Identification of Autophagy-Related Genes in the Potato Psyllid, Bactericera cockerelli and Their Expression Profile in Response to ‘Candidatus Liberibacter Solanacearum’ in the Gut
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Colonies and Tomato Plants
2.2. Identification and Validation of Autophagy-Related Genes
2.3. Expression of Autophagy-Related Genes
2.4. Data Analysis
3. Results
3.1. Autophagy-Related Genes in Potato Psyllid and Other Hemipteran Insects
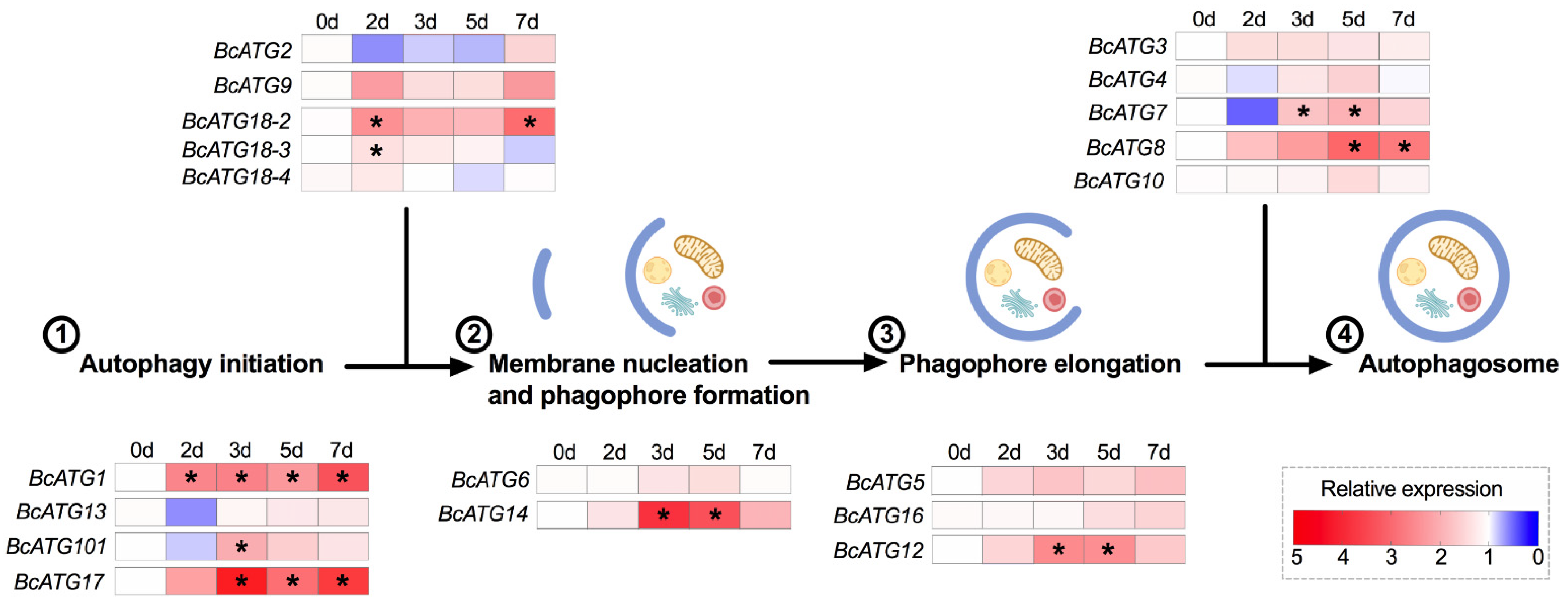
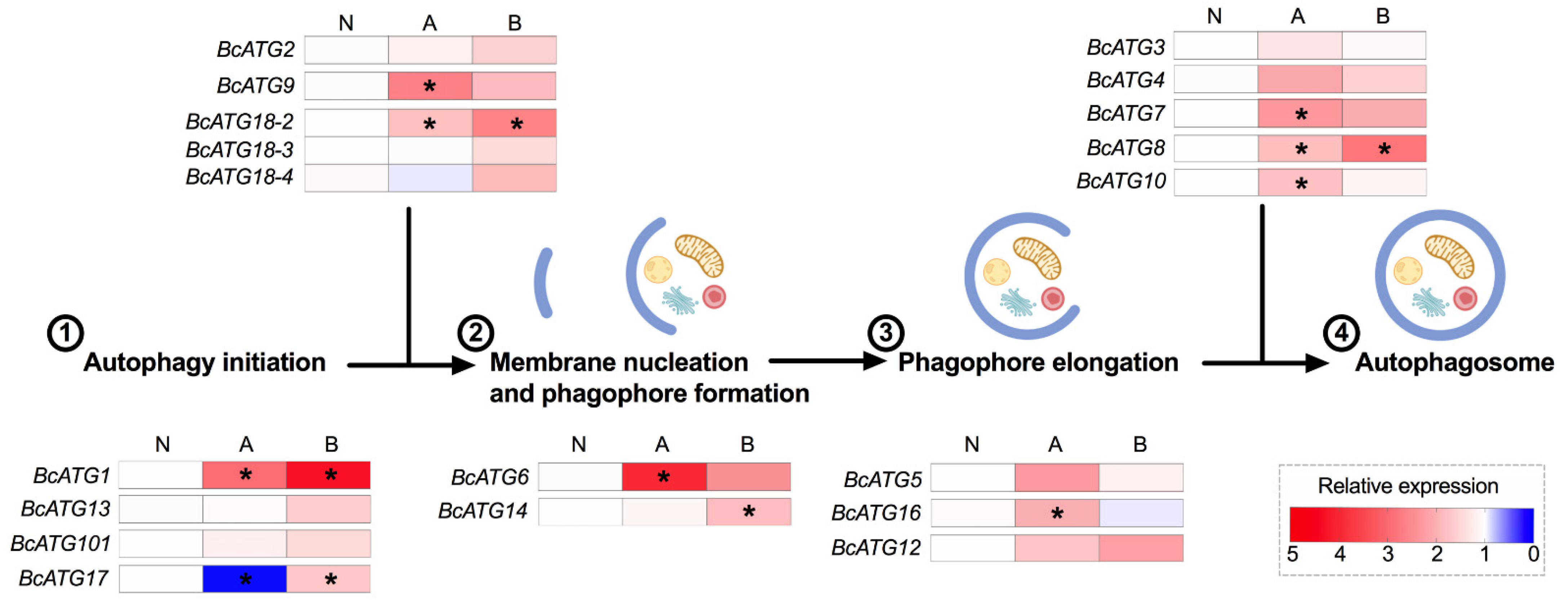
3.2. Expression of Autophagy-Related Genes upon Lso Infection
3.3. Expression of Autophagy-Related Genes in Response to Persistent Lso Infection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mizushima, N.; Levine, B.; Cuervo, A.M.; Klionsky, D.J. Autophagy fights disease through cellular self-digestion. Nature 2008, 451, 1069. [Google Scholar] [CrossRef] [Green Version]
- Khandia, R.; Dadar, M.; Munjal, A.; Dhama, K.; Karthik, K.; Tiwari, R.; Yatoo, M.; Iqbal, H.; Singh, K.P.; Joshi, S.K. A Comprehensive Review of Autophagy and Its Various Roles in Infectious, Non-Infectious, and Lifestyle Diseases: Current Knowledge and Prospects for Disease Prevention, Novel Drug Design, and Therapy. Cells 2019, 8, 674. [Google Scholar] [CrossRef] [Green Version]
- Yano, T.; Kurata, S. Intracellular recognition of pathogens and autophagy as an innate immune host defence. J. Biochem. 2011, 150, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Akioka, M.; Kondo-Kakuta, C.; Yamamoto, H.; Ohsumi, Y. Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J. Cell Sci. 2013, 126, 2534–2544. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Klionsky, D.J. Mammalian autophagy: Core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 2010, 22, 124–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zirin, J.; Perrimon, N. Drosophila as a Model System to Study Autophagy. Semin. Immunopathol. 2010, 32, 363–372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levine, B.; Kroemer, G. Biological functions of autophagy genes: A disease perspective. Cell 2019, 176, 11–42. [Google Scholar] [CrossRef] [Green Version]
- Nakatogawa, H.; Suzuki, K.; Kamada, Y.; Ohsumi, Y. Dynamics and diversity in autophagy mechanisms: Lessons from yeast. Nat. Rev. Mol. Cell Biol. 2009, 10, 458. [Google Scholar] [CrossRef] [Green Version]
- Mizushima, N.; Yoshimori, T.; Ohsumi, Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 2011, 27, 107–132. [Google Scholar] [CrossRef]
