Ecology and Genetic Structure of the Parasitoid Phobocampe confusa (Hymenoptera: Ichneumonidae) in Relation to Its Hosts, Aglais Species (Lepidoptera: Nymphalidae)
Abstract
1. Introduction
2. Materials and Methods
2.1. Host Butterflies
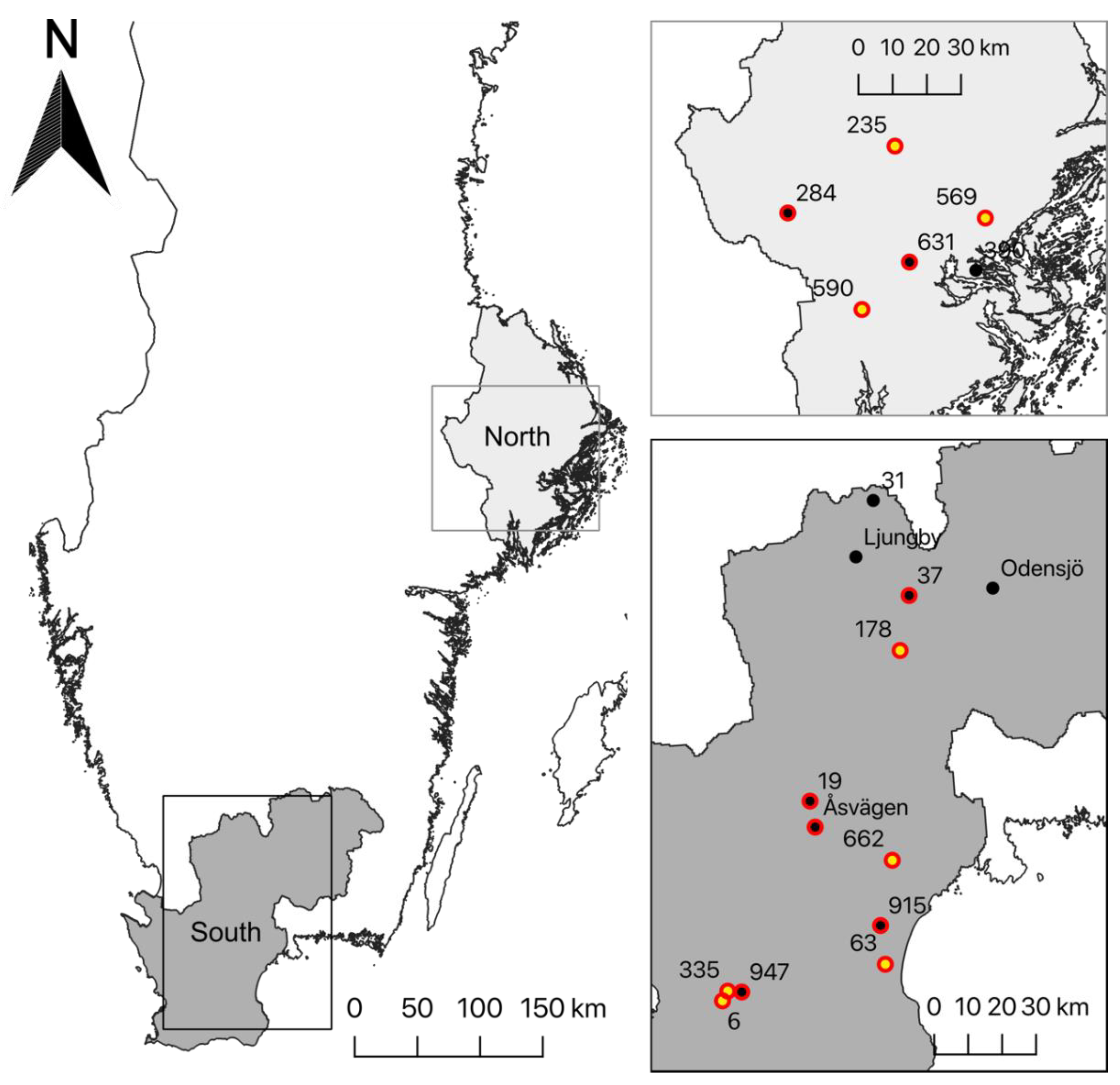
2.2. Study Area and Data Collection
2.3. Phenological Overlap Between P. confusa and Its Hosts
2.4. Pattern of Attack
2.5. Habitat
2.6. Genetic Structure of P. confusa and of A. urticae
2.6.1. DNA Extraction
2.6.2. Mitochondrial Genetic Variation
2.6.3. Nuclear Genetic Variation
3. Results
3.1. Patterns of Occurrence of P. confusa
3.2. Phenological Overlap Between P. confusa and Its Hosts
3.3. Pattern of Attack
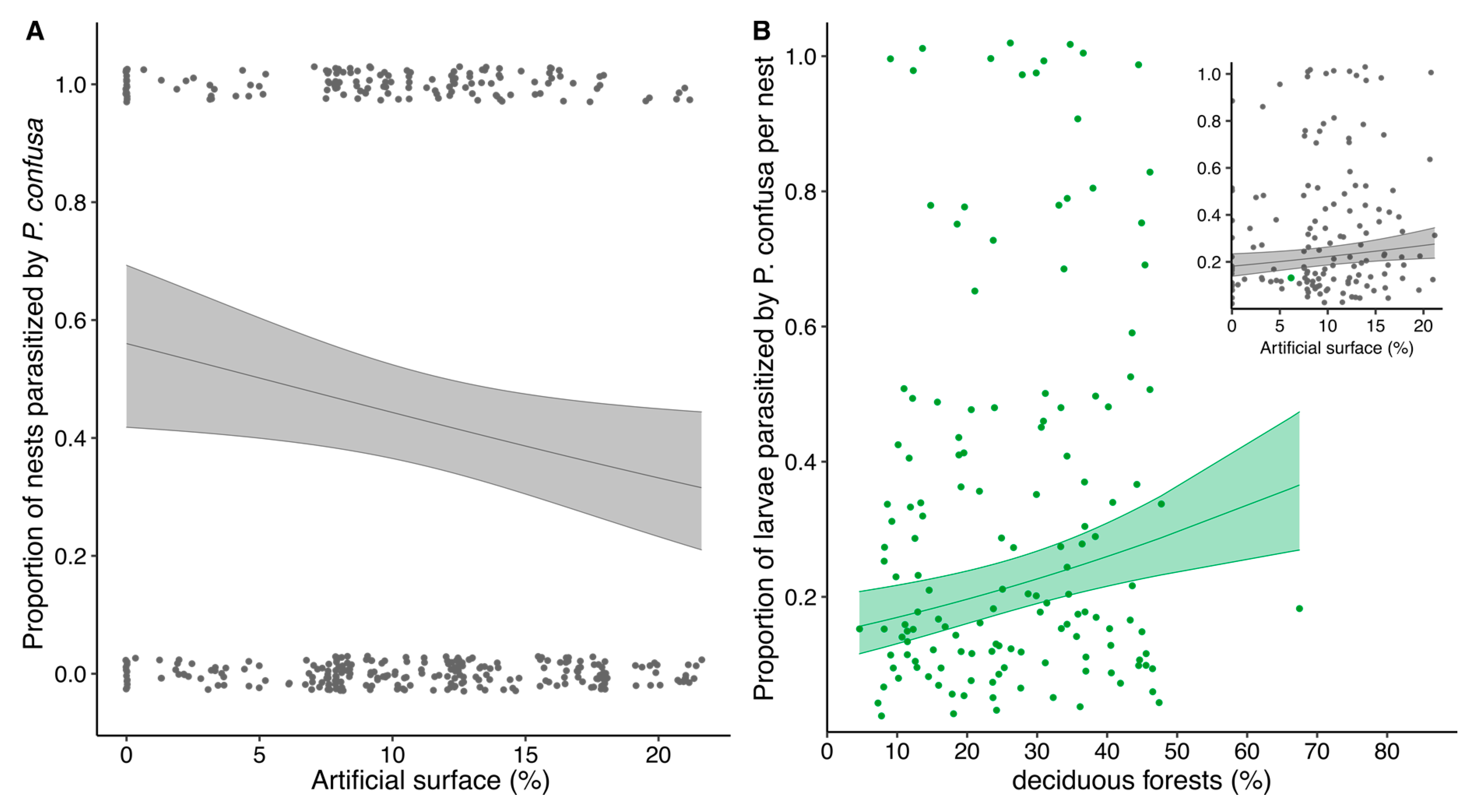
3.4. Habitat
3.5. Genetic Structure of P. confusa and of A. urticae
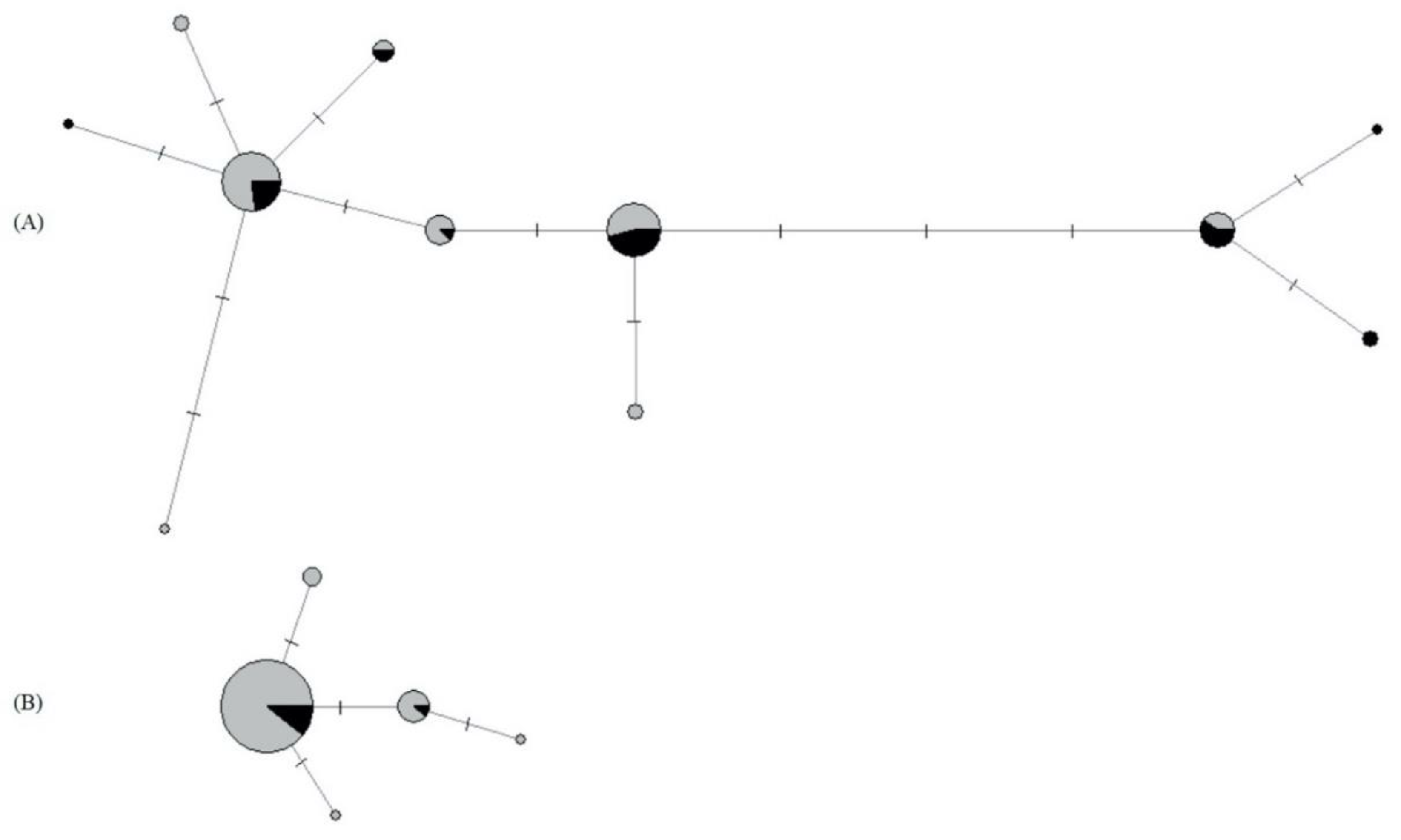
3.5.1. Mitochondrial and Nuclear Genetic Variation of P. confusa
3.5.2. Mitochondrial and Nuclear Genetic Variation of A. urticae
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gauld, I.D.; Wahl, D.B.; Broad, G.R. The suprageneric groups of the Pimplinae (Hymenoptera: Ichneumonidae): A cladistic re-evaluation and evolutionary biological study. Zool. J. Linn. Soc. 2002, 136, 421–485. [Google Scholar] [CrossRef]
- LaSalle, J.; Gauld, I.D. Parasitic Hymenoptera and the biodiversity crisis. Redia 1991, 74, 315–334. [Google Scholar]
- Shaw, M.R.; Hochberg, M.E. The neglect of parasitic Hymenoptera in insect conservation strategies: The British fauna as a prime example. J. Insect Conserv. 2001, 5, 253–263. [Google Scholar] [CrossRef]
- Shaw, M.R. Habitat considerations for parasitic wasps (Hymenoptera). J. Insect Conserv. 2006, 10, 117–127. [Google Scholar] [CrossRef]
- LaSalle, J.; Gauld, I.D. Hymenoptera: Their biodiversity, and their impact on the diversity of other organisms. Hymenopt. Biodivers. 1993, 1–26. [Google Scholar]
- Shaw, M.R.; Stefanescu, C.; Van Nouhuys, S. Parasitoids of European butterflies. In Ecology of Butterflies in Europe; Settele, J., Shreeve, T.G., Konvicka, M., van Dyck, H., Eds.; Cambridge University Press: Cambridge, UK, 2009; pp. 130–156. [Google Scholar]
