data. Vet. Med. Sci. 2021, 7, 2198–2208. [Google Scholar] [CrossRef]
- Barker, S.C.; Walker, A.R. Ticks of Australia. The Species that Infest Domestic Animals and Humans. Zootaxa 2014, 3816, 1–144. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.R.; Bouattour, A.; Camicas, J.-L.; Estrada-Peña, A.; Horak, I.G.; Latif, A.A.; Pegram, R.G.; Preston, P.M. Ticks of Domestic Animals in Africa: A Guide to Identification of Species; Bioscience Reports: Edinburgh Scotland, UK, 2003; ISBN 095451730X. [Google Scholar]
- Xu, G.; Pearson, P.; Dykstra, E.; Andrews, E.S.; Rich, S.M. Human-Biting Ixodes Ticks and Pathogen Prevalence from California, Oregon, and Washington. Vector-Borne Zoonotic Dis. 2019, 19, 106–114. [Google Scholar] [CrossRef] [Green Version]
- Hemdan, E.E.D.; Shouman, M.A.; Karar, M.E. COVIDX-Net: A Framework of Deep Learning Classifiers to Diagnose COVID-19 in X-ray Images. arXiv 2020, arXiv:2003.11055. [Google Scholar]
- Chilamkurthy, S.; Ghosh, R.; Tanamala, S.; Biviji, M.; Campeau, N.G.; Venugopal, V.K.; Mahajan, V.; Rao, P.; Warier, P. Deep learning algorithms for detection of critical findings in head CT scans: A retrospective study. Lancet 2018, 392, 2388–2396. [Google Scholar] [CrossRef]
- Wen, D.; Wei, Z.; Zhou, Y.; Li, G.; Zhang, X.; Han, W. Deep learning methods to process fmri data and their application in the diagnosis of cognitive impairment: A brief overview and our opinion. Front. Neuroinform. 2018, 12, 23. [Google Scholar] [CrossRef] [Green Version]
- Shifat-E-Rabbi, M.; Yin, X.; Fitzgerald, C.E.; Rohde, G.K. Cell Image Classification: A Comparative Overview. Cytom. Part A 2020, 97, 347–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamilton, N.A.; Pantelic, R.S.; Hanson, K.; Teasdale, R.D. Fast automated cell phenotype image classification. BMC Bioinform. 2007, 8, 110. [Google Scholar] [CrossRef] [Green Version]
- Høye, T.T.; Ärje, J.; Bjerge, K.; Hansen, O.L.P.; Iosifidis, A.; Leese, F.; Mann, H.M.R.; Meissner, K.; Melvad, C.; Raitoharju, J. Deep learning and computer vision will transform entomology. Proc. Natl. Acad. Sci. USA 2021, 118, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Xia, D.; Chen, P.; Wang, B.; Zhang, J.; Xie, C. Insect detection and classification based on an improved convolutional neural network. Sensors 2018, 18, 4169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spiesman, B.J.; Gratton, C.; Hatfield, R.G.; Hsu, W.H.; Jepsen, S.; McCornack, B.; Patel, K.; Wang, G. Assessing the potential for deep learning and computer vision to identify bumble bee species from images. Sci. Rep. 2021, 11, 7580. [Google Scholar] [CrossRef] [PubMed]
- Okayasu, K.; Yoshida, K.; Fuchida, M.; Nakamura, A. Vision-Based Classification of Mosquito Species: Comparison of Conventional and Deep Learning Methods. Appl. Sci. 2019, 9, 3935. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Abd-Elrahman, A.; Morton, J.; Wilhelm, V.L. Comparing fully convolutional networks, random forest, support vector machine, and patch-based deep convolutional neural networks for object-based wetland mapping using images from small unmanned aircraft system. GISci. Remote Sens. 2018, 55, 243–264. [Google Scholar] [CrossRef]
- O’Mahony, N.; Campbell, S.; Carvalho, A.; Harapanahalli, S.; Hernandez, G.V.; Krpalkova, L.; Riordan, D.; Walsh, J. Deep Learning vs. Traditional Computer Vision. Adv. Intell. Syst. Comput. 2020, 943, 128–144. [Google Scholar] [CrossRef] [Green Version]
- Ciregan, D.; Meier, U.; Schmidhuber, J. Multi-column deep neural networks for image classification. In Proceedings of the 2012 IEEE Conference on Computer Vision and Pattern Recognition, Providence, RI, USA, 16–21 June 2012; pp. 3642–3649. [Google Scholar] [CrossRef] [Green Version]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Delving Deep into Rectifiers: Surpassing Human-Level Performance on ImageNet Classificatio. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Santiago, Chile, 7–13 December 2015; pp. 1026–1034. [Google Scholar] [CrossRef] [Green Version]
- Amarathunga, D.C.K.; Grundy, J.; Parry, H.; Dorin, A. Methods of insect image capture and classification: A Systematic literature review. Smart Agric. Technol. 2021, 1, 100023. [Google Scholar] [CrossRef]
- Akbarian, S.; Cawston, T.; Moreno, L.; Patel, S.; Allen, V.; Dolatabadi, E. A computer vision approach to combat lyme disease. arXiv 2020, arXiv:2009.11931. [Google Scholar]