- He, C.; Klionsky, D.J. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 2009, 43, 67–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shelly, S.; Lukinova, N.; Bambina, S.; Berman, A.; Cherry, S. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity 2009, 30, 588–598. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.-L.; Wang, X.-R.; Wei, X.-M.; Huang, H.; Wu, J.-X.; Chen, X.-X.; Liu, S.-S.; Wang, X.-W. The autophagy pathway participates in resistance to tomato yellow leaf curl virus infection in whiteflies. Autophagy 2016, 12, 1560–1574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Chen, Q.; Li, M.; Mao, Q.; Chen, H.; Wu, W.; Jia, D.; Wei, T. Autophagy pathway induced by a plant virus facilitates viral spread and transmission by its insect vector. PLoS Pathog. 2017, 13, e1006727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voronin, D.; Cook, D.A.; Steven, A.; Taylor, M.J. Autophagy regulates Wolbachia populations across diverse symbiotic associations. Proc. Natl. Acad. Sci. USA 2012, 109, E1638–E1646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vigneron, A.; Masson, F.; Vallier, A.; Balmand, S.; Rey, M.; Vincent-Monégat, C.; Aksoy, E.; Aubailly-Giraud, E.; Zaidman-Rémy, A.; Heddi, A. Insects recycle endosymbionts when the benefit is over. Curr. Biol. 2014, 24, 2267–2273. [Google Scholar] [CrossRef] [Green Version]
- Hansen, A.; Trumble, J.; Stouthamer, R.; Paine, T. New Huanglongbing (HLB) Candidatus species,” Ca. Liberibacter psyllarous” found to infect tomato and potato is vectored by the psyllid Bactericera cockerelli. Appl. Environ. Microbiol. 2008, 73, 7531–7535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glynn, J.; Islam, M.; Bai, Y.; Lan, S.; Wen, A.; Gudmestad, N.; Civerolo, E.; Lin, H. Multilocus sequence typing of ‘Candidatus Liberibacter solanacearum’isolates from North America and New Zealand. J. Plant. Pathol. 2012, 94, 223–228. [Google Scholar]
- Haapalainen, M.L.; Wang, J.; Latvala, S.; Lehtonen, M.T.; Pirhonen, M.; Nissinen, A.I. Genetic variation of ‘Candidatus Liberibacter solanacearum’haplotype C and identification of a novel haplotype from Trioza urticae and stinging nettle. Phytopathology 2018, 108, 925–934. [Google Scholar] [CrossRef] [Green Version]
- Lin, H.; Islam, M.S.; Bai, Y.; Wen, A.; Lan, S.; Gudmestad, N.C.; Civerolo, E.L. Genetic diversity of ‘Cadidatus Liberibacter solanacearum’strains in the United States and Mexico revealed by simple sequence repeat markers. Eur. J. Plant. Pathol. 2012, 132, 297–308. [Google Scholar] [CrossRef] [Green Version]
- Nelson, W.R.; Sengoda, V.G.; Alfaro-Fernandez, A.O.; Font, M.I.; Crosslin, J.M.; Munyaneza, J.E. A new haplotype of “Candidatus Liberibacter solanacearum” identified in the Mediterranean region. Eur. J. Plant. Pathol. 2013, 135, 633–639. [Google Scholar] [CrossRef]
- Swisher Grimm, K.D.; Garczynski, S.F. Identification of a new haplotype of ‘Candidatus Liberibacter solanacearum’in Solanum tuberosum. Plant. Dis. 2018, 103, 468–474. [Google Scholar] [CrossRef] [Green Version]
- Mauck, K.E.; Sun, P.; Meduri, V.R.; Hansen, A.K. New Ca. Liberibacter psyllaurous haplotype resurrected from a 49-year-old specimen of Solanum umbelliferum: A native host of the psyllid vector. Sci. Rep. 2019, 9, 9530. [Google Scholar] [CrossRef] [PubMed]
