Simple Summary
Insect pest resistance to synthetic insecticides is a major problem that limits efficient management and thus decreases productivity for farmers and increases the use of harmful materials that pollute the environment and endanger humans and beneficial organisms. A major approach for resistance management is understanding how insect pest field populations develop resistance at molecular levels. To provide a comprehensive insight into the resistance mechanisms of Spodoptera frugiperda larvae to lambda-cyhalothrin 5%, we investigated the molecular basis of resistance mechanism in field collected population of fall armyworm (Spodoptera frugiperda) to lambda-cyhalothrin 5% insecticide, a pyrethroid insecticide by using de novo transcriptomics analysis. We found that resistance to lambda-cyhalothrin 5% can be metabolic by increasing the levels of detoxifying enzymes such as P450, GST and UGT and related genes to insecticide resistance in the field population. The obtained transcriptome information provides large gene resources available for further studying the resistance development of Spodoptera frugiperda to pesticides. The DGE data provide comprehensive insights into the gene expression profiles of fall armyworm (Spodoptera frugiperda) to lambda-cyhalothrin 5% and will facilitate the study of the role of each gene in lambda-cyhalothrin resistance development.
Abstract
The fall armyworm, Spodoptera frugiperda (J.E. Smith) (Lepidoptera: Noctuidae), is a polyphagous, invasive insect pest which causes significant losses in important crops wherever it has spread. The use of pesticides in agriculture is a key tool in the management of many important crop pests, including S. frugiperda, but continued use of insecticides has selected for various types of resistance, including enzyme systems that provide enhanced mechanisms of detoxification. In the present study, we analyzed the de novo transcriptome of S. frugiperda larvae exposed to Noposion Yihaogong® 5% emulsifiable concentrate (EC) insecticide focusing on detoxification genes and related pathways. Results showed that a total of 1819 differentially expressed genes (DEGs) were identified in larvae after being treated with Noposion Yihaogong® 5% EC insecticide, of which 863 were up- and 956 down-regulated. Majority of these differentially expressed genes were identified in numerous Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, including metabolism of xenobiotics and drug metabolism. Furthermore, many of S. frugiperda genes involved in detoxification pathways influenced by lambda-cyhalothrin stress support their predicted role by further co-expression network analysis. Our RT-qPCR results were consistent with the DEG’s data of transcriptome analysis. The comprehensive transcriptome sequence resource attained through this study enriches the genomic platform of S. frugiperda, and the identified DEGs may enable greater molecular underpinnings behind the insecticide-resistance mechanism caused by lambda-cyhalothrin.
1. Introduction
The fall armyworm, Spodoptera frugiperda (J.E. Smith) (Lepidoptera: Noctuidae), is an important insect pest of major crops occurring mainly in the tropical and subtropical regions of the American continent [1,2]. In recent years, it has invaded West and Central Africa, where it has been responsible for substantial crop losses and threatens food security in the region [3]. S. frugiperda has continued to spread globally and has recently been found in India [4] and southwest China (southwest Yunnan province), where, in January 2019, larvae were found in corn [4,5,6]. The larvae of this insect are highly polyphagous, feeding on a diverse range of host plant species (more than 80 plant species) [7]. Among its host plants are economically important food crops such as maize, rice, sorghum [8], and infrequently, cotton crop [9]. Maize is the preferred host of S. frugiperda with yield losses of between 15% and 73% during an outbreak [9,10,11]. China is the world’s second-largest producer of maize, and the crop is planted over large areas in all provinces, and maize serves as food, feed, and industrial material [12,13].
For decades, synthetic insecticides have been used as the main tool for the control of S. frugiperda, but increased insecticide resistance hinders our ability to control this destructive insect in the field, and thus threatens agricultural crop production wherever the species is now found [14,15,16]. Nevertheless, increased spread and incidence of S. frugiperda has led to increasing and intensive applications of synthetic pesticides to mitigate losses [17]. However, the long-term and widespread use of pesticides has resulted in significant damage to the environment as well as the development of high levels of resistance in insect pest populations [18,19,20,21,22]. The first report of insecticide resistance in S. frugiperda was to the carbamate insecticide, carbaryl [23]. Since then, high levels of resistance to pyrethroid, organophosphate, and diamide insecticides has been documented in field populations from different countries [14,18,21,22,24], and even laboratory-selected populations of S. frugiperda showed significant resistance ratios of more than 40-fold to lambda-cyhalothrin, a pyrethroid [18].
Resistance to synthetic insecticides mainly consists of two mechanisms: target-site resistance and metabolic resistance [25]. Metabolic resistance has been identified in a range of insect species with resistance to different insecticides due to the gene amplification, overexpression, and/or modification of gene-encoding members of the detoxification enzymes, like cytochrome P-450s (P-450s), glutathione S-transferases (GSTs), and carboxylesterases (CEs) [26,27]. In a previous study, the biochemical characterization of resistance in S. frugiperda to a different group of insecticides suggested that resistance was due to both insensitivity of the target site and detoxification of insecticides by metabolic enzymes [14]. In recent years, the induced expression of certain enzymes’ genes related to resistance in insect pests have been reported. For example, several genes (CYP6B8, and CYP321A1, CYP321A7, and CYP321A9) from the cytochrome P-450 family can be induced to metabolize several allelochemicals and insecticides in Spodoptera litura and Helicoverpa zea [28,29,30]. Similarly, the transcription level of many CYP450, GST, and UDP-glucuronosyltransferases (UGT) genes are significantly upregulated in Bombyx mori, Spodoptera exigua, and S. frugiperda after exposure to different allelochemicals and insecticides [27,31,32].
In recent years, researchers have used new technologies and advances in genomic research to identify mechanisms for identifying and potentially controlling pest insecticide resistance. The development of novel next-generation sequencing (NGS) has brought tremendous progress to genomic research in a large number of non-model organisms. RNA-seq data in insect pests would further serve as a useful resource to study aspects of pest control [33,34,35]. Although insecticide resistance mechanisms have been reported before, further research is needed, particularly on the common resistance mechanisms of pests to different insecticides. In this study, we used the high-throughput Illumina HiSeq4000 (Illumina, San Diego, CA, USA) platform to acquire whole-body de novo transcriptome analysis of S. frugiperda. Our focus was to quantify the expression levels of key detoxification genes and related pathways in S. frugiperda. This can be used as a resource to provide insights into insecticide-resistance mechanisms following treatment with lambda-cyhalothrin. The data generated in this study provide abundant resources based on directed sequencing that will be useful to our understanding of the molecular insecticide resistance of S. frugiperda larvae and provide us with new pathways into pest management.
2. Material and Methods
2.1. Insect Collection and Rearing
The field populations of the Spodoptera frugiperda of different larval stages were collected in August 2019 from two different cornfields (Ping Hu, Zhejiang China). Larvae were reared on an artificial diet [36] in a climate control chamber at 25 ± 2 °C with a 14:10 h light:dark photoperiod in the Institute of Plant protection and Microbiology, Zhejiang Academy of Agricultural Sciences Hangzhou, China, for two generations to obtain a more homogenous population before they were used for experiments. Following pupation, newly emerged adults were sexed with mating pairs placed together in cages and provided with 10% honey solution as a food source. A laboratory-susceptible (not true susceptible) population was collected from a maize research field, Yunnan province, in Feb-2019 and maintained on an artificial diet without exposure to insecticides. The early third-instar larvae of the F2 generation from the Zhejiang field population were used for toxicity bioassays and unselected larvae of the same population of the F2 generation were used as a control treatment.