- Schwarzfeld, M.D. Ichneumonidae (Hymenoptera) of the Canadian prairies ecozone: A Review. Arthropods Can. Grassl. 2014, 4, 317–397. [Google Scholar]
- Cronin, J.T.; Reeve, J.D. Host–parasitoid spatial ecology: A plea for a landscape-level synthesis. Proc. R. Soc. B Biol. Sci. 2005, 272, 2225–2235. [Google Scholar] [CrossRef]
- Wei, S.-J.; Zhou, Y.; Fan, X.-L.; Hoffmann, A.A.; Cao, L.-J.; Chen, X.-X.; Xu, Z.-F. Different genetic structures revealed resident populations of a specialist parasitoid wasp in contrast to its migratory host. Ecol. Evol. 2017, 7, 5400–5409. [Google Scholar] [CrossRef]
- Ewers, R.M.; Didham, R.K. Confounding factors in the detection of species responses to habitat fragmentation. Biol. Rev. 2006, 81, 117–142. [Google Scholar] [CrossRef]
- Quicke, D.L. Part III Ecology and Diversity. In The Braconid and Ichneumonid Parasitoid Wasps: Biology, Systematics, Evolution and Ecology; John Wiley & Sons: Chichester, UK, 2015; pp. 467–471. ISBN 1-118-90705-1. [Google Scholar]
- Laliberté, E.; Tylianakis, J.M. Deforestation homogenizes tropical parasitoid–host networks. Ecology 2010, 91, 1740–1747. [Google Scholar] [CrossRef]
- Kruess, A.; Tscharntke, T. Habitat fragmentation, species loss, and biological control. Science 1994, 264, 1581–1584. [Google Scholar] [CrossRef]
- Kruess, A.; Tscharntke, T. Species richness and parasitism in a fragmented landscape: Experiments and field studies with insects on Vicia sepium. Oecologia 2000, 122, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Van Nouhuys, S.; Hanski, I. Colonization rates and distances of a host butterfly and two specific parasitoids in a fragmented landscape. J. Anim. Ecol. 2002, 71, 639–650. [Google Scholar] [CrossRef]
- Couchoux, C.; Seppä, P.; Van Nouhuys, S. Strong dispersal in a parasitoid wasp overwhelms habitat fragmentation and host population dynamics. Mol. Ecol. 2016, 25, 3344–3355. [Google Scholar] [CrossRef] [PubMed]
