A New Albomycin-Producing Strain of Streptomyces globisporus subsp. globisporus May Provide Protection for Ants Messor structor
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ant Colony Rearing and Microbial Isolation
2.2. 16S rRNA Phylogeny of Isolated Strains
2.3. Genome Features and Phylogenomic Analysis
2.4. Analysis of Bioactive Compound Biosynthetic Gene Clusters
2.5. Phenotypic Characterization
2.6. Biological Activity Testing
2.6.1. Screening of the Antimicrobial Potential
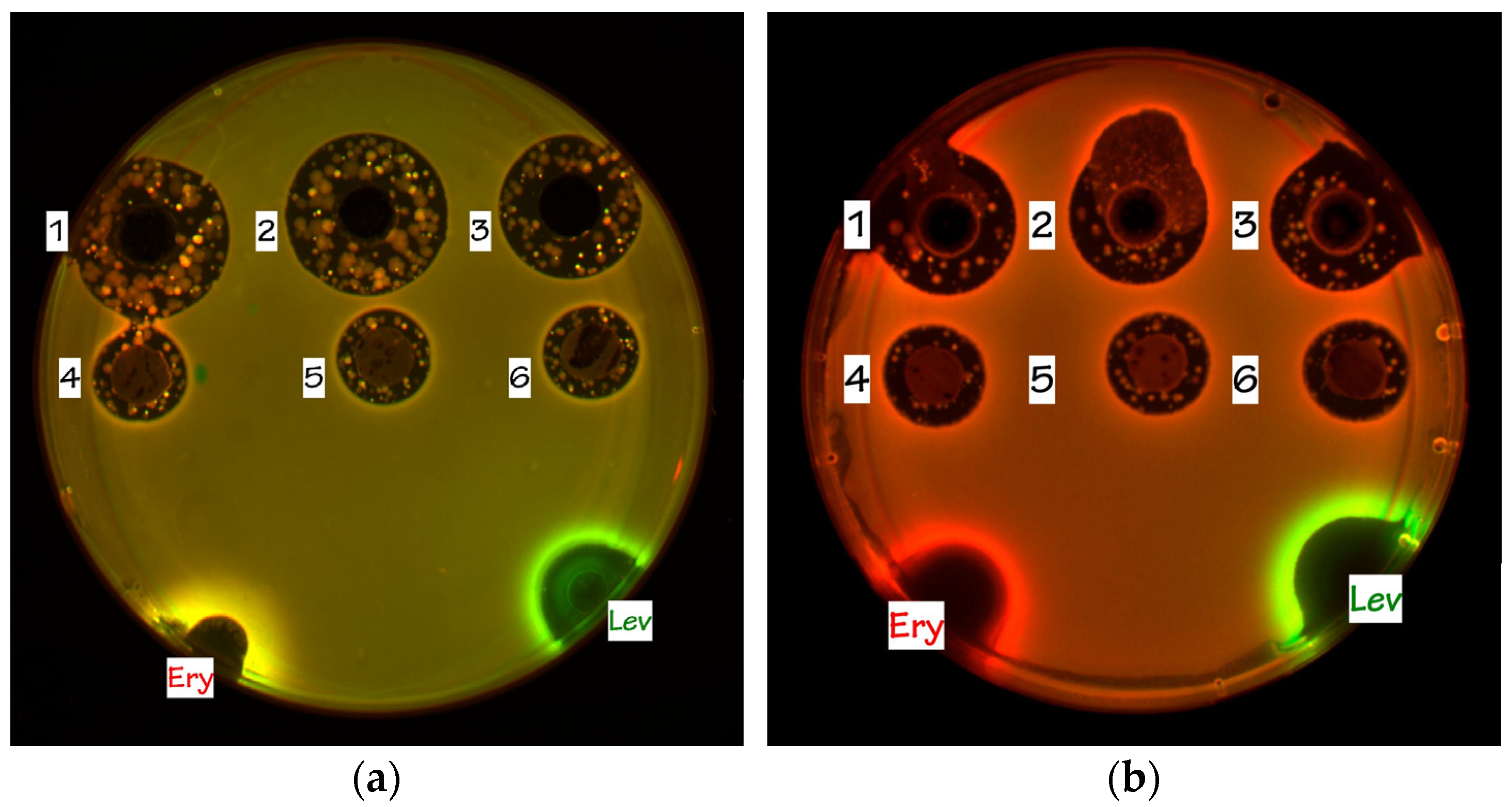
2.6.2. Reporter Assays on Agar Plates
2.6.3. Determination of Minimal Inhibitory Concentration (MIC)
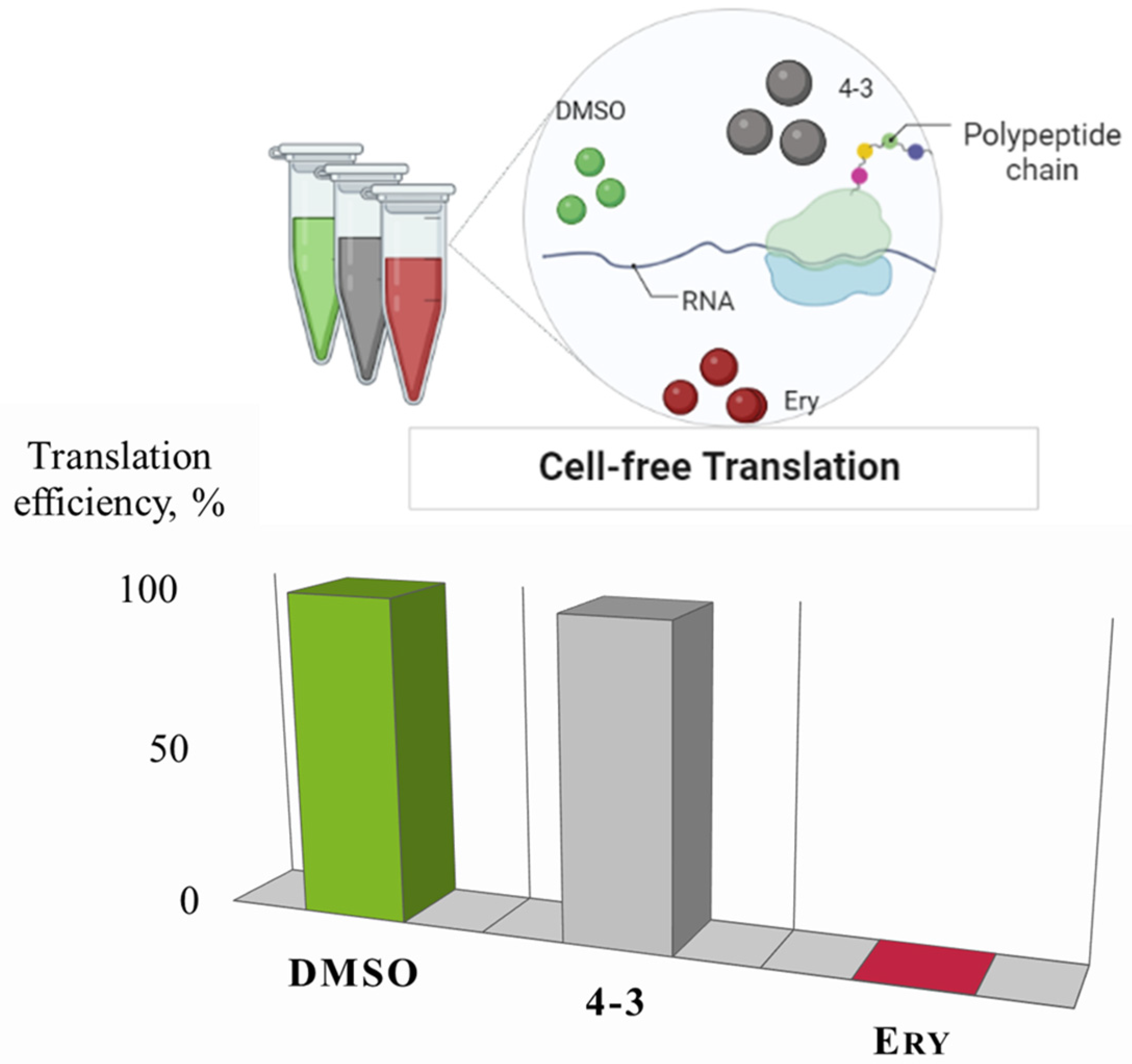
2.6.4. Cell-free Translation
2.7. Purification and Identification of Albomycin
3. Results
3.1. Isolation of Actinobacteria Strains, Associated with Messor Structor Ants
3.2. Genome Features and Phylogenomic Analysis of Streptomyces sp. Strain 4-3
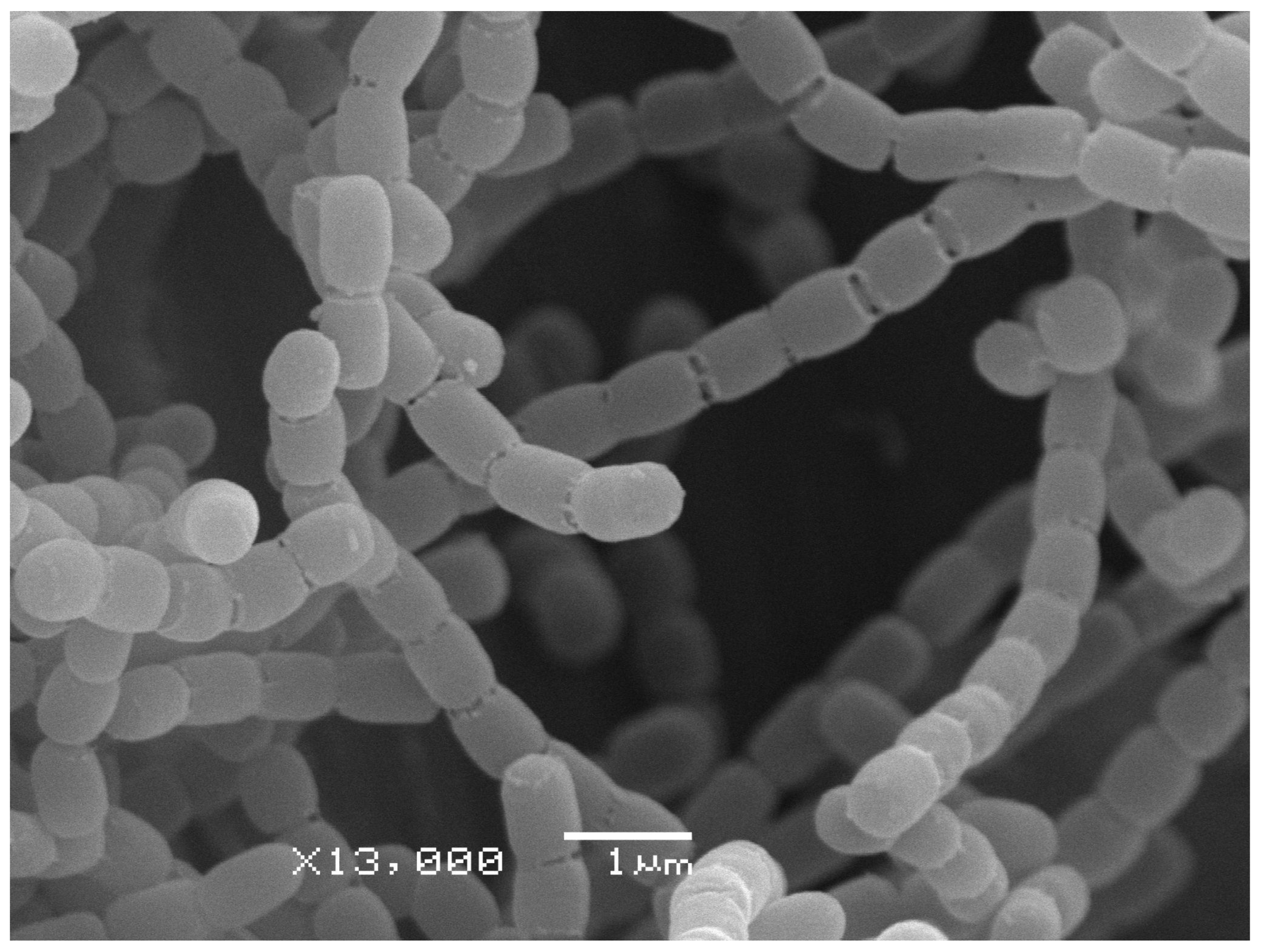
3.3. Phenotypic Characterization of Streptomyces sp. Strain 4-3
3.4. Analysis of 4-3 Bioactive Compound Biosynthetic Gene Clusters
3.5. Screening Antimicrobial Activity
3.6. Analysis of Bioactive Compounds: Cell-free Translation
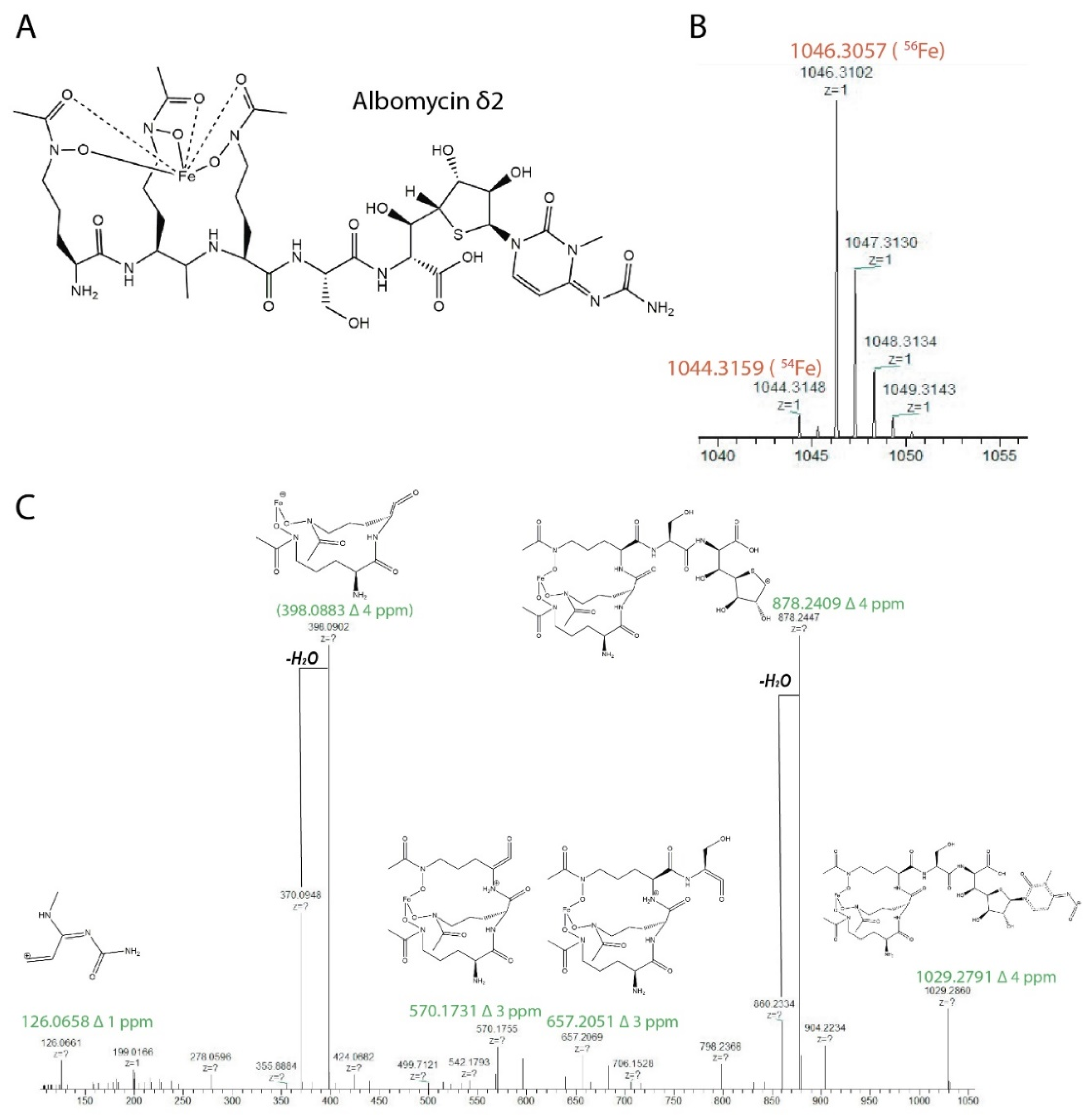
3.7. Purification and Identification of Albomycin
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Levy, S.B.; Marshall, B. Antibacterial resistance worldwide: Causes, challenges and responses. Nat. Med. 2004, 10, S122–S129. [Google Scholar] [CrossRef] [PubMed]
- Larsson, D.G.J.; Flach, C.-F. Antibiotic resistance in the environment. Nat. Rev. Microbiol. 2022, 20, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Razek, A.S.; El-Naggar, M.E.; Allam, A.; Morsy, O.M.; Othman, S.I. Microbial natural products in drug discovery. Processes 2020, 8, 470. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef]