2.2. Insecticide and Toxicity Bioassays for Dose Selection
The commercial insecticide product used in the bioassay according to previously reported methods [16,37] was: lambda-cyhalothrin (Noposion Yihaogong® 5% emulsifiable concentrate (EC) insecticide) Shenzhen Noposion Agrochemical Co. Ltd., Shenzhen, China. Composition: lambda-cyhalothrin: 5%, co-solvent: 8% ethyl acetate, solvent: 72% water, stabilizer: 3% glycerol (antifreeze) and trade secret: 12%. A diet-incorporation method for feeding bioassays was used to calculate the dose-mortality response of the early third-instar S. frugiperda larvae based on methodology previously reported by Carvalho et al. [20]. Artificial diet-incorporation assays were conducted in small transparent Petri dishes as described previously by Bolzan et al. [38]. The stock solution of Noposion Yihaogong® 5% EC insecticide was prepared and then six serial dilutions ranging from 4.686 to 150 and 0.156 to 5.00 mg-L−1 for field population and for lab populations (each concentration was replicated three times) were mixed thoroughly into the semisynthetic diet before solidification of agar (40–45 °C) [39]. Distilled water in the semisynthetic diet was used as a control. Diet supplemented with serial concentrations of test insecticide was cut into small cubes and placed into the small transparent Petri dishes (5-cm diameter), and three small transparent Petri dishes were used for each concentration, including control treatment. A total of 420 third-instar larvae were then transferred onto the contaminated diet (twenty larva per Petri plate), including control treatment. The toxicity bioassays were performed at 25 ± 2 °C with a 14:10 h light:dark photoperiod. Mortality was calculated after 72 h. Larvae were considered alive if they were able to show movement in a coordinated manner when touched with a small soft brush. Control mortality was less than 10%. Probit analysis was done to calculate the lethal concentrations LC50 and LC25 values for each assay of insecticide using the POLO-PC software package (LeOra Software, Berkeley, CA, USA) [40].
2.3. Samples’ Preparation for RNA-Sequence
The early third-instar healthy larvae of S. frugiperda were exposed to a sublethal concentration (LC25) of Noposion Yihaogong® 5% EC (15.871 mg-L−1) for 72 h. Larvae taken from the same population fed on diet without Noposion Yihaogong® 5% EC insecticide served as a control treatment. After 72 h post-exposure, 36 surviving larvae (whole-body) were collected from the treated and untreated group and transferred to micro-tubes (Eppendorf, Hamburg, Germany) on ice and put into liquid nitrogen for 5 min, then stored at −80 °C until RNA extraction. Total RNA was extracted for transcriptomic analyses from three independent biological replicates (12 larvae in each replicate) for each treatment, including untreated as a control.
2.4. Total RNA Isolation, cDNA Library Construction and Illumina Sequencing
Total RNA from Noposion Yihaogong® 5% EC-treated and unselected groups was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA) following the protocol according to manufacturer’s instructions. The total RNA quantity and integrity was analyzed using a Bioanalyzer 2100 device and RNA 1000 Nano Lab Chip Kit (Agilent, Santa Clara, CA, USA) with RIN number > 7.0. Poly (A) RNA was purified from total RNA (5 μg) using poly-T oligo-attached magnetic beads using two rounds of purification. Following purification, the mRNA was fragmented into small pieces using divalent cations under an elevated temperature. The cleaved RNA fragments were reverse-transcribed to create the final library of the cDNA in accordance with the protocol for the RNA-Seq sample preparation kit (Illumina, San Diego, CA, USA). The average insert size for the paired-end libraries was 300 bp (±50 bp). The paired-end sequencing was performed on an Illumina Hiseq4000 at the LC Sciences, Houston, TX, USA.
2.5. De Novo Assembly, Unigene Annotation and Functional Classification
Cutadapt [41] and perl scripts in-house were used to remove the reads that contained adaptor contamination, low-quality bases and undetermined bases. Following this, sequence quality was verified using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), including the Q20, Q30 and GC-content of the clean data. All downstream analyses were based on clean data of high quality. De novo assembly of the transcriptome was performed with Trinity 2.4.0 [42]. Trinity groups transcripts into clusters based on shared sequence content. Transcript clusters of this nature were very loosely referred to as a ‘gene’. The trinity assemblies were further examined using TransDecoder (https://github.com/TransDecoder/TransDecoder) to remove false assembled transcripts. The longest transcript in the cluster was chosen as the ‘gene’ sequence (aka Unigene). All assembled Unigenes were aligned against the non-redundant (Nr) protein database (http://www.ncbi.nlm.nih.gov/), Gene Ontology (GO) (http://www.geneontology.org), Swiss Prot (http://www.expasy.ch/sprot/), Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg/) and eggnog (http://eggnogdb.embl.de/) databases using DIAMOND, with a threshold E-value of 1 × 10−5 [43].
2.6. Differentially Expressed Gene (DEGs) Analysis
Salmon [44] was used to perform expression level for Unigenes by calculating Transcripts Per Kilobase Million (TPM) [45]. The differentially expressed Unigenes were selected with log2 (fold change) > 1 or log2 (fold change) < −1 and with statistical significance (p-value < 0.05) by R package edgeR [46]. Next, GO and KEGG enrichment analyses were performed on the differentially expressed Unigenes by Perl scripts in-house.
2.7. Study of Gene Interaction Network with Relation to Detoxification Pathways
To study the correlation of genes with respect to five detoxification pathways under lambda-cyhalothrin stress, co-expression network analysis was performed using Cytoscape software (version 3.3.0, Cytoscape, San Diego, CA, USA) to construct a co-expression regulation network of the genes and to determine the relationships among them according to the method described previously [46]. The co-expression network map was made with p n > 0.95 as the threshold [47]. Furthermore, we identified major detoxification DEGs from five different pathways from transcriptome under lambda-cyhalothrin stress.
2.8. The RT-qPCR Validation of Differentially Expressed Candidate Genes
RT-qPCR was performed to compare the relative mRNA abundance of candidate genes. Total RNAs from treated and untreated samples were extracted using Trizol Reagent (Takara, Kyoto, Japan) as described above. TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGene Biotech.co., Ltd. Haidian District, Beijing, China 100192 were used to synthesize cDNA from 1 μg of total RNA template. The reaction mixture of 20 μL consisted of Anchored Oligo(dT)18 Primer (0.5 μg/μL) 1 μL, 2× TS Reaction Mix 10 μL, TransScript® RT/RI Enzyme Mix 1 μL, gDNA Remover 1 μL and RNase-free Water 5 μL. The primers were designed using Premier 5 software, and the sequences of the primers used for candidate genes are listed in Supplementary Table S1. The RT-qPCR analysis was performed with three biological (12 larvae in each replicate) and three technical replicates in each treatment, including control, by using a CFX96™ Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) with SsoFast EvoGreen® SuperMix (BIO RAD, Hercules, CA, USA). The RT-qPCR mixture consisted of 20 μL of total volume containing 10 μL of 2× PCR mixture (Sangdon Biotech, Shanghai, China), 0.5 μL of each sense and antisense primers (10 μM), 3 μL of cDNA template and 6 μL of dd H2O. The protocol for qRT-PCR was used as follows: 95 °C for 30 s, followed by 40 cycles at 95 °C for 5 s and at 60 °C for 30 s. GADPH (GenBank: KC262638.1) and the ribosomal protein S30 (NCBI locus AF400225) were used as the internal control gene for normalization. All reactions were run in triplicate and fold changes were calculated using the 2−ΔΔCT method by Livak and Schmittgen [48].
