Comparative Analysis of Mitogenomes among Five Species of Filchnerella (Orthoptera: Acridoidea: Pamphagidae) and Their Phylogenetic and Taxonomic Implications
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample and DNA Extraction
2.2. PCR Amplification and Sequencing
2.3. Sequence Assembly, Annotations and Analysis
2.4. Phylogenetic Analysis
3. Results and Discussion
3.1. Genome Structure
3.2. Nucleotide Composition
3.3. Protein-Coding Genes and Codon Usage
3.4. Transfer and Ribosomal RNA Genes
3.5. Ka/Ks of 12 Grassoppers of Filchnerella
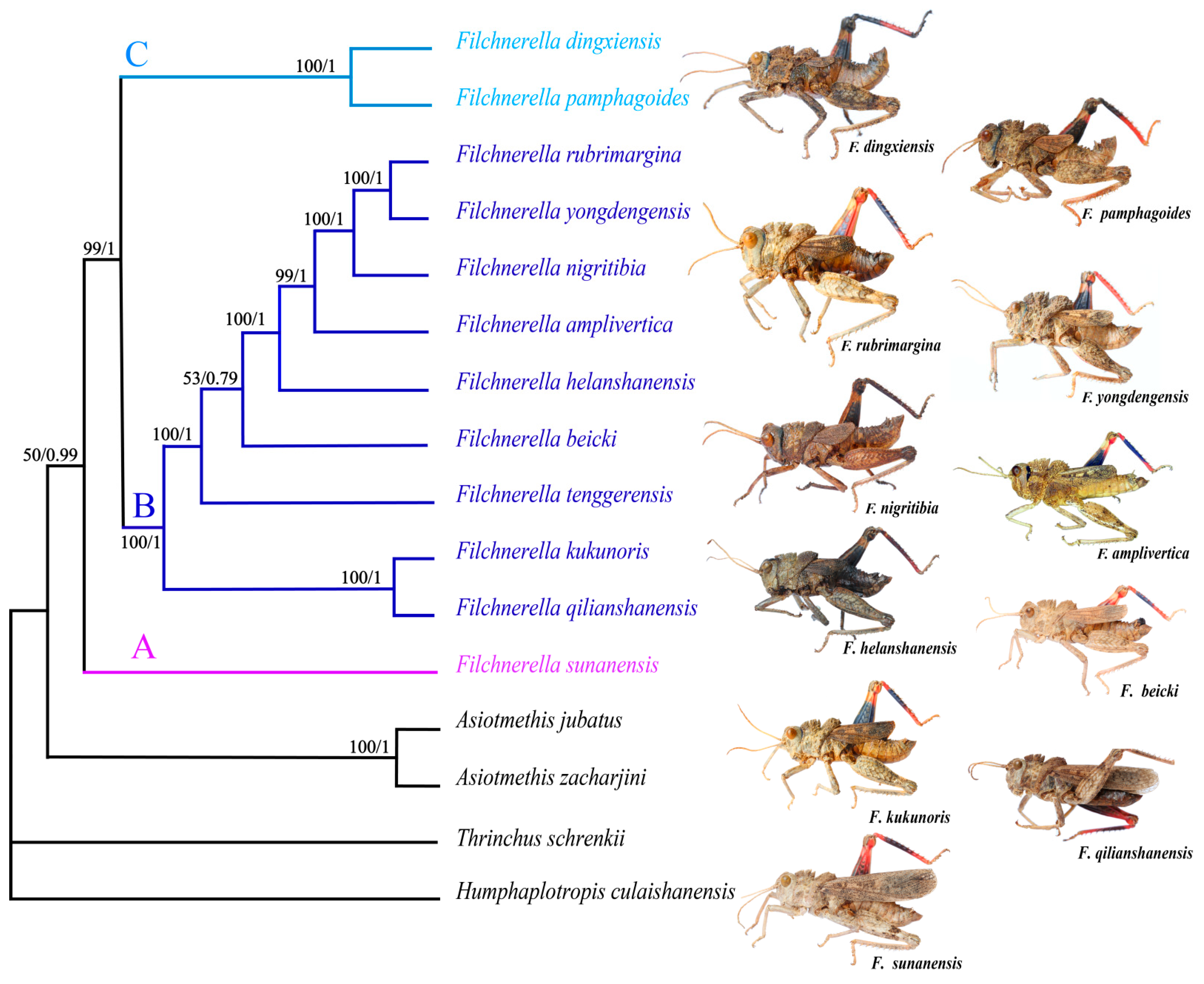
3.6. Phylogenetic Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cao, C.Q.; Li, X.J.; Yin, Z. A new species and key to all known species of the genus Filchnerella Karny, 1908 from China (Orthoptera: Acridoidea, Pamphagidae). Zootaxa 2018, 4413, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.H.; Qiu, Z.Y.; Yuan, H.; Wang, X.Y.; Li, X.J.; Sun, H.M.; Guo, X.Q.; Lu, Y.C.; Feng, X.L.; Majid, M.; et al. Evolutionary rates of and selective constraints on the mitochondrial genomes of Orthoptera insects with different wing types. Mol. Phylogenet. Evol. 2020, 145, 106734. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.C. On the taxonomic system of Acridoidea from China. Acta Biol. Plateau Sin. 1982, 1, 69–99. [Google Scholar]
- Chen, L.P.; Zheng, F.Y.; Bai, J.; Wang, J.M.; Lv, C.Y.; Li, X.; Zhi, Y.C.; Li, X.J. Comparative analysis of mitogenomes among six species of grasshoppers (Orthoptera: Acridoidea: Catantopidae) and their phylogenetic implications in wing-type evolution. Int. J. Biol. Macromol. 2020, 159, 1062–1072. [Google Scholar] [CrossRef]
- Vladimir, H.; Ivan, C.; Marek, E. Was the mitochondrion necessary to start eukaryogenesis? Trends Microbiol. 2019, 27, 96–104. [Google Scholar]
- Flook, P.K.; Rowell, C.H.F.; Gellissen, G. The sequence, organization, and evolution of the Locusta migratoria mitochondrial genome. J. Mol. Evol. 1995, 41, 928–941. [Google Scholar] [CrossRef]
- Zhang, D.X.; Hewitt, G.M. Insect mitochondrial control region: A review of its structure, evolution and usefulness in evolutionary studies. Biochem. Syst. Ecol. 1997, 25, 99–120. [Google Scholar] [CrossRef]
- Curole, J.P.; Kocher, T.D. Mitogenomics: Digging deeper with complete mitochondrial genomes. Trends Ecol. Evol 1999, 14, 394–398. [Google Scholar] [CrossRef]
- Wang, K.; Li, X.K.; Ding, S.M.; Wang, N.; Mao, M.; Wang, M.Q.; Yang, D. The complete mitochondrial genome of the Atylotus miser (Diptera: Tabanomorpha: Tabanidae), with mitochondrial genome phylogeny of lower Brachycera (Orthorrhapha). Gene 2016, 586, 184–196. [Google Scholar] [CrossRef]
- Cheng, X.F.; Zhang, L.P.; Yu, D.N.; Storey, K.B.; Zhang, J.Y. The complete mitochondrial genomes of four cockroaches (Insecta: Blattodea) and phylogenetic analyses within cockroaches. Gene 2016, 586, 115–122. [Google Scholar] [CrossRef]
- Liu, D.F.; Jing, G. The application of nuclear genes sequences in insect molecular systematics. Acta Zootaxono Sin. 2005, 30, 484–492. [Google Scholar]
- Khalid, M.; Hussain, T.; Farooq, Z.; Abbas, K.; Babar, M.E. Mitochondrial Cyt-b and Cox1 genes based molecular diversity and phylogenetic analysis of chukar partridge (Alectoris chukar). Pak. J. Zool. 2020, 52, 1627–1630. [Google Scholar] [CrossRef]
- Ibis, O.; Selcuk, A.Y.; Sacks, B.N.; Yildiz, B.; Ozcan, S.; Kefelioglu, H.; Tez, C. Whole mitochondrial genome of long-clawed mole vole (Prometheomys schaposchnikowi) from Turkey, with its phylogenetic relationships. Genomics 2020, 112, 3247–3255. [Google Scholar] [CrossRef]
- Duan, D.Y.; Tang, J.M.; Chen, Z.; Liu, G.H.; Cheng, T.Y. Mitochondrial genome of Amblyomma javanense: A hard tick parasite of the endangered Malayan pangolin (Manis javanica). Med. Vet. Entomol. 2020, 34, 229–235. [Google Scholar] [CrossRef]