- Dai, J.; Han, R.; Xu, Y.; Li, N.; Wang, J.; Dan, W. Recent progress of antibacterial natural products: Future antibiotics candidates. Bioorg. Chem. 2020, 101, 103922. [Google Scholar] [CrossRef]
- Rosenberg, E.; Zilber-Rosenberg, I. Microbes drive evolution of animals and plants: The hologenome concept. mBio 2016, 7, e01395-15. [Google Scholar] [CrossRef]
- Chevrette, M.G.; Carlson, C.M.; Ortega, H.E.; Thomas, C.; Ananiev, G.E.; Barns, K.J.; Book, A.J.; Cagnazzo, J.; Carlos, C.; Flanigan, W.; et al. The antimicrobial potential of streptomyces from insect microbiomes. Nat. Commun. 2019, 10, 516. [Google Scholar] [CrossRef]
- Wein, T.; Picazo, D.R.; Blow, F.; Woehle, C.; Jami, E.; Reusch, T.B.H.; Martin, W.F.; Dagan, T. Currency, exchange, and inheritance in the evolution of symbiosis. Trends Microbiol. 2019, 27, 836–849. [Google Scholar] [CrossRef]
- Oren, A.; Garrity, G.M.Y. 2021 Valid publication of the names of forty-two phyla of prokaryotes. Int. J. Syst. Evol. Microbiol. 2021, 71, 005056. [Google Scholar] [CrossRef]
- Clay, K. Defensive symbiosis: A microbial perspective. Funct. Ecol. 2014, 28, 293–298. [Google Scholar] [CrossRef]
- Flórez, L.V.; Biedermann, P.H.W.; Engl, T.; Kaltenpoth, M. Defensive symbioses of animals with prokaryotic and eukaryotic microorganisms. Nat. Prod. Rep. 2015, 32, 904–936. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, S.R.; Wojdak, J.M.; Belden, L.K. Defensive symbionts mediate host–parasite interactions at multiple scales. Trends Parasitol. 2017, 33, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Arnam, E.B.V.; Currie, C.R.; Clardy, J. Defense contracts: Molecular protection in insect-microbe symbioses. Chem. Soc. Rev. 2018, 47, 1638–1651. [Google Scholar] [CrossRef] [PubMed]