- Liefting, L.W.; Sutherland, P.W.; Ward, L.I.; Paice, K.L.; Weir, B.S.; Clover, G.R. A new ‘Candidatus Liberibacter’species associated with diseases of solanaceous crops. Plant. Dis. 2009, 93, 208–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamborindeguy, C.; Huot, O.B.; Ibanez, F.; Levy, J. The influence of bacteria on multi-trophic interactions among plants, psyllids, and pathogen. Insect Sci. 2017, 24, 961–974. [Google Scholar] [CrossRef]
- Wamonje, F.; Zhou, N.; Bamrah, R.; Wist, T.; Prager, S. Detection and identification of a ‘Candidatus Liberibacter solanacerum’species from ash tree infesting psyllids. Phytopathology 2021, in press. [Google Scholar] [CrossRef] [PubMed]
- Kwak, Y.; Sun, P.; Meduri, V.R.; Percy, D.M.; Mauck, K.E.; Hansen, A.K. Uncovering Symbionts Across the Psyllid Tree of Life and the Discovery of a New Liberibacter Species,“Candidatus” Liberibacter capsica. Front. Microbiol. 2021, 12, 739763. [Google Scholar] [CrossRef]
- Munyaneza, J.E. Zebra chip disease of potato: Biology, epidemiology, and management. Am. J. Potato Res. 2012, 89, 329–350. [Google Scholar] [CrossRef] [Green Version]
- Cicero, J.; Fisher, T.; Brown, J.K. Localization of ‘Candidatus Liberibacter solanacearum’and Evidence for Surface Appendages in the Potato Psyllid Vector. Phytopathology 2016, 106, 142–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cicero, J.M.; Fisher, T.W.; Qureshi, J.A.; Stansly, P.A.; Brown, J.K. Colonization and Intrusive Invasion of Potato Psyllid by ‘Candidatus Liberibacter solanacearum’. Phytopathology 2017, 107, 36–49. [Google Scholar] [CrossRef] [Green Version]
- Cooper, W.R.; Sengoda, V.G.; Munyaneza, J.E. Localization of ‘Candidatus Liberibacter solanacearum’(Rhizobiales: Rhizobiaceae) in Bactericera cockerelli (Hemiptera: Triozidae). Ann. Entomol. Soc. Am. 2014, 107, 204–210. [Google Scholar] [CrossRef] [Green Version]
- Tang, X.-T.; Fortuna, K.; Mendoza Herrera, A.; Tamborindeguy, C. Liberibacter, a preemptive bacterium: Apoptotic response repression in the host gut at the early infection to facilitate its acquisition and transmission. Front. Microbiol. 2020, 11, 3348. [Google Scholar] [CrossRef]
- Tang, X.-T.; Jing, X.; Lu, M.-X.; Du, Y.-Z. No evidence for an effect of Wolbachia on mtDNA variation and evolution in natural populations of Sesamia inferens (Lepidoptera: Noctuidae). J. Integr. Agric. 2019, 18, 1050–1063. [Google Scholar] [CrossRef]
- Nachappa, P.; Levy, J.; Tamborindeguy, C. Transcriptome analyses of Bactericera cockerelli adults in response to “Candidatus Liberibacter solanacearum” infection. Mol. Genet. Genom. 2012, 287, 803–817. [Google Scholar] [CrossRef] [PubMed]
- Simonet, P.; Gaget, K.; Balmand, S.; Lopes, M.R.; Parisot, N.; Buhler, K.; Duport, G.; Vulsteke, V.; Febvay, G.; Heddi, A. Bacteriocyte cell death in the pea aphid/Buchnera symbiotic system. Proc. Natl. Acad. Sci. USA 2018, 115, E1819–E1828. [Google Scholar] [CrossRef] [Green Version]