- Di, X.C.; Shan, L.C.Y.; Luo, H.; Zhang, B. Complete mitochondrial genome of Rhytidodus viridiflavus (Hemiptera: Cicadellidae: Idiocerinae). Mitochondrial DNA Part B 2020, 5, 1321–1322. [Google Scholar] [CrossRef] [Green Version]
- Flook, P.K.; Rowell, C.H.F. The effectiveness of mitochondrial rRNA gene sequences for the reconstruction of the phylogeny of an insect order (Orthoptera). Mol. Phylogenet. Evol. 1997, 8, 177–192. [Google Scholar] [CrossRef]
- Flook, P.K.; Klee, S.; Rowell, C.H.F. Combined molecular phylogenetic analysis of the Orthoptera (Arthropoda, Insecta) and implications for their higher systematics. Syst. Biol. 1999, 48, 233–253. [Google Scholar] [CrossRef] [Green Version]
- Li, C.X.; Ma, E.B.; Zheng, X.Y. Genetic differentiation of different populations of four locust species in China. Acta Genet. Sin. 2003, 30, 234–244. [Google Scholar]
- Zhang, D.C.; Li, X.J.; Wang, W.Q.; Yin, H.; Yin, Z.; Yin, X.C. Molecular phylogeny of some genera of Pamphagidae (Acridoidea, Orthoptera) from China based on mitochondrial 16S rDNA sequences. Zootaxa 2005, 1103, 41–49. [Google Scholar] [CrossRef]
- Zhang, D.C.; Han, H.Y.; Yin, H.; Li, X.J.; Yin, X.C. Molecular phylogeny of Pamphagidae (Acridoidea, Orthoptera) from China based on mitochondrial cytochrome oxidase II sequences. Insect Sci. 2011, 18, 234–244. [Google Scholar] [CrossRef]
- Zhang, H.L.; Zeng, H.H.; Huang, Y.; Zheng, Z.M. The complete mitochondrial genomes of three grasshoppers, Asiotmethis zacharjini, Filchnerella helanshanensis and Pseudotmethis rubimarginis (Orthoptera: Pamphagidae). Gene 2013, 517, 89–98. [Google Scholar] [CrossRef]
- Li, X.J.; Zhang, D.C.; Wang, W.Q.; Yin, X.C. Molecular phylogeny of six species of Filchnerella based on analyses of the mitochondrial 16S rDNA sequences (Orthoptera: Acridoidea). Acta Zool. Sin. 2005, 51, 138–142. [Google Scholar]
- Zhang, D.C.; Xu, M.Y.; Li, X.J.; Zheng, J.Y. Analyses and Studies of Gene Sequences on the 8 species of Filchnerella based on the Mitochondrial Cytochrome Oxidase II Gene (Orthoptera: Acridoidea: Pamphagidae). Sichuan J. Zool. 2008, 27, 761–763. [Google Scholar]
- Tamura, K.; Aotsuka, T. Rapid isolation method of animal mito-chondrial DNA by the alkaline lysis procedure. Biochem. Genet. 1988, 26, 815–819. [Google Scholar] [CrossRef]
- Aljanabi, S.M.; Martinez, I. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 1997, 25, 4692–4693. [Google Scholar] [CrossRef]
- Ma, C.; Liu, C.X.; Yang, P.C.; Kang, L. The complete mitochondrial genomes of two band-winged grasshoppers, Gastrimargus marmoratus and Oedaleus asiaticus. BMC Genom. 2009, 10, 156–207. [Google Scholar] [CrossRef] [Green Version]
- Li, X.J.; Zhi, Y.C.; Lang, L.; Yin, X.C. The complete mitochondrial genome of Filchnerella beicki Ramme, 1931 (Orthoptera: Acridoidea: Pamphagidae). Mitochondrial DNA 2014, 25, 348–349. [Google Scholar] [CrossRef]
- Li, X.J.; Zhi, Y.C.; Yin, Z.; Yin, X.C. The complete mitochondrial genome of Humphaplotropis culaishanensis sp. nov. (Orthoptera: Acridoidea: Pamphagidae: Pamphaginae). Mitochondrial DNA Part A 2016, 27, 132–133. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.C.; Zhi, Y.C.; Yin, H.; Li, X.J.; Yin, X.C. The complete mitochondrial genome of Thrinchus schrenkii (Orthoptera: Caelifera, Acridoidea, Pamphagidae). Mol. Biol. Rep. 2011, 38, 611–619. [Google Scholar] [CrossRef]
- Li, X.J.; Zhi, Y.C.; Liu, G.J.; Yin, X.C.; Zhang, D.C. The complete mitochondrial genome of Asiotmethis jubatus (Uvarov, 1926) (Orthoptera: Acridoidea: Pamphagidae). Mitochondrial DNA 2015, 26, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The Clustal X window interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, V.K.; Mangalam, A.K.; Dwivedi, S. Primer premier, program for design of degenerate primers from a protein sequence. Biotechniques 1998, 24, 318–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lynnon Corporation, Bioinformatics Solutions. 2011. Available online: http://www.lynnon.com (accessed on 15 December 2017).