- Audusseau, H.; Ryrholm, N.; Stefanescu, C.; Tharel, S.; Jansson, C.; Champeaux, L.; Shaw, M.R.; Raper, C.; Lewis, O.T.; Janz, N.; et al. Altered parasitism of a butterfly assemblage associated with a range-expanding species. Biorxiv 2020. [Google Scholar] [CrossRef]
- Shaw, M.R.; Horstmann, K.; Whiffin, A.L. Two hundred and twenty-five species of reared western Palaearctic Campopleginae (Hymenoptera: Ichneumonidae) in the National Museums of Scotland, with descriptions of new species of Campoplex and Diadegma, and records of fifty-five species new to Britain. Entomol. Gaz. 2016, 66, 245–247. [Google Scholar]
- Wiemers, M.; Chazot, N.; Wheat, C.W.; Schweiger, O.; Wahlberg, N. A complete time-calibrated multi-gene phylogeny of the European butterflies. ZooKeys 2020, 844175. [Google Scholar]
- Eliasson, C.U.; Ryrholm, N.; Gärdenfors, U. Nationalnyckeln till Sveriges flora och fauna: Fjärilar. Dagfjärilar: (Hesperiidae-Nymphalidae); Swedish University of Agricultural Sciences: Uppsala, Sweden, 2005. [Google Scholar]
- Lindén, A. Adaptive and nonadaptive changes in phenological synchrony. Proc. Natl. Acad. Sci. USA 2018, 115, 5057–5059. [Google Scholar] [CrossRef]
- Hartig, F. DHARMa: Residual Diagnostics for Hierarchical (Multi-Level/Mixed) Regression Models. R Package Version 0.3.0. Available online: http://florianhartig.github.io/DHARMa/ (accessed on 25 May 2020).
- Pebesma, E. Simple features for R: Standardized support for spatial vector data. R J. 2018, 10, 439–446. [Google Scholar] [CrossRef]
- Hijmans, R.J. Raster: Geographic Data Analysis and Modeling. R Package Version 3.0-12. Available online: https://CRAN.R-project.org/package=raster (accessed on 25 May 2020).
- Vos, P.; Hogers, R.; Bleeker, M.; Reijans, M.; van de Lee, T.; Hornes, M.; Friters, A.; Pot, J.; Paleman, J.; Kuiper, M. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 1995, 23, 4407–4414. [Google Scholar] [CrossRef]