2.9. Statistical Analysis
Concentration-mortality data was analyzed by Probit analysis [49] with POLO 2.0 program LeOra Software (www.leorasoftware.com) to determine the LC50 value, standard errors, slopes and 95% fiducial limit (FL). All data including RT-qPCR were analyzed using the statistical software SPSS (version 19.0, SPSS Inc., Chicago, IL, USA). The statistically significant mean values of the treatments were calculated using analysis of variance (ANOVA), and the important variations among the treatments were determined using the Student’s t test (p < 0.05).
3. Results
3.1. Toxicity of Noposion Yihaogong® 5% EC insecticide
Lethal and sublethal concentrations (LC50 and LC25) of Noposion Yihaogong® 5% EC treatment of the field-collected population for early third-instar larvae of S. frugiperda and the lab-susceptible strain at 72 h were identified (Table 1). The results showed that the toxicity (LC50 and LC25 values) was estimated to be approximately 40.62 and 15.87 µg-g−1 for the field-collected population and for the lab-susceptible population was 1.3 and 0.54 µg-g−1 (Table 1). The estimated sublethal concentration (LC25) was further applied for de novo transcriptome analysis Noposion Yihaogong® 5% EC treatment compared with unselected as a control from the same field-collected population of Zhejiang province.

Table 1.
Toxicity of Noposion Yihaogong® 5% EC insecticide to early third-instar Spodoptera frugiperda larvae.
3.2. Illumina Sequencing and De Novo Assembly
To investigate the effect of Noposion Yihaogong® 5% EC on the third-instar S. frugiperda larvae, Illumina Hiseq4000 technology was used to sequence six cDNA libraries from the whole body of the S. frugiperda larvae exposed to a sublethal concentration of Noposion Yihaogong® 5% EC insecticide and for the unselected group. In total, raw reads for Noposion Yihaogong® 5% EC-1, Noposion Yihaogong® 5% EC-2, Noposion Yihaogong® 5% EC-3, unselected-1, unselected-2 and unselected-3 were 55,781,304, 57,178,320, 44,957,636, 53,871,434, 54,235,678 and 56,970,122, respectively. After mapping to the reference genome (https://bipaa.genouest.org/v3.1) and the junction database, a total number of 53,123,060, 54,393,760, 42,904,598, 51,566,052, 51,465,482 and 54,229,496 clean reads were generated with more than 90% validity (Table 2). Altogether, all libraries were good quality with an average percentage of Q20 (97.98%) and Q30 (93.75%) containing an average GC percentage of 50.07%, respectively (Table 2). Furthermore, a total of 66,501 transcripts were assembled from all the transcriptome data and the clean reads were assembled into 26,814 unigenes. The minimum and maximum size of unigenes were 201 and 12,042 bp, with mean size of 516 bp, and GC content and N50 were 40.78 bp and 1333, respectively (Supplementary Table S2).

Table 2.
Summary of the sequencing data.
3.3. Functional Annotation of the S. frugiperda-Treated Larvae Unigenes
The functional annotation of 26,814 unigene sequences were all aligned against six different public databases, including nr, GO, KEGG, Pfam, Swiss-Prot and eggNOG. The results from functional annotation unigene sequences showed that 13,585 (50.66%), 9115 (33.99%), 9105 (33.96%), 9434 (35.18%), 8055 (30.04%) and 12,465 (46.49%) unigenes matched to nr, GO, KEGG, Pfam, Swiss-Prot and eggNOG protein databases, respectively (Table 3). After nr database annotation, the species distribution of the best-match result for each sequence demonstrated that 73% of the unigenes had highest homology against sequences of Spodoptera litura, followed by Helicoverpa armigera (6.47%), Heliothis virescens (2.97%) Trichoplusia ni (2.96%) and Drosophila melanogaster (1.71) (Figure 1).

Table 3.
Unigenes annotation against six public databases (DB).
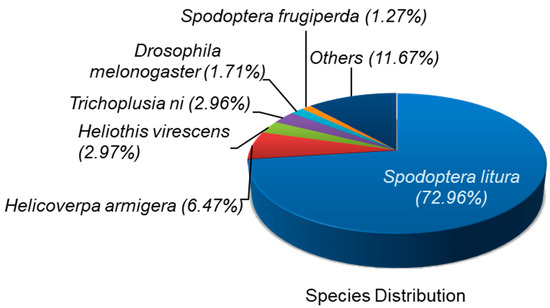
Figure 1.
De novo transcriptome analysis of unselected and Noposion Yihaogong® 5% EC insecticide-exposed larvae. Pie chart showing the result from annotation gene ontology annotation and classification of the Spodoptera frugiperda transcriptome.
3.4. Differentially Expressed Unigenes in Noposion Yihaogong® 5% EC Insecticide-Treated S. frugiperda Larvae
A total of 1819 differentially expressed unigenes (DEGs) were found between Noposion Yihaogong® 5% EC insecticide-treated and the untreated group. Of these, 863 (47.44%) were upregulated and 956 (52.56%) were downregulated (Figure 2A). To ascertain differential gene expression in control as compared to Noposion Yihaogong® 5% EC-exposed samples, a volcano plot was constructed with a log2 fold-change > 1 for upregulated genes and < −1 for downregulated genes (Figure 2B).
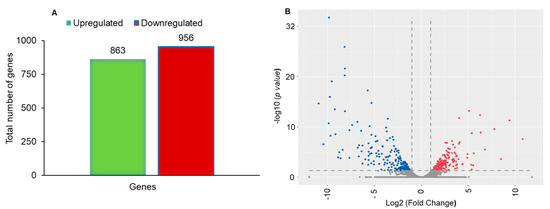
Figure 2.
De novo transcriptome analysis of unselected and Noposion Yihaogong® 5% EC insecticide-exposed larvae. (A) Bar graph showing total number of up- and down-regulated differentially expressed genes (DEGs). (B) Volcano plot analysis of differentially regulated genes in untreated and Noposion Yihaogong® 5% EC insecticide-exposed sample. DeSeq was used for determining upregulation and downregulation of genes based upon relative Fragments Per Kilobase Million (FPKM) counts (x-axis: log2fold change; y-axis: −log (p-value)).
3.5. Gene Ontology (GO) Enrichment Analysis
Gene ontology (GO) assignments were used to functionally classify the predicted unigenes from the whole body of S. frugiperda. According to the sequence similarity, 4045 unigenes (15.12%) were annotated and classified into 50 functional groups of 3 main ontologies: the biological process was the largest class followed by cellular components and molecular function class (Figure 3). Based on the GO, we found that the majority of genes were enriched for the three largest terms, such as oxidation-reduction process, chitin-based cuticle development and proteolysis. The main DEGs of GO terms were mainly enriched for cellular components class, such as extracellular space, cytoplasm and extracellular space matrix. In the category of molecular function, the three subcategories of DEGs enriched for GO terms “structural constituent of cuticle”, “oxide of reductase activity” and “structural constituent of chitin-based larval cuticle” were the most enriched terms (Figure 3).
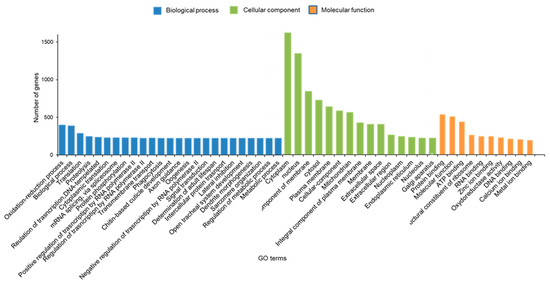
Figure 3.