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [Green Version]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating Maximum-Likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovli´c, I.; Zhou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2019, 20, 348–355. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Li, H.X.; Yu, X.F.; Yang, M.F. Characterization of two complete mitochondrial genomes of Atkinsoniella (Hemiptera: Cicadellidae: Cicadellinae) and the phylogenetic implications. Insects 2021, 12, 338. [Google Scholar] [CrossRef]
- Yang, M.S.; Song, L.; Zhou, L.; Shi, Y.X.; Song, N.; Zhang, Y.L. Mitochondrial genomes of four satyrine butterflies and phylogenetic relationships of the family Nymphalidae (Lepidoptera: Papilionoidea). Int. J. Biol. Macromol. 2020, 145, 272–281. [Google Scholar] [CrossRef]
- Li, X.Y.; Yan, L.P.; Pape, T.; Gao, Y.Y.; Zhang, D. Evolutionary insights into bot flies (Insecta: Diptera: Oestridae) from comparative analysis of the mitochondrial genomes. Int. J. Biol. Macromol. 2020, 149, 371–380. [Google Scholar] [CrossRef]
- Oliveira, D.C.S.G.; Raychoudhury, R.; Lavrov, D.V.; Werren, J.H. Rapidly evolving mitochondrial genome and directional selection in mitochondrial genes in the parasitic wasp Nasonia (Hymenoptera: Pteromalidae). Mol. Biol. Evol. 2008, 25, 2167–2180. [Google Scholar] [CrossRef] [Green Version]
- Hurst, L.D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 2002, 18, 486–487. [Google Scholar] [CrossRef]
- Yang, Z.; Bielawski, J.R. Statistical methods for detecting molecular adaptation. Trends. Ecol. Evol. 2000, 15, 496–503. [Google Scholar]
- Li, X.J.; Zhang, D.P.; Yin, H.X. Comparative analysis of mitogenomes among three species of Haplotropidini grasshoppers and a new species of the genus Sulcotropis (Orthoptera: Acridoidea: Pamphagidae) from China. Zootaxa 2020, 4802, 541–555. [Google Scholar] [CrossRef]
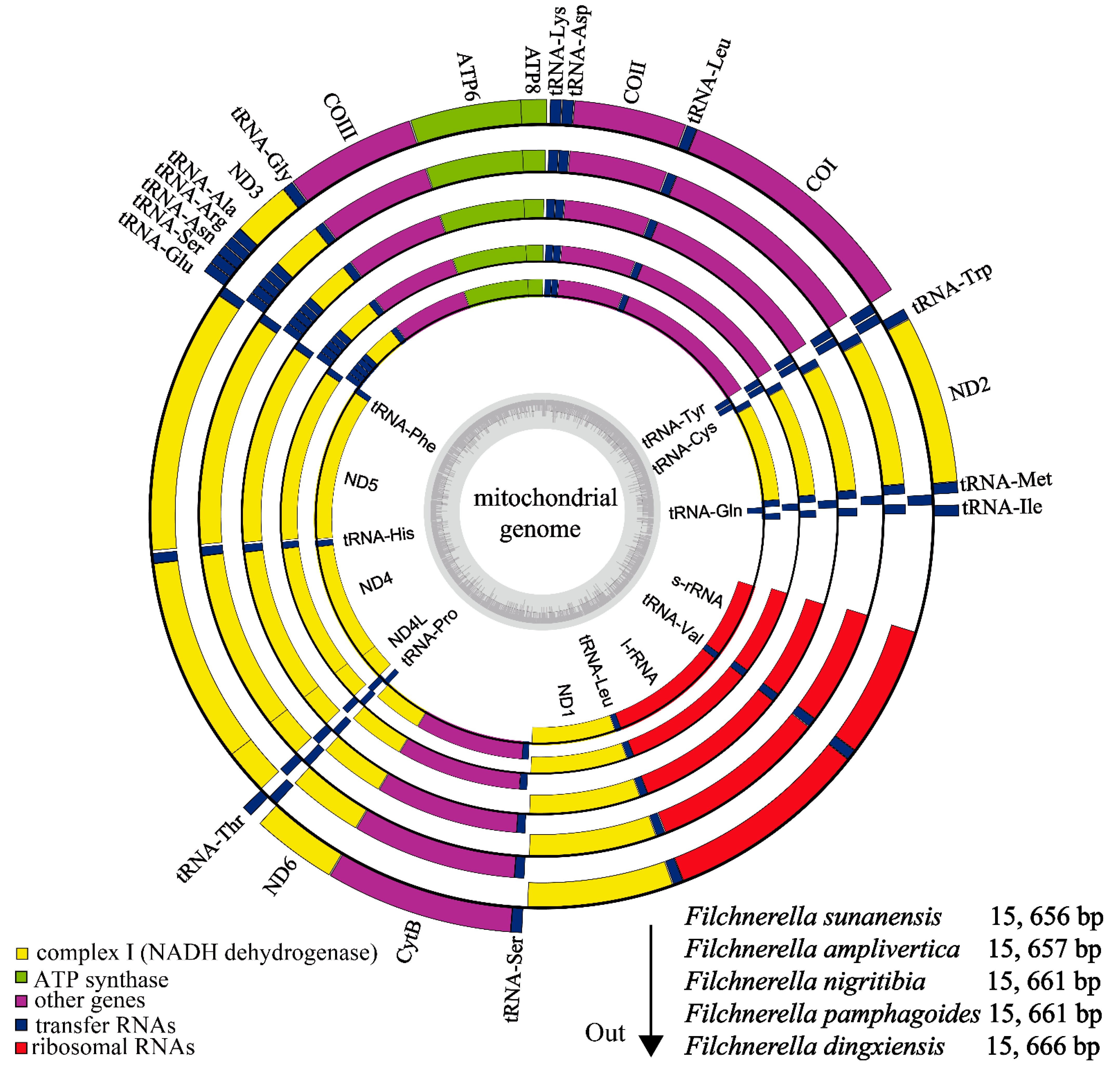
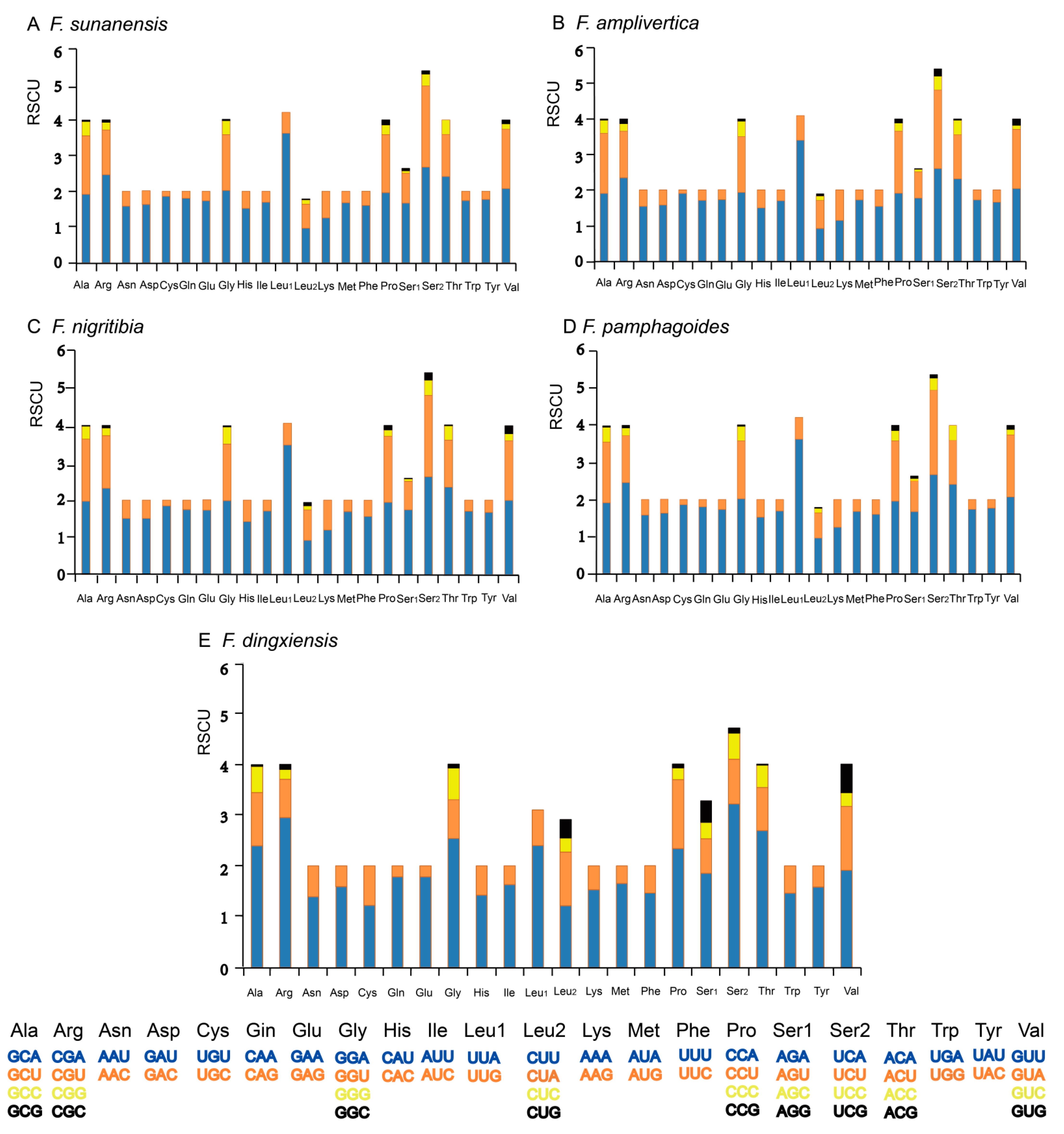
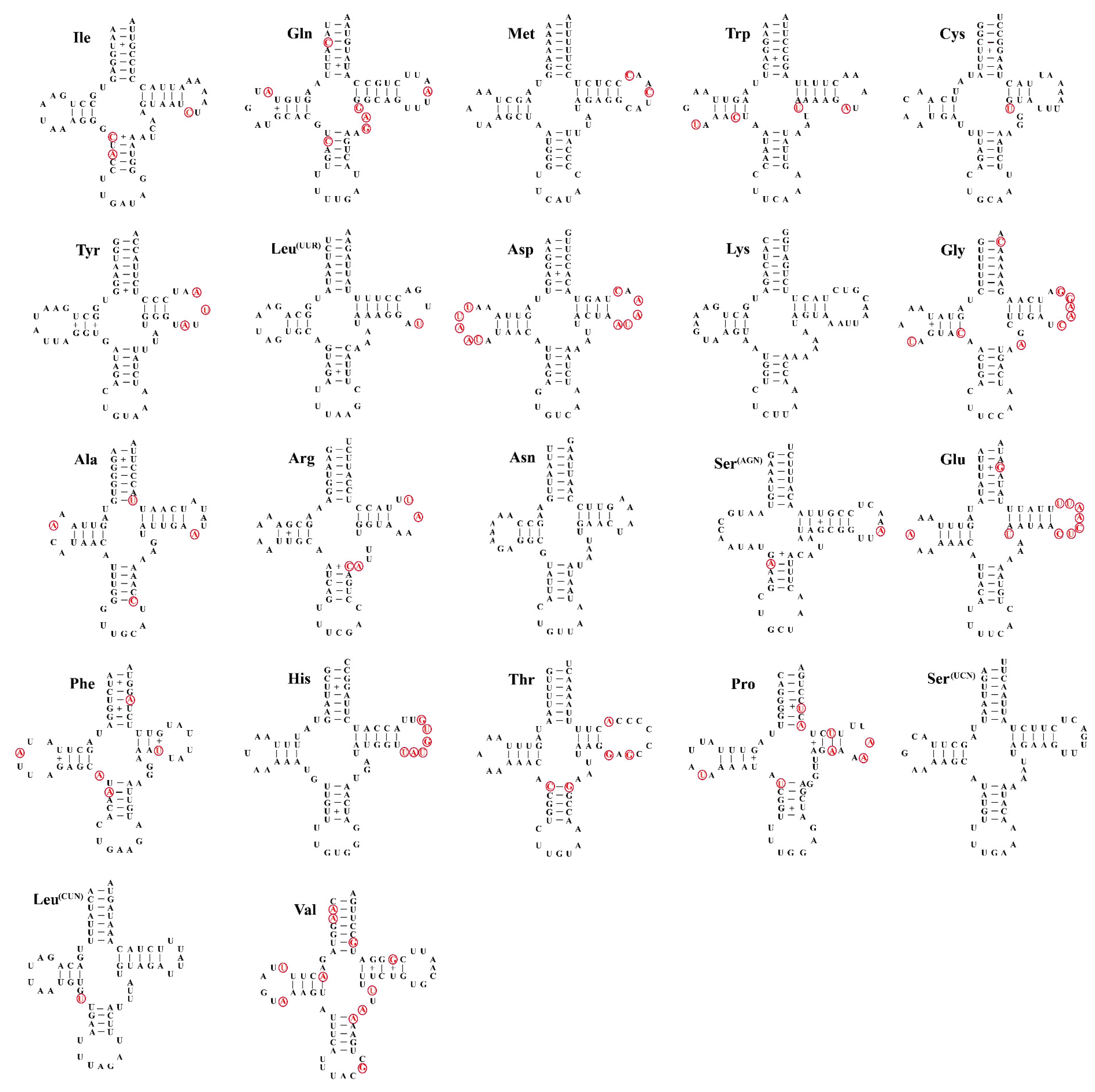
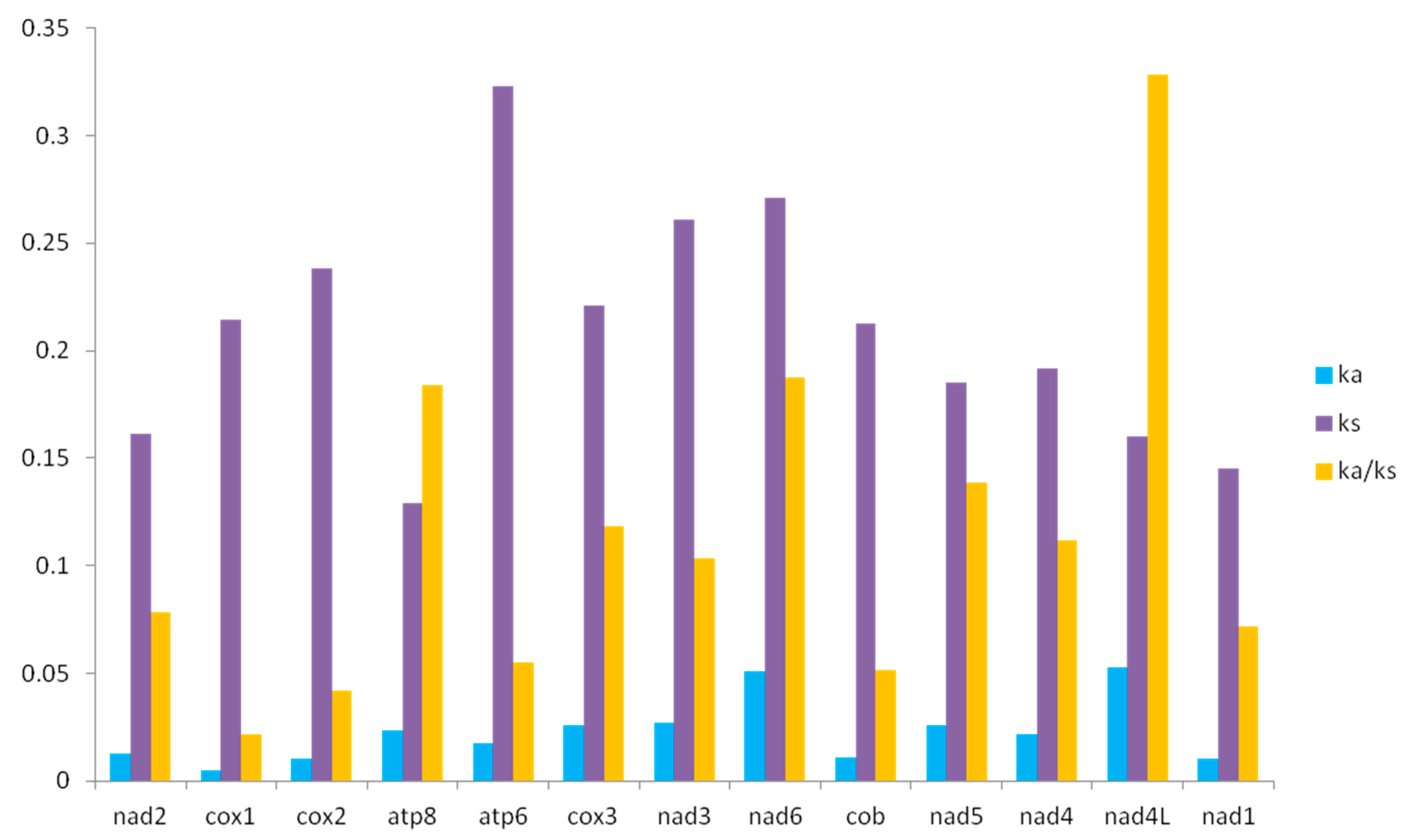
| Species | Collecting Time | Collecting Sites | Collector |
|---|---|---|---|
| Filchnerella amplivertica Li, Zhang & Yin, 2009 | July 2003 | Zhongwei, Ningxia | LI Xin-Jiang, WANG Wen-Qiang |
| Filchnerella pamphagoides Karny, 1908 | September 2003 | Lanzhou, Gansu | ZHANG Dao-Chuan, LI Xin-Jiang |
| Filchnerella sunanensis Liu, 1982 | July 2006 | Sunan, Gansu | LI Xin-Jiang, ZHENG Jin-Yu |
| Filchnerella nigritibia Zheng, 1992 | July 2006 | Dawukou, Ningxia | ZHANG Dao-Chuan, LI Xin-Jiang |