- Gerardo, N.M.; Altincicek, B.; Anselme, C.; Atamian, H.; Barribeau, S.M.; De Vos, M.; Duncan, E.J.; Evans, J.D.; Gabaldón, T.; Ghanim, M. Immunity and other defenses in pea aphids, Acyrthosiphon pisum. Genome Biol. 2010, 11, R21. [Google Scholar] [CrossRef]
- Chen, W.; Hasegawa, D.K.; Kaur, N.; Kliot, A.; Pinheiro, P.V.; Luan, J.; Stensmyr, M.C.; Zheng, Y.; Liu, W.; Sun, H. The draft genome of whitefly Bemisia tabaci MEAM1, a global crop pest, provides novel insights into virus transmission, host adaptation, and insecticide resistance. BMC Biol. 2016, 14, 110. [Google Scholar] [CrossRef]
- Xie, W.; Chen, C.; Yang, Z.; Guo, L.; Yang, X.; Wang, D.; Chen, M.; Huang, J.; Wen, Y.; Zeng, Y. Genome sequencing of the sweetpotato whitefly Bemisia tabaci MED/Q. GigaScience 2017, 6, gix018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hunter, W.B.; Reese, J. International Psyllid Genome Consortium. The Asian citrus psyllid genome (Diaphorina citri, Hemiptera). J. Citrus Pathol. 2014, 1, 1. [Google Scholar] [CrossRef]
- Yao, J.; Saenkham, P.; Levy, J.; Ibanez, F.; Noroy, C.; Mendoza, A.; Huot, O.; MEYER, D.F.; Tamborindeguy, C. Interactions ‘Candidatus Liberibacter solanacearum’–Bactericera cockerelli: Haplotype effect on vector fitness and gene expression analyses. Front. Cell. Infect. Microbiol. 2016, 6, 62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nachappa, P.; Levy, J.; Pierson, E.; Tamborindeguy, C. Correlation between “Candidatus Liberibacter solanacearum” infection levels and fecundity in its psyllid vector. J. Invertebr. Pathol. 2014, 115, 55–61. [Google Scholar] [CrossRef]
- Li, W.; Abad, J.A.; French-Monar, R.D.; Rascoe, J.; Wen, A.; Gudmestad, N.C.; Secor, G.A.; Lee, M.; Duan, Y.; Levy, L. Multiplex real-time PCR for detection, identification and quantification of ‘Candidatus Liberibacter solanacearum’in potato plants with zebra chip. J. Microbiol. Methods 2009, 78, 59–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Érdi, B.; Nagy, P.; Zvara, Á.; Varga, Á.; Pircs, K.; Ménesi, D.; Puskás, L.G.; Juhász, G. Loss of the starvation-induced gene Rack1 leads to glycogen deficiency and impaired autophagic responses in Drosophila. Autophagy 2012, 8, 1124–1135. [Google Scholar] [CrossRef] [Green Version]
- Ibanez, F.; Hancock, J.; Tamborindeguy, C. Identification and expression analysis of aquaporins in the potato psyllid, Bactericera cockerelli. PLoS ONE 2014, 9, e111745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Ibanez, F.; Tamborindeguy, C. Selection of reference genes for expression analysis in the potato psyllid, Bactericera cockerelli. Insect Mol. Biol. 2016, 25, 227–238. [Google Scholar] [CrossRef]