Gene Ontology (GO) enriched terms of differentially expressed genes (DEGs) of Spodoptera frugiperda larvae after exposure to a Noposion Yihaogong® 5% EC (TR) versus the control group (unselected). The x-axis lists the sub-GO terms under categories of biological process, cellular component and molecular function. The y-axis is the number of DEGs involved in each term.
3.6. KEGG Pathway Analysis
Subsequently, the KEGG pathway assignment was also performed on unigenes to identify the biological pathways, including metabolic and regulatory pathways, that will be actively involved in the S. frugiperda larvae. The results of KEGG pathways enrichment showed that there were five major KEGG pathways categories, in which Metabolism was the most dominant category of the KEGG pathway (2486), followed by Genetic Information Processing (1646), Cellular Processes (861), Environmental Information Processing (767) and Organismal Systems (410), that are actively involved in different functions (Figure 4). The genes involved in the major subcategories of KEGG pathways included: carbohydrates metabolism (502), xenobiotics biodegradation and metabolism (169), translation (710), folding, sorting and degradation (476), signal transduction (558), transport and catabolism (676) and cell growth and death (131) (Figure 4). This result indicated that in S. frugiperda larvae, the three main categories of biological process, cellular component and molecular function were all somehow affected by Noposion Yihaogong® 5% EC insecticide exposure. In the present study, the KEGG pathway assignment will be helpful to further research specific biological processes, functions and pathways that exist in the S. frugiperda larvae. The most significant KEGG pathway in the DEGs analysis (KEGG) pathways of S. frugiperda after exposure to a Noposion Yihaogong® 5% EC insecticide are shown in Supplementary Figure S1.
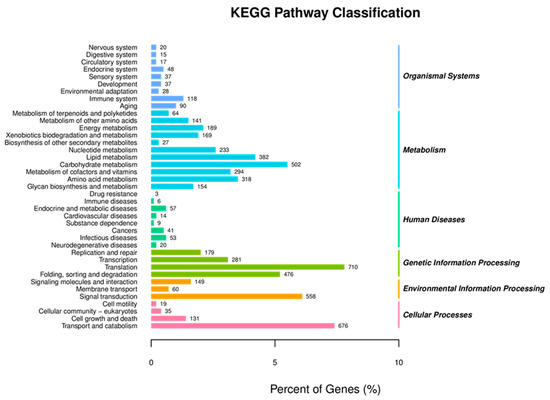
Figure 4.
Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation and pathways of the Spodoptera frugiperda transcriptome.
3.7. Identification of Key Genes and Pathways Involved in Detoxification
In this study, GO/KEGG enrichment analyses were used to identify metabolic enzymes and related genes involved in detoxification of insecticides. We found several upregulated genes harboring important detoxification enzymes. In recent studies, a number of detoxification genes have been identified from the de novo transcriptome analysis of S. frugiperda larvae, with the majority of these genes belonging to the important class of detoxification enzymes, cytochrome P450 (P450s). Cytochrome P450s are an extensively distributed protein superfamily that play a key part in the metabolism and detoxification of a wide range of plant secondary metabolites and synthetic chemicals. Cytochrome P450 enzymes have been divided into four different clades, including CYP2, CYP3, CYP4 and mitochondria. In the present study, upregulated detoxification genes have been identified to include twelve genes relating to cytochrome P450s, three genes from esterase and two genes from UDP-glucuronosyltransferases (UGT) (Figure 5).
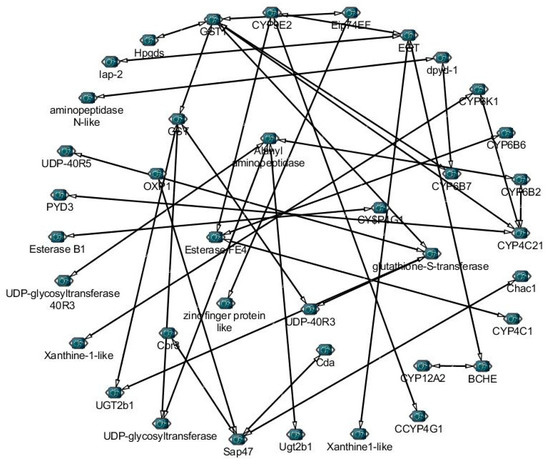
Figure 5.
The correlation networks of 52 detoxification genes from different families involved in pathways under Noposion Yihaogong® 5% EC insecticide stress treatment were established based on the Pearson correlation coefficients of these gene pairs using RNA-Seq data. The PCC of co-relation gene pairs was considered significant (p < 0.05).
3.8. Study of Gene Interaction Network with Relation to Detoxification Pathways
To further understand the roles of detoxification genes, we identified 52 genes from transcriptome under Noposion Yihaogong® 5% EC insecticide stress and analyzed the correlation network based on the Pearson correlation coefficients (PCCs) (Figure 5). Results showed the 33 positive and 32 negative correlations of selected genes involved in detoxification pathways under Noposion Yihaogong® 5% EC insecticide stress. Taking all together, study of the co-expression network show that detoxification genes are greatly influenced under insecticide stress (Figure 5). Furthermore, our results show the important role of major detoxification DEGs involved in five different pathways from transcriptome under Noposion Yihaogong® 5% EC insecticide stress (Figure 6).
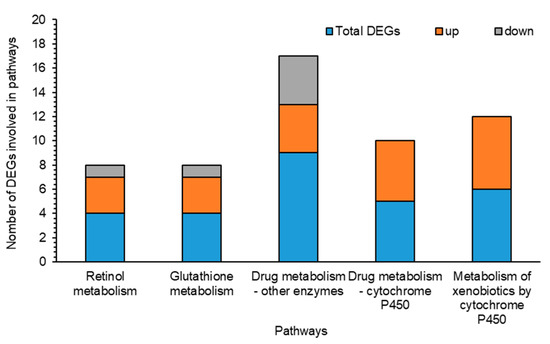
Figure 6.
Comparison of up- and down-regulated genes of differentially expressed genes (DEGs) involved in different pathways of Spodoptera frugiperda after exposure of its larvae to a sublethal concentration of Noposion Yihaogong® 5% EC insecticide versus the unselected group (CK). The x-axis lists the major detoxification pathways’ terms under categories of retinol metabolism, glutathione metabolism and drug metabolism by other enzymes, drug metabolism by cytochrome P450 and metabolism of xenobiotics by cytochrome P450. The y-axis is the number of DEGs involved in each pathway under Noposion Yihaogong® 5% EC insecticide stress.
3.9. Validation of Differentially Expressed Genes (DEGs) Using RT-qPCR
To further explore the expression level of the identified detoxification enzyme genes after the treatment of S. frugiperda larvae with Noposion Yihaogong® 5% EC insecticide, RT-qPCR analysis was performed to validate the DEGs databases acquired from RNA-sequencing analysis. Twelve DEGs in cytochrome P450s were significantly upregulated in the whole body of Noposion Yihaogong® 5% EC insecticide-treated larvae (Figure 7). This result showed that these detoxification enzyme genes, especially from cytochrome P450, were potentially involved in the detoxification of Noposion Yihaogong® 5% EC insecticide.
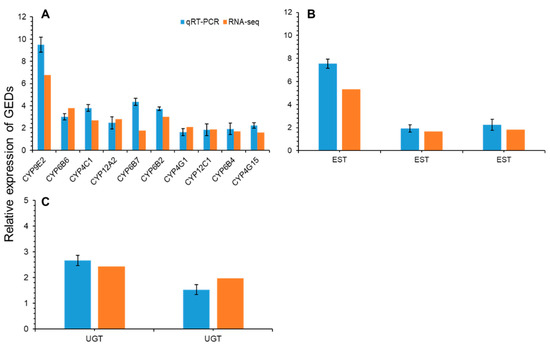
Figure 7.