- Meudt, H.M.; Clarke, A.C. Almost forgotten or latest practice? AFLP applications, analyses and advances. Trends Plant Sci. 2007, 12, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Hebert, P.D.N.; Ratnasingham, S.; de Waard, J.R. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. London. Ser. B: Biol. Sci. 2003, 270, S96–S99. [Google Scholar] [CrossRef] [PubMed]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Bandelt, H.J.; Forster, P.; Sykes, B.C.; Richards, M.B. Mitochondrial portraits of human populations using median networks. Genetics 1995, 141, 743–753. [Google Scholar]
- Ajmone-Marsan, P.; Vecchiotti-Antaldi, G.; Bertoni, G.; Valentini, A.; Cassandro, M.; Kuiper, M. AFLP™ markers for DNA fingerprinting in cattle. Anim. Genet. 1997, 28, 418–426. [Google Scholar] [CrossRef]
- Baus, E.; Darrock, D.J.; Bruford, M.W. Gene-flow patterns in Atlantic and Mediterranean populations of the Lusitanian sea star Asterina gibbosa. Mol. Ecol. 2005, 14, 3373–3382. [Google Scholar] [CrossRef]
- King, R.A.; Tibble, A.L.; Symondson, W.O. Opening a can of worms: Unprecedented sympatric cryptic diversity within British lumbricid earthworms. Mol. Ecol. 2008, 17, 4684–4698. [Google Scholar] [CrossRef]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987; ISBN 0-231-06321-0. [Google Scholar]
- Ehrich, D. aflpdat: A collection of r functions for convenient handling of AFLP data. Mol. Ecol. Notes 2006, 6, 603–604. [Google Scholar] [CrossRef]
- Guillot, G.; Santos, F. Using AFLP markers and the Geneland program for the inference of population genetic structure. Mol. Ecol. Resour. 2010, 10, 1082–1084. [Google Scholar] [CrossRef]
- Guillot, G.; Mortier, F.; Estoup, A. Geneland: A computer package for landscape genetics. Mol. Ecol. Notes 2005, 5, 712–715. [Google Scholar] [CrossRef]
- Hance, T.; van Baaren, J.; Vernon, P.; Boivin, G. Impact of Extreme Temperatures on Parasitoids in a Climate Change Perspective. Annu. Rev. Entomol. 2007, 52, 107–126. [Google Scholar] [CrossRef] [PubMed]
- Stocker, T.F.; Qin, D.; Plattner, G.-K.; Tignor, M.M.B.; Allen, S.K.; Boschung, J.; Nauels, A.; Xia, Y.; Bex, V.; Midgley, P.M. IPCC 2013: The physical science basis. In Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; Cambridge University Press: Cambridge, UK, 2013. [Google Scholar]