- Gil, R.; Latorre, A. Unity makes strength: A review on mutualistic symbiosis in representative insect clades. Life 2019, 9, 21. [Google Scholar] [CrossRef] [PubMed]
- Kaltenpoth, M. Actinobacteria as mutualists: General healthcare for insects? Trends Microbiol. 2009, 17, 529–535. [Google Scholar] [CrossRef] [PubMed]
- Seipke, R.F.; Kaltenpoth, M.; Hutchings, M.I. Streptomyces as symbionts: An emerging and widespread theme? FEMS Microbiol. Rev. 2012, 36, 862–876. [Google Scholar] [CrossRef]
- Kaltenpoth, M.; Engl, T. Defensive microbial symbionts in Hymenoptera. Funct. Ecol. 2014, 28, 315–327. [Google Scholar] [CrossRef]
- van der Meij, A.; Worsley, S.F.; Hutchings, M.I.; van Wezel, G.P. Chemical ecology of antibiotic production by Actinomycetes. FEMS Microbiol. Rev. 2017, 41, 392–416. [Google Scholar] [CrossRef]
- Currie, C.R.; Wong, B.; Stuart, A.E.; Schultz, T.R.; Rehner, S.A.; Mueller, U.G.; Sung, G.-H.; Spatafora, J.W.; Straus, N.A. Ancient tripartite coevolution in the Attine ant-microbe symbiosis. Science 2003, 299, 386–388. [Google Scholar] [CrossRef]
- Gause, G.F.; Preobrazhenskaya, T.P.; Sveshnikova, M.A.; Terekhova, L.P.; Maksimova, T.S. Guide for Determination of Actinomycetes: Genera Streptomyces, Streptoverticillium, and Chainia; Nauka: Moscow, Russia, 1983. [Google Scholar]
- Prauser, H.; Falta, R. Phagensensibilität, zellwand-zusammensetzung und taxonomie von actinomyceten. J. Basic Microbiol. 1968, 8, 39–46. [Google Scholar] [CrossRef]
- Zakalyukina, Y.V.; Osterman, I.A.; Wolf, J.; Neumann-Schaal, M.; Nouioui, I.; Biryukov, M.V. Amycolatopsis camponoti sp. nov., new tetracenomycin-producing actinomycete isolated from carpenter ant Camponotus vagus. Antonie Van Leeuwenhoek 2022, 115, 533–544. [Google Scholar] [CrossRef] [PubMed]
- Shirling, E.B.; Gottlieb, D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 1966, 16, 313–340. [Google Scholar] [CrossRef]