| Filchnerella dingxiensis Zhang, Xiao & Zhi, 2011 | July 2006 | Dingxi, Gansu | ZHANG Dao-Chuan, ZHI Yong-Chao |
| Species | Size (bp) | Accession No | Reference |
|---|---|---|---|
| Filchnerella nigritibia Zheng, 1992 | 15,661 | MZ433420 | This study |
| Filchnerella amplivertica Li, Zhang & Yin, 2009 | 15,657 | MZ433418 | This study |
| Filchnerella pamphagoides Karny, 1908 | 15,661 | MZ433417 | This study |
| Filchnerella sunanensis Liu, 1982 | 15,656 | MZ433421 | This study |
| Filchnerella dingxiensis Zhang, Xiao & Zhi, 2011 | 15,666 | MZ433419 | This study |
| Filchnerella beicki Ramme, 1931 | 15,658 | NC_024923 | [27] |
| Filchnerella helanshanensis Zheng, 1992 | 15,657 | NC_020329 | [21] |
| Filchnerella qilianshanensis Xi & Zheng, 1984 | 15,661 | NC_046558 | Unpublished |
| Filchnerella tenggerensis Zheng & Fu, 1989 | 15,659 | NC_046559 | Unpublished |
| Filchnerellarubrimargina Zheng, 1992 | 15,661 | NC_052733 | Unpublished |
| Filchnerellayongdengensis Xi& Zheng, 1984 | 15674 | MK_903560 | [2] |
| Filchnerellakukunoris Bey-Bienko, 1948 | 15662 | MK_903590 | [2] |
| Humphaplotropis culaishanensis Li, Cao & Yin, 2014 | 15,659 | NC_023535 | [28] |
| Thrinchus schrenkii Fischer von Waldheim, 1846 | 15,672 | NC_014610 | [29] |
| Asiotmethis jubatus (Uvarov, 1926) | 15,669 | NC_025904 | [30] |
| Asiotmethis zacharjini (Bey-Bienko, 1926) | 15,660 | NC_020328 | [21] |
| Gene | Coding Strand | Nucleotide Number | Size (bp) | Intergenic Length | Anticodon | Initiation Codon | Termination Codon |
|---|---|---|---|---|---|---|---|
| tRNAIle | J | 1–68/./././. | 68/./././. | 0/./././. | GAT/./././. | ||
| tRNAGln | N | 69–137/./././. | 69/./././. | −1/./././. | TTG/./././. | ||
| tRNAMet | J | 137–205/./././. | 69/./././. | 0/./././. | CAT/./././. | ||
| ND2 | J | 206–1228/./././. | 1023/./././. | 2/./././. | ATG/./././. | TAG/./././. | |
| tRNATrp | J | 1231–1297/././1231–1298/. | 67/././68/. | −8/./././. | TCA/./././. | ||
| tRNACys | N | 1290–1354/././1291–1355/. | 65/./././. | 8/11/9/8/. | GCA/./././. | ||
| tRNATyr | N | 1363–1428/1364–1429/././. | 66/./././. | 1/./././. | GTA/./././. | ||
| COI | J | 1430–2963/1431–2964/././. | 1534/./././. | 0/./././. | CCG/./././. | T/./././. | |
| tRNALeu(UUR) | J | 2964–3029/2965–3030/././. | 66/./././. | 4/././5/4 | TAA/./././. | ||
| COII | J | 3034–3717/3035–3718 /3035–3718/3036–3719/. | 684/./././. | 3/./././. | ATG/./././. | TAA/./././T | |
| tRNAAsp | J | 3721–3789/3722–3791/./3723–3793/. | 69/70/./71/. | 2/./././. | GTC/./././. | ||
| tRNALys | J | 3792–3862/3794–3864/3794–3864 /3796–3866/3796–3866 | 71/./././. | 17/./././. | CTT/./././. | ||