- Stireman, J.O.; Dyer, L.A.; Janzen, D.H.; Singer, M.S.; Lill, J.T.; Marquis, R.J.; Ricklefs, R.E.; Gentry, G.L.; Hallwachs, W.; Coley, P.D. Climatic unpredictability and parasitism of caterpillars: Implications of global warming. Proc. Natl. Acad. Sci. USA 2005, 102, 17384–17387. [Google Scholar] [CrossRef]
- Jeffs, C.T.; Lewis, O.T. Effects of climate warming on host–parasitoid interactions. Ecol. Entomol. 2013, 38, 209–218. [Google Scholar] [CrossRef]
- Deutsch, C.A.; Tewksbury, J.J.; Huey, R.B.; Sheldon, K.S.; Ghalambor, C.K.; Haak, D.C.; Martin, P.R. Impacts of climate warming on terrestrial ectotherms across latitude. Proc. Natl. Acad. Sci. USA 2008, 105, 6668–6672. [Google Scholar] [CrossRef]
- Schreven, S.J.; Frago, E.; Stens, A.; De Jong, P.W.; Van Loon, J.J. Contrasting effects of heat pulses on different trophic levels, an experiment with a herbivore-parasitoid model system. PLoS ONE 2017, 12, e0176704. [Google Scholar] [CrossRef]
- Tougeron, K.; Damien, M.; Le Lann, C.; Brodeur, J.; van Baaren, J. Rapid responses of winter aphid-parasitoid communities to climate warming. Front. Ecol. Evol. 2018, 6, e173. [Google Scholar] [CrossRef]
- Dyer, L.A.; Richards, L.A.; Short, S.A.; Dodson, C.D. Effects of CO2 and Temperature on tritrophic interactions. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Jeffries, M.J.; Lawton, J.H. Enemy free space and the structure of ecological communities. Biol. J. Linn. Soc. 1984, 23, 269–286. [Google Scholar] [CrossRef]
- Keane, R.M.; Crawley, M.J. Exotic plant invasions and the enemy release hypothesis. Trends Ecol. Evol. 2002, 17, 164–170. [Google Scholar] [CrossRef]
- Taylor, K. Biological flora of the British Isles: Urtica dioica L. J. Ecol. 2009, 97, 1436–1458. [Google Scholar] [CrossRef]
- Corcos, D.; Cerretti, P.; Caruso, V.; Mei, M.; Falco, M.; Marini, L. Impact of urbanization on predator and parasitoid insects at multiple spatial scales. PLoS ONE 2019, 14. [Google Scholar] [CrossRef] [PubMed]