- Zakalyukina, Y.V.; Birykov, M.V.; Lukianov, D.A.; Shiriaev, D.I.; Komarova, E.S.; Skvortsov, D.A.; Kostyukevich, Y.; Tashlitsky, V.N.; Polshakov, V.I.; Nikolaev, E.; et al. Nybomycin-producing streptomyces isolated from carpenter ant Camponotus vagus. Biochimie 2019, 160, 93–99. [Google Scholar] [CrossRef]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes de novo assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef] [PubMed]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The Neighbor-Joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: Oxford, UK, 2000; ISBN 0-19-513584-9. [Google Scholar]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Volynkina, I.A.; Zakalyukina, Y.V.; Alferova, V.A.; Belik, A.R.; Yagoda, D.K.; Nikandrova, A.A.; Buyuklyan, Y.A.; Udalov, A.V.; Golovin, E.V.; Kryakvin, M.A.; et al. Mechanism-based approach to new antibiotic producers screening among actinomycetes in the course of civil science. Antibiotics 2022, 11, 1198. [Google Scholar] [CrossRef] [PubMed]
- Osterman, I.A.; Komarova, E.S.; Shiryaev, D.I.; Korniltsev, I.A.; Khven, I.M.; Lukyanov, D.A.; Tashlitsky, V.N.; Serebryakova, M.V.; Efremenkova, O.V.; Ivanenkov, Y.A.; et al. Sorting out antibiotics’ mechanisms of action: A double fluorescent protein reporter for high-throughput screening of ribosome and DNA biosynthesis inhibitors. Antimicrob. Agents Chemother. 2016, 60, 7481–7489. [Google Scholar] [CrossRef] [PubMed]
- Osterman, I.A.; Wieland, M.; Maviza, T.P.; Lashkevich, K.A.; Lukianov, D.A.; Komarova, E.S.; Zakalyukina, Y.V.; Buschauer, R.; Shiriaev, D.I.; Leyn, S.A.; et al. Tetracenomycin X inhibits translation by binding within the ribosomal exit tunnel. Nat. Chem. Biol. 2020, 16, 1071–1077. [Google Scholar] [CrossRef] [PubMed]