| ATP8 | J | 3880–4041/3882–4043/./3884–4045/. | 162/./././. | −7/./././. | ATC/./././. | TAA/./././. | |
| ATP6 | J | 4035–4712/4037–4714/./4039–4716/. | 678/./././. | 3/./././4 | ATG/./././. | TAA/./././. | |
| COIII | J | 4716–5507/4718–5509/. /4720–5511/4721–5512 | 792/./././. | 3/./././. | ATG/./././. | TAA/./././. | |
| tRNAGly | J | 5511–5577/5513–5580/./5515–5581/5516–5582 | 67/68/./67/. | 0/./././. | TCC/./././. | ||
| ND3 | J | 5578–5931/5581–5932/5581–5934 /5582–5935/5583–5936 | 354/352/ 354/./. | −2/0/−2/./. | ATC/./././. | TAG/T /TAG/./. | |
| tRNAAla | J | 5930–5995/5933–5998/. /5934–5999/5935–6000 | 66/./././. | 0/3/././. | TGC/./././. | ||
| tRNAArg | J | 5996–6059/6002–6064/6002–6065 /6003–6066/6004–6067 | 64/63/64/./. | 2/1/./2/. | TCG/./././. | ||
| tRNAAsn | J | 6062–6126/6066–6130/6067–6131 /6069–6133/6070–6134 | 65/./././. | 0/./././. | GTT/./././. | ||
| tRNASer (AGN) | J | 6127–6193/6131–6197/6132–6198 /6134–6200/6135–6201 | 67/./././. | 1/./././. | GCT/./././. | ||
| tRNAGlu | J | 6195–6263/6199–6266/6200–6267 /6202–6268/6203–6269 | 69/68/./67/. | 1/./././. | TTC/./././. | ||
| tRNAPhe | N | 6265–6329/6268–6333/6269–6334 /6270–6335/6271–6336 | 65/66/././. | 0/./././. | GAA/./././. | ||
| ND5 | N | 6330–8046/6334–8050/6335–8051 /6336–8052/6337–8053 | 1717/./././. | 15/./././. | ATT/./././. | T/./././. | |
| tRNAHis | N | 8062–8131/8066–8132/8067–8133 /8068–8135/8069–8136 | 70/67/./68/. | 0/./−1/./. | GTG/./././. | ||
| ND4 | N | 8132–9465/8133–9466/8133–9467 /8135–9469/8136–9470 | 1334/./1335/./. | −7/./././. | ATG/./././. | TA/./TAG/./. | |
| ND4L | N | 9459–9752/9460–9753/9461–9754 /9463–9756/9464–9757 | 294/./././. | 2/./././. | ATG/./././. | TAA/./././. | |
| tRNAThr | J | 9755–9822/9756–9823/9757–9826 /9759–9826/9760–9827 | 68/./70/68/. | 0/./././. | TGT/./././. | ||
| tRNAPro | N | 9823–9888/9824–9889/9827–9893 /./9828–9894 | 66/./67/./. | 2/./././. | TGG/./././. | ||
| ND6 | J | 9891–10412/9892–10413/9896–10417 /9896–10417/9897–10418 | 522/./././. | 3/./././. | GTG/./././. | TAA/./././. | |
| Cytb | J | 10416–11558/10417–11557/10421–11563 /./10422–11564 | 1143/1141/ 1143/./. | −2/0/−2/./. | ATG/./././. | TAG/T /TAG/./. | |
| tRNASer (UCN) | J | 11557–11626/11558–11627/11562–11631/ /./11563–11632 | 70/./././. | 26/./././25 | TGA/./././. | ||
| ND1 | N | 11653–12597/11654–12598/11658–12602 /./. | 945/./././. | 3/./././4 | ATA/./././. | TAG/./././. | |
| tRNALeu(CUN) | N | 12601–12666/12602–12667/12606–12671 /./12607–12672 | 66/./././. | 0/././3/0 | TAG/./././. | ||
| lrRNA | N | 12667–13986/12668–13987/12672–13991 /12675–13988/12673–13993 | 1320/./. /1314/1321 | 0/././2/0 | |||