- Quicke, D.L. Part I Morphology and Biology, chapter 8 Host location, associative learning and host assessment. In The Braconid and Ichneumonid Parasitoid Wasps: Biology, Systematics, Evolution and Ecology; John Wiley & Sons: Chichester, UK, 2015; pp. 127–136. ISBN 1-118-90705-1. [Google Scholar]
- Werren, J.H. The evolution of inbreeding in haplodiploid organisms. In The Natural History of Inbreeding and Outbreeding; University of Chicago Press: Chicago, IL, USA, 1993; pp. 42–59. [Google Scholar]
- Charlesworth, D.; Charlesworth, B. Inbreeding depression and its evolutionary consequences. Annu. Rev. Ecol. Syst. 1987, 18, 237–268. [Google Scholar] [CrossRef]
- Henter, H.J. Inbreeding depression and haplodiploidy: Experimental measures in a parasitoid and comparisons across diploid and haplodiploid insect taxa. Evolution 2003, 57, 1793–1803. [Google Scholar] [CrossRef]
- Vandewoestijne, S.; Nève, G.; Baguette, M. Spatial and temporal population genetic structure of the butterfly Aglais urticae L. (Lepidoptera, Nymphalidae). Mol. Ecol. 1999, 8, 1539–1543. [Google Scholar] [CrossRef]
- Vandewoestijne, S.; Baguette, M.; Brakefield, P.M.; Saccheri, I.J. Phylogeography of Aglais urticae (Lepidoptera) based on DNA sequences of the mitochondrial COI gene and control region. Mol. Phylogenet. Evol. 2004, 31, 630–646. [Google Scholar] [CrossRef]
- Davey, J.W.; Blaxter, M.L. RADSeq: Next-generation population genetics. Brief. Funct. Genom. 2010, 9, 416–423. [Google Scholar] [CrossRef]
- Gaston, K.J. The magnitude of global insect species richness. Conserv. Biol. 1991, 5, 283–296. [Google Scholar] [CrossRef]
- Pyornila, M. Parasitism in Aglais urticae (L.) (Lep., Nymphalidae). III. Parasitism of larval stages by ichneumonids. Ann. Entomol. Fenn. 1976, 42, 156–161. [Google Scholar]

| Year | Region/Host | A. urticae | A. io | A. levana | Total by Region |
|---|---|---|---|---|---|
| 2017 | North | 65/22/374/57 | 81/30/589/70 | - | 146/52/963/127 |
| South | 82/34/612/68 | 27/10/605/45 | 2/1/712/69 | 111/45/1929/182 | |
| Total by species | 147/56/986/125 | 108/40/1194/115 | 2/1/712/69 | 257/97/2892/309 | |
| 2018 | North | 6/4/598/58 | 11/3/379/26 | - | 17/7/977/84 |