- Nouioui, I.; Carro, L.; García-López, M.; Meier-Kolthoff, J.P.; Woyke, T.; Kyrpides, N.C.; Pukall, R.; Klenk, H.-P.; Goodfellow, M.; Göker, M. Genome-based taxonomic classification of the phylum Actinobacteria. Front. Microbiol. 2018, 9, 2007. [Google Scholar] [CrossRef] [PubMed]
- Ciufo, S.; Kannan, S.; Sharma, S.; Badretdin, A.; Clark, K.; Turner, S.; Brover, S.; Schoch, C.L.; Kimchi, A.; DiCuccio, M. Using average nucleotide identity to improve taxonomic assignments in prokaryotic genomes at the NCBI. Int. J. Syst. Evol. Microbiol. 2018, 68, 2386–2392. [Google Scholar] [CrossRef]
- Zeng, Y.; Kulkarni, A.; Yang, Z.; Patil, P.B.; Zhou, W.; Chi, X.; Van Lanen, S.; Chen, S. Biosynthesis of albomycin δ2 provides a template for assembling siderophore and aminoacyl-tRNA synthetase inhibitor conjugates. ACS Chem. Biol. 2012, 7, 1565–1575. [Google Scholar] [CrossRef]
- Ushimaru, R.; Chen, Z.; Zhao, H.; Fan, P.; Liu, H. Identification of the enzymes mediating the maturation of the seryl-tRNA synthetase inhibitor SB-217452 during the biosynthesis of albomycins. Angew. Chem. Int. Ed. 2020, 59, 3558–3562. [Google Scholar] [CrossRef]
- Zeng, Y.; Roy, H.; Patil, P.B.; Ibba, M.; Chen, S. Characterization of two seryl-tRNA synthetases in albomycin-producing Streptomyces sp. strain ATCC 700974. Antimicrob. Agents Chemother. 2009, 53, 4619–4627. [Google Scholar] [CrossRef]
- Gause, G.F. Recent studies on albomycin, a new antibiotic. Br. Med. J. 1955, 2, 1177–1179. [Google Scholar] [CrossRef]
- Travin, D.Y.; Severinov, K.; Dubiley, S. Natural Trojan horse inhibitors of aminoacyl-tRNA synthetases. RSC Chem. Biol. 2021, 2, 468–485. [Google Scholar] [CrossRef]
- Stapley, E.O.; Ormond, R.E. Similarity of albomycin and grisein. Science 1957, 125, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Maehr, H. antibiotics and other naturally occurring hydroxamic acids and hydroxamates. Pure Appl. Chem. 1971, 28, 603–636. [Google Scholar] [CrossRef]