| tRNAVal | N | 13987–14056/13988–14058/13992–14062 /13991–14061/13994–14064 | 70/71/././. | 0/./././. | TAC/./././. | ||
| srRNA | N | 14057–14908/14059–14909/14063–14914 /14062–14914/14065–14917 | 852/851/852/853/. | 0/./././. | |||
| A+T rich | 14909–15656/14910–15657/14915–15661 /./14918–15666 | 748/./747/./. |
| Species | Regions | T% | C% | A% | G% | Size (bp) | A+T% | AT-Skew | GC-Skew |
|---|---|---|---|---|---|---|---|---|---|
| F. sunanensis | All genes | 30.6 | 16.1 | 42.3 | 11.0 | 15,656.0 | 72.9 | 0.16 | −0.19 |
| rRNAgenes | 29.7 | 16.9 | 43.2 | 10.3 | 2172.0 | 72.9 | 0.19 | −0.24 | |
| tRNA genes | 31.1 | 16.5 | 38.7 | 13.6 | 1482.0 | 69.8 | 0.11 | −0.10 | |
| A+T-rich region | 37.7 | 9.4 | 45.6 | 7.4 | 748.0 | 83.3 | 0.09 | −0.12 | |
| PCGs | 41.4 | 14.0 | 31.2 | 13.5 | 11,182.0 | 72.6 | −0.14 | −0.02 | |
| All codons | |||||||||
| 1st | 35 | 13.4 | 31.7 | 20.3 | 3729.0 | 66.7 | −0.05 | 0.20 | |
| 2nd | 45 | 20.4 | 19.7 | 14.6 | 3727.0 | 64.7 | −0.39 | −0.17 | |
| 3rd | 44 | 8.1 | 42.1 | 5.6 | 3726.0 | 86.1 | −0.02 | −0.18 | |
| Genes on J-strand | |||||||||
| 1st | 28 | 15.8 | 35.1 | 20.6 | 2298.0 | 63.1 | 0.11 | 0.13 | |
| 2nd | 43 | 22.6 | 20.5 | 13.5 | 2297.0 | 63.5 | −0.35 | −0.25 | |
| 3rd | 32 | 12.0 | 53.2 | 2.4 | 2297.0 | 85.2 | 0.25 | −0.67 | |
| Total | 34.8 | 16.8 | 36.3 | 12.2 | 6892.0 | 71.1 | 0.02 | −0.16 | |
| Genes on N-strand | |||||||||
| 1st | 45 | 9.5 | 26.1 | 19.7 | 1431 | 71.1 | −0.27 | 0.35 | |
| 2nd | 48 | 16.9 | 18.5 | 16.4 | 1430 | 66.5 | −0.44 | −0.02 | |
| 3rd | 63 | 1.8 | 24.3 | 10.6 | 1429 | 87.3 | −0.44 | 0.71 | |
| Total | 52.1 | 9.4 | 23.0 | 15.6 | 4290 | 75.1 | −0.39 | 0.25 | |
| F. amplivertica | All genes | 30.3 | 16.4 | 42.1 | 11.2 | 15,657.0 | 72.4 | 0.16 | −0.19 |
| rRNA genes | 29.7 | 16.9 | 43.2 | 10.3 | 2171.0 | 72.9 | 0.19 | −0.24 | |
| tRNA genes | 31.4 | 16.5 | 38.4 | 13.8 | 1482.0 | 69.8 | 0.10 | −0.09 | |
| A+T-rich region | 36.9 | 9.9 | 46.5 | 6.7 | 748.0 | 83.4 | 0.12 | −0.19 | |
| PCGs | 41.1 | 14.3 | 30.7 | 13.9 | 11,178.0 | 71.8 | −0.14 | −0.01 | |
| All codons | |||||||||
| 1st | 34 | 13.6 | 31.2 | 20.8 | 3729.0 | 65.2 | −0.04 | 0.21 | |
| 2nd | 45 | 20.5 | 19.6 | 14.7 | 3725.0 | 64.6 | −0.39 | −0.16 | |
| 3rd | 44 | 8.8 | 41.4 | 6.2 | 3724.0 | 85.4 | −0.03 | −0.17 | |
| Genes on J-strand | |||||||||
| 1st | 28 | 16.2 | 34.6 | 21.1 | 2298.0 | 62.6 | 0.11 | 0.13 | |
| 2nd | 43 | 22.7 | 20.3 | 13.8 | 2295.0 | 63.3 | −0.36 | −0.24 | |
| 3rd | 31 | 13.4 | 52.1 | 3.4 | 2295.0 | 83.1 | 0.25 | −0.60 | |
| Total | 34.2 | 17.4 | 35.7 | 12.8 | 6888.0 | 69.9 | 0.02 | −0.15 | |
| Genes on N-strand | |||||||||