| South | 78/23/669/66 | 76/19/685/63 | 0/0/871/98 | 154/42/2225/227 | |
| Total by species | 84/27/1267/124 | 87/22/1064/89 | 0/0/871/98 | 171/49/3202/311 |
| Variables | Sum sq. | Df | F | p |
|---|---|---|---|---|
| Host | 0.802 | 2 | 6.17 | 0.004 |
| Region | 0.003 | 1 | 0.038 | 0.85 |
| Year | 0.347 | 1 | 5.34 | 0.024 |
| Region × host | 0.458 | 1 | 7.04 | 0.010 |
| Residuals | 4.031 | 62 |
| Variables | Proportion of Nest Parasitized | Proportion of Larvae Parasitized Per Nest | ||||
|---|---|---|---|---|---|---|
| LR Chisq | Df | p | LR Chisq | Df | p | |
| Phenological overlap | 14.40 | 1 | <0.001 | 10.40 | 1 | 0.001 |
| Phenological overlap2 | 5.57 | 1 | 0.018 | 9.83 | 1 | 0.002 |
| Instar at collection | 25.78 | 3 | <0.001 | 8.38 | 3 | 0.039 |
| Butterfly species | - | - | - | 5.08 | 1 | 0.024 |
| Year | - | - | - | 5.25 | 1 | 0.022 |
| Week of sampling | - | - | - | 7.90 | 1 | 0.005 |
| Week of sampling2 | - | - | - | 7.36 | 1 | 0.007 |
| Region | - | - | - | 0.47 | 1 | 0.49 |
| Region × year | - | - | - | 11.57 | 1 | <0.001 |
| Species | Region | Site | Molecular Data | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CO1 | AFLPs | |||||||||
| NCO1 | NH | NPS | Hd | π | NAFLP | VM% | Gdiv | |||
| A. urticae | North | 235 | 15 | 4 | 6 | 0.714 | 0.00370 | 15 | 54.3 | 0.147 |
| 569 | 4 | 3 | 7 | 0.833 | 0.00608 | 5 | 26.3 | 0.124 | ||
| 590 | 12 | 5 | 6 | 0.833 | 0.00440 | 11 | 49.0 | 0.138 | ||
| Total | 31 | 8 | 9 | 0.800 | 0.00428 | 31 | 72.0 | 0.144 | ||
| South | 6 | 24 | 6 | 7 | 0.688 | 0.00296 | 24 | 65.8 | 0.142 | |
| 63 | 6 | 2 | 3 | 0.533 | 0.00265 | 6 | 31.7 | 0.132 | ||
| 178 | 3 | 2 | 3 | 0.667 | 0.00199 | 3 | 16.5 | 0.110 | ||
| 335 | 7 | 3 | 2 | 0.762 | 0.00174 | 7 | 38.3 | 0.152 | ||
| 662 | 15 | 5 | 7 | 0.748 | 0.00272 | 15 | 53.1 | 0.139 | ||
| Total | 55 | 8 | 10 | 0.741 | 0.00277 | 55 | 87.2 | 0.143 | ||
| P. confusa | North | 235 | 2 | 1 | 0 | - | - | 2 | 3.66 | 0.036 |
| 284 | 1 | 1 | - | - | - | 0 | - | - | ||
| 569 | 2 | 1 | 0 | - | - | 1 | - | - | ||
| 590 | 3 | 1 | 0 | - | - | 2 | 4.88 | 0.049 | ||