- Lin, Z.; Xu, X.; Zhao, S.; Yang, X.; Guo, J.; Zhang, Q.; Jing, C.; Chen, S.; He, Y. Total synthesis and antimicrobial evaluation of natural albomycins against clinical pathogens. Nat. Commun. 2018, 9, 3445. [Google Scholar] [CrossRef]
- Benz, G.; Schröder, T.D.; Kurz, J.; Wünsche, C.; Karl, W.; Steffens, G.; Pfitzner, J.D.; Schmidt, D. Constitution of the deferriform of the albomycins Δ1, Δ2 and ϵ. Angew. Chem. 1982, 21, 527–528. [Google Scholar] [CrossRef]
- Li, X.; Lei, X.; Zhang, C.; Jiang, Z.; Shi, Y.; Wang, S.; Wang, L.; Hong, B. Complete genome sequence of Streptomyces globisporus C-1027, the producer of an enediyne antibiotic lidamycin. J. Biotechnol. 2016, 222, 9–10. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-R.; Cho, G.; Jeon, C.-W.; Weller, D.M.; Thomashow, L.S.; Paulitz, T.C.; Kwak, Y.-S. A Mutualistic interaction between streptomyces bacteria, strawberry plants and pollinating bees. Nat. Commun. 2019, 10, 4802. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Ou, P.; Liu, L.; Jin, X. Anti-MRSA Activity of Actinomycin X2 and Collismycin a produced by Streptomyces globisporus WA5-2-37 from the intestinal tract of american cockroach (Periplaneta americana). Front. Microbiol. 2020, 11, 555. [Google Scholar] [CrossRef] [PubMed]
- Arnoldi, K.V. survey of harvester ants of the genus Messor (Hymenoptera, Formicidae) of the fauna of the USSR. Zool. Zh. 1977, 56, 1637–1648. [Google Scholar] [CrossRef]
- Kipyatkov, V.E. The World of Social Insects, 2nd ed.; Leningrad University Press: Leningrad, Russia, 1991; ISBN 5-288-00376-9. [Google Scholar]
- Pramanik, A.; Stroeher, U.H.; Krejci, J.; Standish, A.J.; Bohn, E.; Paton, J.C.; Autenrieth, I.B.; Braun, V. Albomycin is an effective antibiotic, as exemplified with Yersinia enterocolitica and Streptococcus pneumoniae. Int. J. Med. Microbiol. 2007, 297, 459–469. [Google Scholar] [CrossRef]