| 1st | 44 | 9.6 | 25.9 | 20.4 | 1431 | 69.9 | −0.26 | 0.36 | |
| 2nd | 48 | 17.1 | 18.5 | 16.3 | 1430 | 66.5 | −0.44 | −0.02 | |
| 3rd | 64 | 1.3 | 24.1 | 10.8 | 1429 | 88.1 | −0.45 | 0.79 | |
| Total | 52.0 | 9.3 | 22.8 | 15.8 | 4290 | 74.8 | −0.39 | 0.26 | |
| F. nigritibia | All genes | 30.3 | 16.5 | 42.0 | 11.3 | 15,661.0 | 72.3 | 0.16 | −0.19 |
| rRNA genes | 29.7 | 16.8 | 43.1 | 10.4 | 2172.0 | 72.8 | 0.18 | −0.24 | |
| tRNA genes | 31.7 | 16.2 | 38.4 | 13.7 | 1486.0 | 70.1 | 0.10 | −0.08 | |
| A+T-rich region | 37.8 | 10.4 | 44.4 | 7.4 | 747.0 | 82.2 | 0.08 | −0.17 | |
| PCGs | 40.9 | 14.4 | 30.9 | 13.9 | 11,183.0 | 71.8 | −0.14 | −0.02 | |
| All codons | |||||||||
| 1st | 34 | 13.7 | 31.3 | 20.7 | 3729.0 | 65.3 | −0.04 | 0.21 | |
| 2nd | 45 | 20.6 | 19.7 | 14.7 | 3727.0 | 64.7 | −0.39 | −0.17 | |
| 3rd | 43 | 8.7 | 41.7 | 6.2 | 3727.0 | 84.7 | −0.02 | −0.17 | |
| Genes on J-strand | |||||||||
| 1st | 28 | 16.4 | 34.5 | 21.1 | 2298.0 | 62.5 | 0.10 | 0.13 | |
| 2nd | 43 | 22.9 | 20.3 | 13.8 | 2297.0 | 63.3 | −0.36 | −0.25 | |
| 3rd | 31 | 13.3 | 52.4 | 3.3 | 2297.0 | 83.4 | 0.26 | −0.60 | |
| Total | 34.0 | 17.5 | 35.8 | 12.7 | 6892.0 | 69.8 | 0.03 | −0.16 | |
| Genes on N-strand | |||||||||
| 1st | 44 | 9.4 | 26.0 | 20.1 | 1431 | 70.0 | −0.26 | 0.36 | |
| 2nd | 48 | 17.1 | 18.7 | 16.2 | 1430 | 66.7 | −0.44 | −0.03 | |
| 3rd | 63 | 1.4 | 24.4 | 10.8 | 1430 | 87.4 | −0.44 | 0.77 | |
| Total | 52.0 | 9.3 | 23.0 | 15.7 | 4291 | 75.0 | −0.39 | 0.26 | |
| F. pamphagoides | All genes | 30.5 | 16.3 | 42.0 | 11.1 | 15,661.0 | 72.5 | 0.16 | −0.19 |
| rRNA genes | 30.0 | 17.2 | 42.1 | 10.7 | 2167.0 | 72.1 | 0.17 | −0.23 | |
| tRNA genes | 30.9 | 16.4 | 38.9 | 13.7 | 1485.0 | 69.8 | 0.11 | −0.09 | |
| A+T-rich region | 36.1 | 10.2 | 46.2 | 7.5 | 747.0 | 82.3 | 0.12 | −0.15 | |
| PCGs | 41.3 | 14.1 | 30.9 | 13.7 | 11,183.0 | 72.2 | −0.14 | −0.01 | |
| All codons | |||||||||
| 1st | 35 | 13.3 | 31.2 | 20.9 | 3729.0 | 66.2 | −0.06 | 0.22 | |
| 2nd | 45 | 20.7 | 19.5 | 14.8 | 3727.0 | 64.5 | −0.40 | −0.17 | |
| 3rd | 44 | 8.3 | 42.0 | 5.4 | 3727.0 | 86.0 | −0.02 | −0.21 | |
| Genes on J-strand | |||||||||
| 1st | 28 | 15.9 | 34.5 | 21.2 | 2298.0 | 62.5 | 0.10 | 0.14 | |
| 2nd | 43 | 22.9 | 20.3 | 13.7 | 2297.0 | 63.3 | −0.36 | −0.25 | |
| 3rd | 32 | 12.9 | 52.2 | 2.8 | 2297.0 | 84.2 | 0.24 | −0.64 | |
| Total | 34.5 | 17.2 | 35.7 | 12.6 | 6892.0 | 70.2 | 0.02 | −0.15 | |
| Genes on N-strand | |||||||||
| 1st | 45 | 9.2 | 25.8 | 20.3 | 1431.0 | 70.8 | −0.27 | 0.38 | |
| 2nd | 48 | 17.2 | 18.3 | 16.4 | 1430.0 | 66.3 | −0.45 | −0.02 | |
| 3rd | 64 | 1.0 | 25.5 | 9.7 | 1430.0 | 89.5 | −0.43 | 0.81 | |