- Levine, B.; Mizushima, N.; Virgin, H.W. Autophagy in immunity and inflammation. Nature 2011, 469, 323. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, G.; Cheng, M.I.; Chen, C.; Nguyen, B.N.; Whiteley, A.T.; Kianian, S.; Cox, J.S.; Green, D.R.; McDonald, K.L.; Portnoy, D.A. Listeria monocytogenes triggers noncanonical autophagy upon phagocytosis, but avoids subsequent growth-restricting xenophagy. Proc. Natl. Acad. Sci. USA 2018, 115, E210–E217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krokowski, S.; Mostowy, S. Interactions between Shigella flexneri and the autophagy machinery. Front. Cell. Infect. Microbiol. 2016, 6, 17. [Google Scholar] [CrossRef]
- Sorbara, M.T.; Ellison, L.K.; Ramjeet, M.; Travassos, L.H.; Jones, N.L.; Girardin, S.E.; Philpott, D.J. The protein ATG16L1 suppresses inflammatory cytokines induced by the intracellular sensors Nod1 and Nod2 in an autophagy-independent manner. Immunity 2013, 39, 858–873. [Google Scholar] [CrossRef] [Green Version]
- Ren, C.; Finkel, S.E.; Tower, J. Conditional inhibition of autophagy genes in adult Drosophila impairs immunity without compromising longevity. Exp. Gerontol. 2009, 44, 228–235. [Google Scholar] [CrossRef] [Green Version]
- Miller, C.; Celli, J. Avoidance and subversion of eukaryotic homeostatic autophagy mechanisms by bacterial pathogens. J. Mol. Biol. 2016, 428, 3387–3398. [Google Scholar] [CrossRef] [Green Version]
- Kuo, C.-J.; Hansen, M.; Troemel, E. Autophagy and innate immunity: Insights from invertebrate model organisms. Autophagy 2018, 14, 233–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gelino, S.; Hansen, M. Autophagy-an emerging anti-aging mechanism. J. Clin. Exp. Pathol. 2012, Suppl. 4, 006. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Brumell, J.H. Bacteria–autophagy interplay: A battle for survival. Nat. Rev. Microbiol. 2014, 12, 101. [Google Scholar] [CrossRef] [PubMed]
- Biering, S.B.; Choi, J.; Halstrom, R.A.; Brown, H.M.; Beatty, W.L.; Lee, S.; McCune, B.T.; Dominici, E.; Williams, L.E.; Orchard, R.C. Viral replication complexes are targeted by LC3-guided interferon-inducible GTPases. Cell Host Microbe 2017, 22, 74–85.e77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kageyama, S.; Omori, H.; Saitoh, T.; Sone, T.; Guan, J.-L.; Akira, S.; Imamoto, F.; Noda, T.; Yoshimori, T. The LC3 recruitment mechanism is separate from Atg9L1-dependent membrane formation in the autophagic response against Salmonella. Mol. Biol. Cell 2011, 22, 2290–2300. [Google Scholar] [CrossRef] [PubMed]
- Bernard, A.; Jin, M.; Xu, Z.; Klionsky, D.J. A large-scale analysis of autophagy-related gene expression identifies new regulators of autophagy. Autophagy 2015, 11, 2114–2122. [Google Scholar] [CrossRef]