| Subject Strain | ANI,% | dDDH (in %) | G + C Content Difference (in %) |
|---|---|---|---|
| Streptomyces globisporus subsp. globisporus DSM 40136 | 99.3 | 96.3 | 0.14 |
| Streptomyces globisporus subsp. globisporus DSM 40199T | 99.4 | 95.9 | 0.12 |
| Streptomyces rubiginosohelvolus DSM 40176T | 96.3 | 66.2 | 0.18 |
| Streptomyces pluricolorescens DSM 40019T | 96.2 | 66.1 | 0.14 |
| Streptomyces parvus NRRL B-1455T | 94.8 | 56.2 | 0.01 |
| Streptomyces parvus JCM 4069T | 94.7 | 55.7 | 0.05 |
| Streptomyces sindenensis JCM 4164T | 94.2 | 52.9 | 0.25 |
| Streptomyces badius JCM 4350T | 94.2 | 52.5 | 0.11 |
| Streptomyces anulatus JCM 4721T | 92.1 | 41.8 | 0.15 |
| Media | 1 | 2 | 3 |
|---|---|---|---|
| Yeast extract-malt extract (ISP 2) | |||
| Growth | good | good | good |
| Aerial spore-mass color | oyster white | oster white | cream |
| Substrate mycelial color | beige | ochre yellow | beige |
| Soluble pigment | none | none | none |
| Oatmeal (ISP 3) | |||
| Growth | good | good | good |
| Aerial spore-mass color | oyster white | oster white | cream |
| Substrate mycelial color | brown beige | ivory | beige |
| Soluble pigment | brown beige | none | none |
| Inorganic salts-starch (ISP 4) | |||
| Growth | good | good | good |
| Aerial spore-mass color | white | light gray | sparse |
| Substrate mycelial color | colorless | green brown | beige |
| Soluble pigment | none | none | none |
| Glycerol-asparagine (ISP 5) | |||
| Growth | good | good | good |
| Aerial spore-mass color | light olive | none | none |
| Substrate mycelial color | sand yellow | ivory | beige |
| Soluble pigment | sand yellow | none | none |
| Peptone-yeast extract iron (ISP 6) | |||
| Growth | good | good | good |