| Total | 52.2 | 9.2 | 23.2 | 15.5 | 4291.0 | 75.4 | −0.38 | 0.26 | |
| F. dingxiensis | All genes | 30.6 | 16.3 | 42.0 | 11.0 | 15,666.0 | 72.7 | 0.16 | −0.19 |
| rRNA genes | 29.9 | 17.2 | 42.5 | 10.3 | 2175.0 | 72.4 | 0.17 | −0.25 | |
| tRNA genes | 30.9 | 16.6 | 38.7 | 13.8 | 1484.0 | 69.6 | 0.11 | −0.09 | |
| A+T-rich region | 37.1 | 9.7 | 45.7 | 7.5 | 750.0 | 82.8 | 0.10 | −0.13 | |
| PCGs | 41.3 | 14.0 | 31.0 | 13.6 | 11,184.0 | 72.4 | −0.14 | −0.01 | |
| All codons | |||||||||
| 1st | 35.8 | 16.3 | 28.8 | 19.0 | 3731.0 | 64.7 | −0.11 | 0.08 | |
| 2nd | 50.9 | 14.4 | 22.1 | 12.6 | 3727.0 | 73.0 | −0.39 | −0.07 | |
| 3rd | 37.3 | 11.3 | 42.2 | 9.3 | 3726.0 | 79.4 | 0.06 | −0.10 | |
| Genes on J-strand | |||||||||
| 1st | 28.4 | 15.7 | 35.4 | 20.5 | 2300.0 | 63.7 | 0.11 | 0.13 | |
| 2nd | 43.3 | 22.6 | 20.5 | 13.7 | 2297.0 | 63.8 | −0.36 | −0.25 | |
| 3rd | 32.8 | 12.5 | 52.3 | 2.5 | 2296.0 | 85.1 | 0.23 | −0.67 | |
| Total | 34.8 | 16.9 | 36.0 | 12.2 | 6893.0 | 70.9 | 0.02 | −0.16 | |
| Genes on N-strand | |||||||||
| 1st | 44.4 | 9.4 | 26.0 | 20.1 | 1431.0 | 70.4 | −0.26 | 0.36 | |
| 2nd | 47.8 | 17.2 | 18.4 | 16.6 | 1430.0 | 66.2 | −0.44 | −0.02 | |
| 3rd | 63.0 | 1.4 | 24.8 | 10.8 | 1430.0 | 87.8 | −0.44 | 0.77 | |
| Total | 51.7 | 9.3 | 23.1 | 15.9 | 4291.0 | 74.8 | −0.38 | 0.26 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, F.-Y.; Shi, Q.-Y.; Ling, Y.; Chen, J.-Y.; Zhang, B.-F.; Li, X.-J. Comparative Analysis of Mitogenomes among Five Species of Filchnerella (Orthoptera: Acridoidea: Pamphagidae) and Their Phylogenetic and Taxonomic Implications. Insects 2021, 12, 605. https://doi.org/10.3390/insects12070605
Zheng F-Y, Shi Q-Y, Ling Y, Chen J-Y, Zhang B-F, Li X-J. Comparative Analysis of Mitogenomes among Five Species of Filchnerella (Orthoptera: Acridoidea: Pamphagidae) and Their Phylogenetic and Taxonomic Implications. Insects. 2021; 12(7):605. https://doi.org/10.3390/insects12070605
Chicago/Turabian StyleZheng, Fang-Yuan, Qiu-Yue Shi, Yao Ling, Jian-Yu Chen, Bo-Fan Zhang, and Xin-Jiang Li. 2021. "Comparative Analysis of Mitogenomes among Five Species of Filchnerella (Orthoptera: Acridoidea: Pamphagidae) and Their Phylogenetic and Taxonomic Implications" Insects 12, no. 7: 605. https://doi.org/10.3390/insects12070605
APA StyleZheng, F.-Y., Shi, Q.-Y., Ling, Y., Chen, J.-Y., Zhang, B.-F., & Li, X.-J. (2021). Comparative Analysis of Mitogenomes among Five Species of Filchnerella (Orthoptera: Acridoidea: Pamphagidae) and Their Phylogenetic and Taxonomic Implications. Insects, 12(7), 605. https://doi.org/10.3390/insects12070605