Results for the qRT-PCR confirmation of the DEGs library. (A) A qRT-PCR analysis of ten upregulated genes of detoxification genes: P450 (cytochrome P450), (B) three upregulated EST (esterases) and (C) two upregulated Noposion Yihaogong® 5% EC insecticide-related detoxification genes, UDP-glucuronosyltransferases (UGT). Bars are means ± SE.
4. Discussion
The use of synthetic insecticides is currently the most common method to control fall armyworm in agricultural crops; however, this is only partially effective due to the development of resistance among field populations [16,37]. The rapid evolution of insecticide resistance of many agricultural pests is attributed to various resistance mechanisms and needs attention. Amplification, overexpression and/or modification of genes that encode a number of the microsomal oxidase such as CYPs, GSTs and CarEs groups have been reported to be responsible for metabolic resistance in a range of insect species to carbamates, pyrethroids and organophosphates insecticides [20,50]. Comprehensive biochemical studies showed that various detoxifying enzymes, such as cytochrome P450s (CYPs), glutathione S-transferases (GSTs) and carboxylesterases (CarEe), are induced by the various groups of insecticides in the resistant field strains of the fall armyworm [14]. Furthermore, it has been frequently reported that pyrethroid resistance of major crops’ insect pests is commonly associated with the overexpression of one or more cytochrome P450 genes [51,52].
In this study, the field population of S. frugiperda showed greater resistance (31.1-fold) to Noposion Yihaogong® 5% EC insecticide than the lab-susceptible population. Similar to our results, previous studies reported high levels of resistance against pyrethroid insecticides, including lambda-cyhalothrin and fenevalerate (with highest LC50 (253.88 and 3000 mg/L) values), from field populations of S. frugiperda in different provinces of China [24,53]. However, conventional synthetic pyrethroids may no longer be effective against S. frugiperda due to developed resistance. Our results (lambda-cyhalothrin; RR50 = 31.2-fold) are consistent with previous studies which also showed that S. frugiperda populations in the Americas have developed resistance to pyrethroid insecticides, e.g., permethrin (RR50 = 220-fold), zeta-cypermethrin (RR50 = 62-fold) and fluvalinate (RR50 = 216.1-fold), respectively [6,14,18,54,55]. Furthermore, the transcriptomes of S. frugiperda have been generated using short-read sequencing generated by Illumina from various developmental stages exposed to different kinds of synthetic and biopesticides [33,37,56]. Based on the transcription data, the differential expression of unigenes suggests that enriched GOs, and many biological pathways, are associated with specific stages of development and insecticide resistance mechanisms. We analyzed the whole-body transcriptional response of S. frugiperda larvae exposed to Noposion Yihaogong® 5% EC insecticide to investigate the differentially expressed detoxification genes and related metabolic pathways. A total of 1819 DEGs were detected between the Noposion Yihaogong® 5% EC insecticide-treated and the unselected group. Of these, 863 (47.44%) DEGs were upregulated and 956 (52.56%) DEGs were shown to be downregulated at significant levels in the Noposion Yihaogong® 5% EC insecticide-treated group as compared with the unselected group. Most of the upregulated genes belonged to the cytochrome P450 superfamily. These results coincide with those documented for S. frugiperda Sf9 cells and diamondback moth, Plutella xylostella, when treated with lufenuron, harmine and chlorantraniliprole [33,37,57]. In the present study, ten cytochrome P450 genes were commonly upregulated in all treated larvae. Transcription of six CYP450 genes: CYP9E2, CYP6B6, CYP4C1, CYP12A2, CYP6B7 and CYP6B2, increased significantly with Log2FC values of 3.0–6.3. Furthermore, transcription of four of the ten CYP450 genes increased significantly with Log2FC values of 1.5–2 (Figure 7). In previous studies, a similar trend has been documented in other insects, including S. frugiperda, in which overexpression of numerous cytochrome P450 genes was observed following exposure to a range of different synthetic insecticides, including non-pyrethroid classes [37,58,59,60]. These cytochrome P450 genes have been reported to play a key role in the metabolism and detoxification with broad substrate specificity that can respond to different insecticides [60,61,62]. Cytochrome P450s have a variety of metabolic functions, with their overexpression acting as the most common insecticide detoxification mechanism in insects [63,64,65]. Therefore, although our results revealed that overexpression of specific cytochrome P450s genes is associated with the exposure to Noposion Yihaogong® 5% EC insecticide in S. frugiperda, whether these P450s can actively metabolize Noposion Yihaogong® 5% EC insecticide requires further investigation.
In the transcriptome, we identified five esterase (three up- and two down-regulated) and four UGT-glucosyltransferase unigenes (two up- and two down-regulated) that were significantly affected by Noposion Yihaogong® 5% EC insecticide treatment in the S. frugiperda larvae. However, no differential expression of GST genes was observed. This indicates that it may be differential expression of the EST genes which is regulated by the insect’s exposure to insecticides and corresponds to the target insect’s tolerance or susceptibility [66]. The upregulation of UGT genes in response to insecticide exposure has been observed in different insecticide systems [59,67]. Interestingly, previous studies have also described downregulation in the transcription level of two UGTs in the antennae of a female Spodoptera littoralis following exposure to semiochemicals [68], demonstrating their varied roles in insects. Our findings indicate that cytochrome P450, UGT and EST genes together play an important role for S. frugiperda larvae in mediating insecticide-induced stress and in the detoxification of Noposion Yihaogong® 5% EC insecticide.
In addition to detoxification, reduced sensitivity and permeability are important at the target site for tolerance to insecticides. In the present investigation, transcription of all the cuticle protein genes were commonly differentially downregulated in the treated S. frugiperda larvae. Cuticle proteins are thought to be the first barrier used by insects to combat the penetration of xenobiotics and regulate water loss. Contrary to our results, several studies have reported differential expressions of cuticular genes in insects following exposure to insecticides [35,59,69,70]. This is identified as a protective mechanism by which the thickening of the cuticle will reduce penetration of the insecticides. However, present findings suggest that downregulation of cuticular genes may contribute to decreasing tolerance in S. frugiperda larvae to Noposion Yihaogong® 5% EC insecticide, which eventually leads to their increased susceptibility to stress induced by insecticides. Furthermore, we identified downregulated transcription of three nose resistance fluoxetine genes (NRF6) in Noposion Yihaogong® 5% EC insecticide-treated larvae. In previous studies, NRF6 has been shown to be involved in the acquisition and transportation of various molecules, including several xenobiotics compounds that exhibited increased expression against xenobiotic-induced stress in various insects [35,71]. In addition, one nuclear hormone receptor (FTZ-F1) gene also exhibited downregulated expression following the insecticide treatment. These nuclear hormone receptors can be used as transcription activators to initiate the expression of particular metabolic enzymes (cytochrome P450s), which promote detoxification [72]. In addition, many annotated DEGs were dynamically involved in key detoxification KEGG pathways, including retinol metabolism, glutathione metabolism, drug metabolism by other enzymes, drug metabolism by cytochrome P450 and metabolism of xenobiotics by cytochrome P450 (Figure 6). This demonstrates their association with mitigating the adverse effects of insecticide exposure for larvae. Many xenobiotic metabolites are lipophilic, making them difficult to excrete from the body. As a result, these lipophilic substances accumulate and are often antagonistic to biological processes in the organism [73]. Our transcriptomic results suggest that S. frugiperda larval tolerance to insecticides does not take place by the regulation of a single gene, but rather as a result of multiparty regulation by multiple detoxification genes.