| Aerial spore-mass color | white | none | none |
| Substrate mycelial color | beige | sand yellow | beige |
| Soluble pigment | none | none | none |
| Tyrosine (ISP 7) | |||
| Growth | weak | good | good |
| Aerial spore-mass color | ivory | none | cream |
| Substrate mycelial color | yellow–red | beige | beige |
| Soluble pigment | none | none | none |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zakalyukina, Y.V.; Pavlov, N.A.; Lukianov, D.A.; Marina, V.I.; Belozerova, O.A.; Tashlitsky, V.N.; Guglya, E.B.; Osterman, I.A.; Biryukov, M.V. A New Albomycin-Producing Strain of Streptomyces globisporus subsp. globisporus May Provide Protection for Ants Messor structor. Insects 2022, 13, 1042. https://doi.org/10.3390/insects13111042
Zakalyukina YV, Pavlov NA, Lukianov DA, Marina VI, Belozerova OA, Tashlitsky VN, Guglya EB, Osterman IA, Biryukov MV. A New Albomycin-Producing Strain of Streptomyces globisporus subsp. globisporus May Provide Protection for Ants Messor structor. Insects. 2022; 13(11):1042. https://doi.org/10.3390/insects13111042
Chicago/Turabian StyleZakalyukina, Yuliya V., Nikolay A. Pavlov, Dmitrii A. Lukianov, Valeria I. Marina, Olga A. Belozerova, Vadim N. Tashlitsky, Elena B. Guglya, Ilya A. Osterman, and Mikhail V. Biryukov. 2022. "A New Albomycin-Producing Strain of Streptomyces globisporus subsp. globisporus May Provide Protection for Ants Messor structor" Insects 13, no. 11: 1042. https://doi.org/10.3390/insects13111042
APA StyleZakalyukina, Y. V., Pavlov, N. A., Lukianov, D. A., Marina, V. I., Belozerova, O. A., Tashlitsky, V. N., Guglya, E. B., Osterman, I. A., & Biryukov, M. V. (2022). A New Albomycin-Producing Strain of Streptomyces globisporus subsp. globisporus May Provide Protection for Ants Messor structor. Insects, 13(11), 1042. https://doi.org/10.3390/insects13111042