- Omodior, O.; Saeedpour-Parizi, M.R.; Rahman, M.K.; Azad, A.; Clay, K. Using convolutional neural networks for tick image recognition—A preliminary exploration. Exp. Appl. Acarol. 2021, 84, 607–622. [Google Scholar] [CrossRef]
- Anderson, J.F.; Magnarelli, L.A. Biology of Ticks. Infect. Dis. Clin. N. Am. 2008, 22, 195–215. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef] [Green Version]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the Ieee Conference On Computer Vision And Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar] [CrossRef] [Green Version]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar] [CrossRef] [Green Version]
- Xu, G.; Mather, T.N.; Hollingsworth, C.S.; Rich, S.M. Passive Surveillance of Ixodes scapularis (Say), Their Biting Activity, and Associated Pathogens in Massachusetts. Vector-Borne Zoonotic Dis. 2016, 16, 520–527. [Google Scholar] [CrossRef] [Green Version]
- Shorten, C.; Khoshgoftaar, T.M. A survey on Image Data Augmentation for Deep Learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Pan, S.J.; Yang, Q. A Survey on Transfer Learning. IEEE Trans. Knowl. Data Eng. 2009, 22, 1345–1359. [Google Scholar] [CrossRef]
- Yosinski, J.; Clune, J.; Bengio, Y.; Lipson, H. How transferable are features in deep neural networks? Adv. Neural Inf. Process. Syst. 2014, 4, 3320–3328. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Li, F.F. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar] [CrossRef] [Green Version]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef] [Green Version]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper with Convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar] [CrossRef] [Green Version]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. MobileNetV2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar] [CrossRef] [Green Version]
- Kingma, D.P.; Ba, J.L. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M.; et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Distributed Systems. arXiv 2016, arXiv:1603.04467. [Google Scholar]
- Raghu, M.; Zhang, C.; Kleinberg, J.; Bengio, S. Transfusion: Understanding transfer learning for medical imaging. arXiv 2019, arXiv:1902.07208. [Google Scholar]
- Chen, Z.; Zhang, T.; Ouyang, C. End-to-end airplane detection using transfer learning in remote sensing images. Remote Sens. 2018, 10, 139. [Google Scholar] [CrossRef] [Green Version]
- Kopsco, H.L.; Xu, G.; Luo, C.Y.; Rich, S.M.; Mather, T.N. Crowdsourced photographs as an effective method for large-scale passivetick surveillance. J. Med. Entomol. 2020, 57, 1955–1963. [Google Scholar] [CrossRef] [PubMed]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Wäldchen, J.; Mäder, P. Machine learning for image based species identification. Methods Ecol. Evol. 2018, 9, 2216–2225. [Google Scholar] [CrossRef]


| Configuration Item | Value |
|---|---|
| Type and specification | LENOVO ThinkStation P720 Workstation |
| CPU | Intel Xeon Silver 4110 2.10 GHz |
| GPU | NVIDIA GeForce GTX 1080 |
| Memory | 80 GB |
| Hard disk | 1 TB |
| Operating system Image acquisition device | Microsoft Windows 10 Pro Leica S9i stereo microscope |
| Programming language | Python 3.8.5 |
| Deep learning framework | Tensorflow 2.4.1 |
| Architectures | Number of Parameters | Accuracy (SD) | Loss |
|---|---|---|---|
| VGG16 | 138 M | 99.37% (±0.29) | 0.02 |
| ResNet50 | 25.6 M | 99.42% (±0.17) | 0.03 |
| InceptionV3 | 23.8 M | 99.5% (±0.15) | 0.01 |
| DenseNet121 | 8 M | 99.2% (±0.29) | 0.03 |
| MobileNetV2 | 3.5 M | 98.73% (±0.37) | 0.04 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, C.-Y.; Pearson, P.; Xu, G.; Rich, S.M. A Computer Vision-Based Approach for Tick Identification Using Deep Learning Models. Insects 2022, 13, 116. https://doi.org/10.3390/insects13020116
Luo C-Y, Pearson P, Xu G, Rich SM. A Computer Vision-Based Approach for Tick Identification Using Deep Learning Models. Insects. 2022; 13(2):116. https://doi.org/10.3390/insects13020116
Chicago/Turabian StyleLuo, Chu-Yuan, Patrick Pearson, Guang Xu, and Stephen M. Rich. 2022. "A Computer Vision-Based Approach for Tick Identification Using Deep Learning Models" Insects 13, no. 2: 116. https://doi.org/10.3390/insects13020116
APA StyleLuo, C.-Y., Pearson, P., Xu, G., & Rich, S. M. (2022). A Computer Vision-Based Approach for Tick Identification Using Deep Learning Models. Insects, 13(2), 116. https://doi.org/10.3390/insects13020116