| 631 | 1 | 1 | - | - | - | 1 | - | - | ||
| Total | 9 | 2 | 1 | 0.222 | 0.00036 | 6 | 8.54 | 0.040 | ||
| South | 6 | 50 | 3 | 2 | 0.251 | 0.00041 | 16 | 13.4 | 0.049 | |
| 19 | 1 | 1 | - | - | - | 1 | - | - | ||
| 37 | 1 | 1 | - | - | - | 1 | - | - | ||
| 63 | 6 | 1 | 0 | 0.000 | 0.00000 | 4 | 8.53 | 0.043 | ||
| 178 | 5 | 3 | 3 | 0.800 | 0.00259 | 2 | 4.87 | 0.049 | ||
| 335 | 10 | 1 | 0 | 0.000 | 0.00000 | 5 | 7.32 | 0.039 | ||
| 662 | 1 | 1 | - | - | - | 0 | - | - | ||
| 915 | 2 | 2 | 1.000 | 0.00162 | 1 | - | - | |||
| 947 | 2 | 1 | 0 | 0.000 | 0.00000 | 2 | 2.44 | 0.024 | ||
| Åsvägen | 1 | 1 | - | - | - | 1 | - | - | ||
| Total | 79 | 5 | 4 | 0.294 | 0.00053 | 33 | 15.8 | 0.045 | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Audusseau, H.; Baudrin, G.; Shaw, M.R.; Keehnen, N.L.P.; Schmucki, R.; Dupont, L. Ecology and Genetic Structure of the Parasitoid Phobocampe confusa (Hymenoptera: Ichneumonidae) in Relation to Its Hosts, Aglais Species (Lepidoptera: Nymphalidae). Insects 2020, 11, 478. https://doi.org/10.3390/insects11080478
Audusseau H, Baudrin G, Shaw MR, Keehnen NLP, Schmucki R, Dupont L. Ecology and Genetic Structure of the Parasitoid Phobocampe confusa (Hymenoptera: Ichneumonidae) in Relation to Its Hosts, Aglais Species (Lepidoptera: Nymphalidae). Insects. 2020; 11(8):478. https://doi.org/10.3390/insects11080478
Chicago/Turabian StyleAudusseau, Hélène, Gaspard Baudrin, Mark R. Shaw, Naomi L. P. Keehnen, Reto Schmucki, and Lise Dupont. 2020. "Ecology and Genetic Structure of the Parasitoid Phobocampe confusa (Hymenoptera: Ichneumonidae) in Relation to Its Hosts, Aglais Species (Lepidoptera: Nymphalidae)" Insects 11, no. 8: 478. https://doi.org/10.3390/insects11080478
APA StyleAudusseau, H., Baudrin, G., Shaw, M. R., Keehnen, N. L. P., Schmucki, R., & Dupont, L. (2020). Ecology and Genetic Structure of the Parasitoid Phobocampe confusa (Hymenoptera: Ichneumonidae) in Relation to Its Hosts, Aglais Species (Lepidoptera: Nymphalidae). Insects, 11(8), 478. https://doi.org/10.3390/insects11080478