In conclusion, the molecular functions of individual S. frugiperda genes and the related signal transduction and metabolic pathways remain largely unclear. Our present results show that Noposion Yihaogong® 5% EC insecticide can induce a significant change in the whole-body transcriptomic profile of S. frugiperda larvae. The upregulated expression of several detoxification-related genes, GO terms and KEGG pathways may be important to pesticide detoxification in S. frugiperda. Together, this transcriptome will serve as a valuable community resource for studies investigating the molecular underpinnings of detoxification genes in fall armyworm and provide a reference for the development of effective strategies to control fall armyworm.
Supplementary Materials
The following are available online at https://www.mdpi.com/2075-4450/12/2/132/s1, Figure S1: The most enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of Spodoptera frugiperda after exposure to a 5% lambda-cyhalothrin. The pathways’ significance are shown together with their q-value (color), rich factor (vertical ordinate), and a number of involved genes (size of circles). Table S1: Total number of reads and Table S2: Primers used for qRT-PCR.
Author Contributions
Conceptualization, M.H. and Y.L.; Methodology, M.H., J.Z. and Z.Z.; Software, M.H. and X.L.; Validation, M.H. and Y.L.; Formal Analysis, M.H. and F.X.; Investigation, M.H., and L.W.; Resources, Y.L. and J.H.; Data Curation, M.H. and X.L.; Writing—Original Draft Preparation, X.L.; Writing—Review & Editing, M.H., M.M.K., S.S., M.P.Z. and G.M.F.-G.; Visualization, M.H.; Supervision, Y.L.; Project Administration, Y.L. and M.H.; Funding Acquisition, M.H., J.Z. and Y.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Key R&D Program of Zhejiang Province (2020C02003), the Shanghai Innovation Project for Agricultural Promotion (2019N3-9), the Joint Agricultural Project between Pinghu County and Zhejiang Academy of Agricultural Sciences (PH20190002) and Project funded by China Postdoctoral Science foundation (233952).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in article
Conflicts of Interest
The authors declare no conflict of interest.
References
- Johnson, S.J. Migration and the life history strategy of the fall armyworm, Spodoptera frugiperda in the western hemisphere. Int. J. Trop. Insect Sci. 1987, 8, 543–549. [Google Scholar] [CrossRef]
- Pitre, H.N. Relationship of Fall Armyworm (Lepidoptera: Noctuidae) from Florida, Honduras, Jamaica, and Mississippi: Susceptibility to Insecticides with Reference to Migration. Fla. Entomol. 1988, 71, 56–61. [Google Scholar] [CrossRef]
- Goergen, G.; Kumar, P.L.; Sankung, S.B.; Togola, A.; Tamò, M. First report of outbreaks of the fall armyworm Spodoptera frugiperda (J E Smith) (Lepidoptera, Noctuidae), a new alien invasive pest in West and Central Africa. PLoS ONE 2016, 11, e0165632. [Google Scholar] [CrossRef] [PubMed]
- Center, N.A.T.E.S. Spodoptera frugiperda harms winter corn in 3 cities and states in southwestern Yunnan. Plant Pathog. Pest Inf. 2019, 2019, 1–31. [Google Scholar]
- Wu, Q.; Jiang, Y.Y.; Wu, K. Analysis of migration routes of the fall armyworm Spodoptera frugiperda (J.E. Smith) from Myanmar to China. Plant Prot. 2019, 45, 1–6. [Google Scholar]
- Li, X.J.; Wu, M.F.; Ma, J.; Gao, B.Y.; Wu, Q.L.; Chen, A.D.; Liu, J.; Jiang, Y.Y.; Zhai, B.P.; Early, R.; et al. Prediction of migratory routes of the invasive fall armyworm in eastern China using a trajectory analytical approach. Pest Manag. Sci. 2020, 76, 454–463. [Google Scholar] [CrossRef]
- Prasanna, B.; Huesing, J.E.; Eddy, R.; Peschke, V.M. Fall Armyworm in Africa: A Guide for Integrated Pest Management; United States Agency for International Development: Washington, DC, USA, 2018.
- Montezano, D.G.; Specht, A.; Gómez, D.R.S.; Specht, V.F.R.; Silva, J.C.S.; Moraes, S.V.P.; Peterson, J.A.; Hunt, T.E. Host Plants of Spodoptera frugiperda (Lepidoptera: Noctuidae) in the Americas. Afr. Entomol. 2018, 26, 286–300. [Google Scholar] [CrossRef]
- Blanco, C.A.; Chiaravalle, W.; Rizza, M.D.; Farias, J.R.; Degano, M.F.G.; Gastaminza, G.; Sánchez, D.M.; Murúa, M.G.; Omoto, C.; Pieralisi, B.K.; et al. Current situation of pests targeted by Bt crops in Latin America. Curr. Opin. Insect Sci. 2016, 15, 131–138. [Google Scholar] [CrossRef]
- Hruska, A.J.; Gladstone, S.M. Effect of Period and Level of Infestation of the Fall Armyworm, Spodoptera frugiperda, on Irrigated Maize Yield. Fla. Entomol. 1988, 71, 249–254. [Google Scholar] [CrossRef]
- Hruska, A.J.; Gould, F. Fall Armyworm (Lepidoptera: Noctuidae) and Diatraea lineolata (Lepidoptera: Pyralidae): Impact of Larval Population Level and Temporal Occurrence on Maize Yield in Nicaragua. J. Econ. Entomol. 1997, 90, 611–622. [Google Scholar] [CrossRef]
- Wennberg, A. Food and Agriculture Organization of the United Nations. In Encyclopedia of Toxicology, 3rd ed.; Academic Press: Cambridge, MA, USA, 2014; ISBN 9780123864543. [Google Scholar]
- Shen, P.; Zhang, Q.; Lin, Y.; Li, W.; Li, A.; Song, Q. Thinking to promote the industrialization of genetically modified corn of our country. China Biotechnol. 2016, 36, 24–29. [Google Scholar]
- Yu, S.J.; Nguyen, S.N.; Elghar, G.E.A. Biochemical characteristics of insecticide resistance in the fall armyworm, Spodoptera frugiperda (J.E. Smith). Pestic. Biochem. Physiol. 2003, 77, 1–11. [Google Scholar] [CrossRef]
- Yu, S.J.; Berry, R.E.; Terriere, L.C. Host plant stimulation of detoxifying enzymes in a phytophagous insect. Pestic. Biochem. Physiol. 1979, 12, 280–284. [Google Scholar] [CrossRef]
- Boaventura, D.; Bolzan, A.; Padovez, F.E.O.; Okuma, D.M.; Omoto, C.; Nauen, R. Detection of a ryanodine receptor target-site mutation in diamide insecticide resistant fall armyworm, Spodoptera frugiperda. Pest Manag. Sci. 2020, 76, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Sisay, B.; Tefera, T.; Wakgari, M.; Ayalew, G.; Mendesil, E. The efficacy of selected synthetic insecticides and botanicals against fall armyworm, Spodoptera frugiperda, in maize. Insects 2019, 10, 45. [Google Scholar] [CrossRef]