- Choy, A.; Dancourt, J.; Mugo, B.; O’Connor, T.J.; Isberg, R.R.; Melia, T.J.; Roy, C.R. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science 2012, 338, 1072–1076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendoza Herrera, A.; Levy, J.; Harrison, K.; Yao, J.; Ibanez, F.; Tamborindeguy, C. Infection by Candidatus Liberibacter solanacearum’haplotypes A and B in Solanum lycopersicum’Moneymaker’. Plant. Dis. 2018, 102, 2009–2015. [Google Scholar] [CrossRef] [Green Version]
- Harrison, K.; Tamborindeguy, C.; Scheuring, D.C.; Herrera, A.M.; Silva, A.; Badillo-Vargas, I.E.; Miller, J.C.; Levy, J.G. Differences in Zebra Chip Severity between ‘Candidatus Liberibacter Solanacearum’Haplotypes in Texas. Am. J. Potato Res. 2019, 96, 86–93. [Google Scholar] [CrossRef]

| Gene Name | Code |
|---|---|
| Serine/threonine-protein kinase ULK2-like isoform 1 | BcATG1 |
| Autophagy-related protein 2-like | BcATG2 |
| Autophagy related protein Atg3-like protein | BcATG3 |
| Cysteine protease ATG4B-like isoform 1 | BcATG4B |
| Autophagy protein 5 | BcATG5 |
| Beclin-1-like protein | BcATG6 |
| Ubiquitin-like modifier-activating enzyme ATG7 | BcATG7 |
| Gamma-aminobutyric acid receptor-associated protein | BcATG8 |
| Autophagy-related protein 9A | BcATG9 |
| Ubiquitin-like-conjugating enzyme ATG10 | BcATG10 |
| Autophagy protein 12-like | BcATG12 |
| Autophagy-related protein 13 homolog | BcATG13 |
| Beclin 1-associated autophagy-related key regulator-like | BcATG14 |
| Autophagy-related protein 16-1-like | BcATG16 |
| RB1-inducible coiled-coil protein 1-like | BcATG17 |
| WD repeat domain phosphoinositide-interacting protein 2 | BcATG18-2 |
| WD repeat domain phosphoinositide-interacting protein 3 | BcATG18-3 |
| WD repeat domain phosphoinositide-interacting protein 4 | BcATG18-4 |
| Autophagy-related protein 101-like isoform 1 | BcATG101 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, X.-T.; Tamborindeguy, C. Identification of Autophagy-Related Genes in the Potato Psyllid, Bactericera cockerelli and Their Expression Profile in Response to ‘Candidatus Liberibacter Solanacearum’ in the Gut. Insects 2021, 12, 1073. https://doi.org/10.3390/insects12121073
Tang X-T, Tamborindeguy C. Identification of Autophagy-Related Genes in the Potato Psyllid, Bactericera cockerelli and Their Expression Profile in Response to ‘Candidatus Liberibacter Solanacearum’ in the Gut. Insects. 2021; 12(12):1073. https://doi.org/10.3390/insects12121073
Chicago/Turabian StyleTang, Xiao-Tian, and Cecilia Tamborindeguy. 2021. "Identification of Autophagy-Related Genes in the Potato Psyllid, Bactericera cockerelli and Their Expression Profile in Response to ‘Candidatus Liberibacter Solanacearum’ in the Gut" Insects 12, no. 12: 1073. https://doi.org/10.3390/insects12121073
APA StyleTang, X.-T., & Tamborindeguy, C. (2021). Identification of Autophagy-Related Genes in the Potato Psyllid, Bactericera cockerelli and Their Expression Profile in Response to ‘Candidatus Liberibacter Solanacearum’ in the Gut. Insects, 12(12), 1073. https://doi.org/10.3390/insects12121073