- Rodríguez, G.I.D.; Omoto, C. Inheritance of lambda-cyhalothrin resistance in Spodoptera frugiperda (J.E. Smith) (Lepidoptera: Noctuidae). Neotrop. Entomol. 2001, 30, 311–316. [Google Scholar] [CrossRef]
- Qiu, X.; Zhu, T.; Li, J.; Pan, H.; Li, Q.; Miao, G.; Gong, J. Organochlorine Pesticides in the Air around the Taihu Lake, China. Environ. Sci. Technol. 2004, 38, 1368–1374. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, R.A.; Omoto, C.; Field, L.M.; Williamson, M.S.; Bass, C. Investigating the Molecular Mechanisms of Organophosphate and Pyrethroid Resistance in the Fall Armyworm Spodoptera frugiperda. PLoS ONE 2013, 8, e62268. [Google Scholar] [CrossRef]
- Banerjee, R.; Hasler, J.; Meagher, R.; Nagoshi, R.; Hietala, L.; Huang, F.; Narva, K.; Jurat-Fuentes, J.L. Mechanism and DNA-based detection of field-evolved resistance to transgenic Bt corn in fall armyworm (Spodoptera frugiperda). Sci. Rep. 2017, 7, 10877. [Google Scholar] [CrossRef]
- Boaventura, D.; Martin, M.; Pozzebon, A.; Sanchez, D.M.; Nauen, R. Monitoring of target-site mutations conferring insecticide resistance in Spodoptera frugiperda. Insects 2020, 11, 545. [Google Scholar] [CrossRef]
- Young, J.R.; McMillian, W.W. Differential Feeding by Two Strains of Fall Armyworm Larvae on Carbaryl Treated Surfaces123. J. Econ. Entomol. 1979, 72, 202–203. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, B.; Zheng, W.; Liu, C.; Zhang, D.; Zhao, S.; Li, Z.; Xu, P.; Wilson, K.; Withers, A.; et al. Genetic structure and insecticide resistance characteristics of fall armyworm populations invading China. Mol. Ecol. Resour. 2020, 20, 1682–1696. [Google Scholar] [CrossRef] [PubMed]
- Liu, N. Insecticide Resistance in Mosquitoes: Impact, Mechanisms, and Research Directions. Annu. Rev. Entomol. 2015, 7, 537–559. [Google Scholar] [CrossRef]
- Feyereisen, R. Insect CYP Genes and P450 Enzymes. In Insect Molecular Biology and Biochemistry; Elsevier: Amsterdam, The Netherlands, 2012; pp. 115–128. ISBN 9780123847478. [Google Scholar]
- Giraudo, M.; Hilliou, F.; Fricaux, T.; Audant, P.; Feyereisen, R.; Le Goff, G. Cytochrome P450s from the fall armyworm (Spodoptera frugiperda): Responses to plant allelochemicals and pesticides. Insect Mol. Biol. 2015, 24, 15–128. [Google Scholar] [CrossRef] [PubMed]
- Sasabe, M.; Wen, Z.; Berenbaum, M.R.; Schuler, M.A. Molecular analysis of CYP321A1, a novel cytochrome P450 involved in metabolism of plant allelochemicals (furanocoumarins) and insecticides (cypermethrin) in Helicoverpa zea. Gene 2004, 338, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Baudry, J.; Berenbaum, M.R.; Schuler, M.A. Structural and functional divergence of insect CYP6B proteins: From specialist to generalist cytochrome P450. Proc. Natl. Acad. Sci. USA 2004, 101, 2939–2944. [Google Scholar] [CrossRef]
- Wang, R.L.; He, Y.N.; Staehelin, C.; Liu, S.W.; Su, Y.J.; Zhang, J.E. Identification of two cytochrome monooxygenase P450 genes, CYP321A7 and CYP321A9, from the tobacco cutworm moth (Spodoptera litura) and their expression in response to plant allelochemicals. Int. J. Mol. Sci. 2017, 18, 2278. [Google Scholar] [CrossRef]
- Zhang, N.; Liu, J.; Chen, S.N.; Huang, L.H.; Feng, Q.L.; Zheng, S.C. Expression profiles of glutathione S-transferase superfamily in Spodoptera litura tolerated to sublethal doses of chlorpyrifos. Insect Sci. 2016, 23, 675–687. [Google Scholar] [CrossRef]
- Liu, N.; Li, M.; Gong, Y.; Liu, F.; Li, T. Cytochrome P450s—Their expression, regulation, and role in insecticide resistance. Pestic. Biochem. Physiol. 2015, 120, 77–81. [Google Scholar] [CrossRef]
- Cui, G.; Sun, R.; Veeran, S.; Shu, B.; Yuan, H.; Zhong, G. Combined transcriptomic and proteomic analysis of harmine on Spodoptera frugiperda Sf9 cells to reveal the potential resistance mechanism. J. Proteom. 2020, 211, 103573. [Google Scholar] [CrossRef]
- Zhou, C.; Yang, H.; Wang, Z.; Long, G.Y.; Jin, D.C. Comparative transcriptome analysis of Sogatella furcifera (Horváth) exposed to different insecticides. Sci. Rep. 2018, 8, 8773. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, M.; Hafeez, M.; Mabubu, J.I.; Dawar, F.U.; Li, X.; Khan, M.M.; Hua, H.; Cai, W. Transcriptomic analysis of differentially expressed genes and related pathways in Harmonia axyridis after sulfoxaflor exposure. Int. J. Biol. Macromol. 2018, 119, 157–165. [Google Scholar] [CrossRef]
- Poitout, S.; Bues, R. Special need of linolenic acid in the artificial rearing of Lepidoptera Noctuidae Quadrifinae Plusiinae: Chrysodeixis chalcifes Esp., Autographa gamma L., Macdunnoughia confusa Stph., Trichoplusia ni Hbn (French). Ann. Nutr. 1974, 28, 173–187. [Google Scholar]
- Nascimento, A.R.B.D.; Fresia, P.; Cônsoli, F.L.; Omoto, C. Comparative transcriptome analysis of lufenuron-resistant and susceptible strains of Spodoptera frugiperda (Lepidoptera: Noctuidae). BMC Genom. 2015, 16, 985. [Google Scholar] [CrossRef] [PubMed]
- Bolzan, A.; Padovez, F.E.O.; Nascimento, A.R.B.; Kaiser, I.S.; Lira, E.C.; Amaral, F.S.A.; Kanno, R.H.; Malaquias, J.B.; Omoto, C. Selection and characterization of the inheritance of resistance of Spodoptera frugiperda (Lepidoptera: Noctuidae) to chlorantraniliprole and cross-resistance to other diamide insecticides. Pest. Manag. Sci. 2019, 75, 2682–2689. [Google Scholar] [CrossRef] [PubMed]
- Hafeez, M.; Liu, S.; Jan, S.; Ali, B.; Shahid, M.; Grandon, G.M.F.; Nawaz, M.; Ahmad, A.; Wang, M. Gossypol-induced fitness gain and increased resistance to deltamethrin in beet armyworm, Spodoptera exigua (Hübner). Pest. Manag. Sci. 2019, 76, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.M.; Robertson, J.L.; Savin, N.E. POLO: A New Computer Program for Probit Analysis. Bull. Entomol. Soc. Am. 1977, 23, 209–213. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 200. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 4, 417–419. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 7, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Pujana, M.A.; Han, J.D.J.; Starita, L.M.; Stevens, K.N.; Tewari, M.; Ahn, J.S.; Rennert, G.; Moreno, V.; Kirchhoff, T.; Gold, B.; et al. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nat. Genet. 2007, 39, 1338. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C (T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Finney, D.J. Probit Analysis, 3rd ed.; Cambridge University Press: Cambridge, MA, USA, 1971. [Google Scholar]
- Li, X.; Schuler, M.A.; Berenbaum, M.R. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu. Rev. Entomol. 2007, 52, 231–253. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.G.; Jiang, S.S.; Sanchez, D.M.; Wang, W.; Li, X.R.; Gao, Y.L.; Lu, X.P.; Yang, X.Q. Cytochrome P450-Mediated λ-Cyhalothrin-Resistance in a Field Strain of Helicoverpa armigera from Northeast China. J. Agric. Food Chem. 2019, 67, 3546–3553. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Chen, S.; Wu, S.; Yue, L.; Wu, Y. Constitutive Overexpression of Multiple Cytochrome P450 Genes Associated with Pyrethroid Resistance in Helicoverpa armigera. J. Econ. Entomol. 2006, 99, 1749–1784. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.X.; Huang, J.M.; Ni, H.; Guo, D.; Yang, F.X.; Wang, X.; Wu, S.F.; Gao, C.F. Susceptibility of fall armyworm, Spodoptera frugiperda (J.E.Smmith), to eight insecticides in China, with special reference to lambda-cyhalothrin. Pestic. Biochem. Physiol. 2020, 168, 104623. [Google Scholar] [CrossRef]
- Moreno, R.G.; Sanchez, D.M.; Blanco, C.A.; Whalon, M.E.; Santofimio, H.T.; Maciel, J.C.R.; Difonzo, C. Field-Evolved Resistance of the Fall Armyworm (Lepidoptera: Noctuidae) to Synthetic Insecticides in Puerto Rico and Mexico. J. Econ. Entomol. 2019, 112, 792–802. [Google Scholar] [CrossRef]
- Yu, S.J. Insecticide resistance in the fall armyworm, Spodoptera frugiperda (J. E. Smith). Pestic. Biochem. Physiol. 1991, 39, 84–91. [Google Scholar] [CrossRef]
- Wei, L.; Cao, L.; Miao, Y.; Wu, S.; Xu, S.; Wang, R.; Du, J.; Liang, A.; Fu, Y. Transcriptome analysis of Spodoptera frugiperda 9 (Sf9) cells infected with baculovirus, AcMNPV or AcMNPV-BmK IT. Biotechnol. Lett. 2017, 39, 1139. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Jin, F.; Hu, Z.; Chen, H.; Yin, F.; Li, Z.; Dong, X.; Zhang, D.; Ren, S.; Feng, X. Transcriptome Analysis of Chlorantraniliprole Resistance Development in the Diamondback Moth Plutella xylostella. PLoS ONE 2013, 8, e0072314. [Google Scholar] [CrossRef] [PubMed]
- Yi, Y.; Dou, G.; Yu, Z.; He, H.; Wang, C.; Li, L.; Zhou, J.; Liu, D.; Shi, J.; Li, G.; et al. Z-Ligustilide exerted hormetic effect on growth and detoxification enzymes of Spodoptera litura larvae. Evid. Based Complement. Altern. Med. 2018, 2018, 7104513. [Google Scholar] [CrossRef] [PubMed]
- Wei, N.; Zhong, Y.; Lin, L.; Xie, M.; Zhang, G.; Su, W.; Li, C.; Chen, H. Transcriptome analysis and identification of insecticide tolerance-related genes after exposure to insecticide in sitobion avenae. Genes 2019, 10, 951. [Google Scholar] [CrossRef] [PubMed]
- Enders, L.S.; Rault, L.C.; Moss, T.M.H.; Siegfried, B.D.; Miller, N.J. Transcriptional responses of soybean aphids to sublethal insecticide exposure. Insect Biochem. Mol. Biol. 2020, 118, 103285. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Kim, K.; Kwon, D.H.; Jeong, I.H.; Clark, J.M.; Lee, S.H. Transcriptome-based identification and characterization of genes commonly responding to five different insecticides in the diamondback moth, Plutella xylostella. Pestic. Biochem. Physiol. 2018, 144, 1–9. [Google Scholar] [CrossRef]
- Hafeez, M.; Liu, S.; Jan, S.; Shi, L.; Grandon, G.M.F.; Gulzar, A.; Ali, B.; Rehman, M.; Wang, M. Knock-down of gossypol-inducing cytochrome p450 genes reduced deltamethrin sensitivity in Spodoptera exigua (hübner). Int. J. Mol. Sci. 2019, 20, 2248. [Google Scholar] [CrossRef]
- Feyereisen, R. Insect P450 Enzymes. Annu. Rev. Entomol. 1999, 44, 507–533. [Google Scholar] [CrossRef]
- Willoughby, L.; Chung, H.; Lumb, C.; Robin, C.; Batterham, P.; Daborn, P.J. A comparison of Drosophila melanogaster detoxification gene induction responses for six insecticides, caffeine and phenobarbital. Insect Biochem. Mol. Biol. 2006, 36, 934–942. [Google Scholar] [CrossRef]
- Heckel, D.G. Insect Detoxification and Sequestration Strategies. In Annual Plant Reviews: Insect-Plant Interactions; Wiley-VCH: Weinheim, Germany, 2014; ISBN 9781118829783. [Google Scholar]
- Weill, M.; Berthomieu, A.; Berticat, C.; Lutfalla, G.; Nègre, V.; Pasteur, N.; Philips, A.; Leonetti, J.P.; Fort, P.; Raymond, M. Insecticide resistance: A silent base prediction. Curr. Biol. 2004, 14, R552–R553. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hu, B.; Zhang, S.H.; Ren, M.M.; Tian, X.R.; Wei, Q.; Mburu, D.K.; Su, J.Y. The expression of Spodoptera exigua P450 and UGT genes: Tissue specificity and response to insecticides. Insect Sci. 2017, 26, 199–216. [Google Scholar] [CrossRef] [PubMed]
- Bozzolan, F.; Siaussat, D.; Maria, A.; Durand, N.; Pottier, M.A.; Chertemps, T.; Coisne, M.M. Antennal uridine diphosphate (UDP)-glycosyltransferases in a pest insect: Diversity and putative function in odorant and xenobiotics clearance. Insect Mol. Biol. 2014, 12, 581–594. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.; Wang, W.; Zhang, D.; Lv, Y.; Zhou, D.; Ma, L.; Shen, B.; Sun, Y.; Zhu, C. The cuticle proteins: A putative role for deltamethrin resistance in Culex pipiens pallens. Parasitol. Res. 2015, 144, 4421–4429. [Google Scholar] [CrossRef]
- Wang, W.; Lv, Y.; Fang, F.; Hong, S.; Guo, Q.; Hu, S.; Zou, F.; Shi, L.; Lei, Z.; Ma, K.; et al. Identification of proteins associated with pyrethroid resistance by iTRAQ-based quantitative proteomic analysis in Culex pipiens pallens. Parasit. Vectors 2015, 9, 95. [Google Scholar] [CrossRef]
- Wang, L.L.; Huang, Y.; Lu, X.P.; Jiang, X.Z.; Smagghe, G.; Feng, Z.J.; Yuan, G.R.; Wei, D.; Wang, J.J. Overexpression of two α-esterase genes mediates metabolic resistance to malathion in the oriental fruit fly, Bactrocera dorsalis (Hendel). Insect Mol. Biol. 2015, 24, 467–479. [Google Scholar] [CrossRef]
- Nadal, A.; Quesada, I.; Tudurí, E.; Nogueiras, R.; Magdalena, P.A. Endocrine-disrupting chemicals and the regulation of energy balance. Nat. Rev. Endocrinol. 2017, 13, 536–546. [Google Scholar] [CrossRef]
- Bussy, U.; Boisseau, R.; Thobie-Gautier, C.; Boujtita, M. Electrochemistry-mass spectrometry to study reactive drug metabolites and CYP450 simulations. TrAC Trends Anal. Chem. 2015, 70, 67–73. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).