Lysine Acetyltransferase p300/CBP Plays an Important Role in Reproduction, Embryogenesis and Longevity of the Pea Aphid Acyrthosiphon pisum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Aphid Rearing
2.2. RNA Extraction, Target Gene Identification
2.3. Synthesis of dsRNA
2.4. RNAi Injection Assays
2.5. Quantitative PCR (qPCR)
2.6. Bioinformatics and Data Analysis
3. Results
3.1. Genomic Sequence, mRNA Sequence and Protein Domain Analysis of A. pisum p300/CBP
3.2. The Effect of RNAi-Mediated Attenuation of p300/CBP on Aphid Life-History Traits
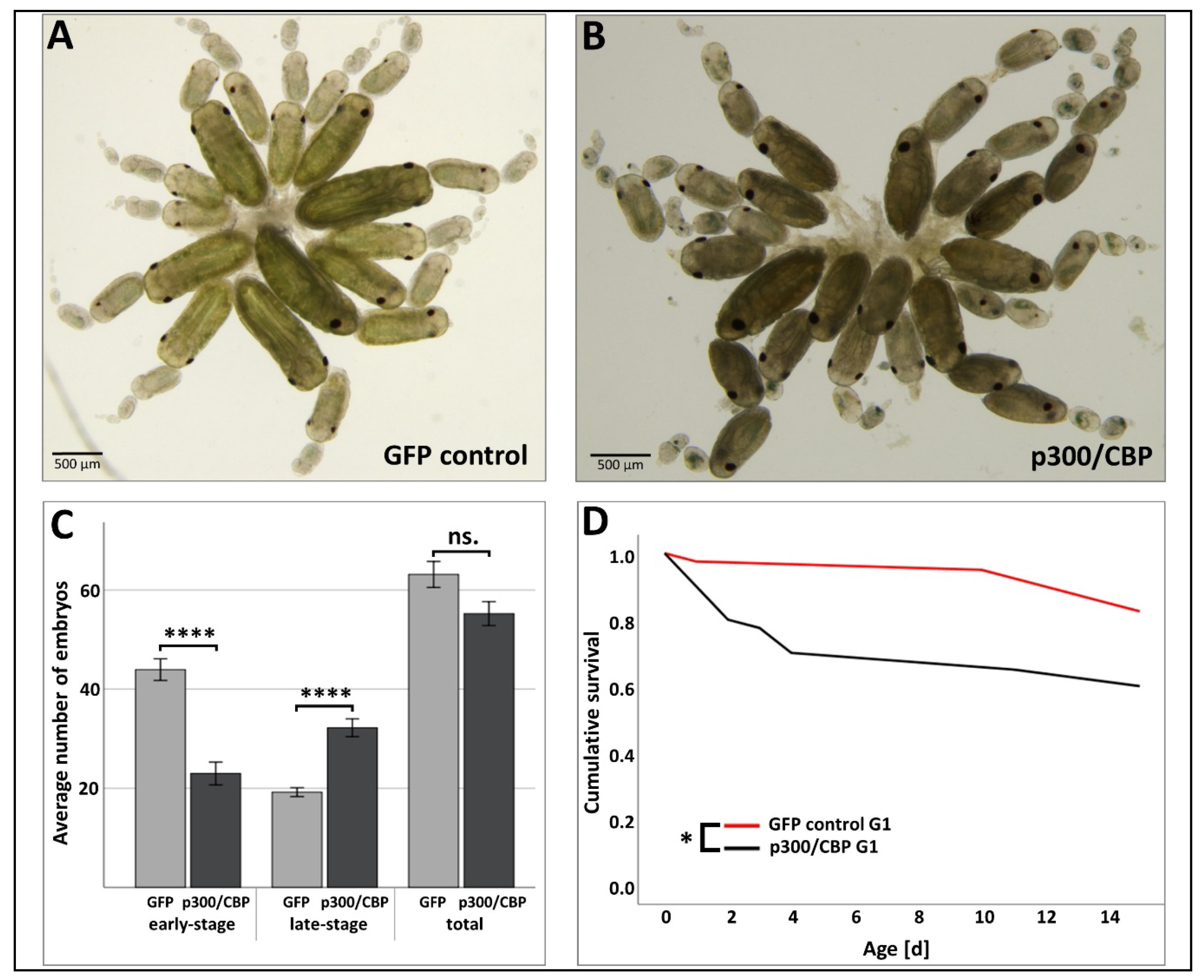
3.3. Effect of RNAi-Mediated Knockdown of p300/CBP on Aphid Embryogenesis and the Transgenerational Silencing Effect in the G1 Generation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Drazic, A.; Myklebust, L.M.; Ree, R.; Arnesen, T. The world of protein acetylation. Biochim. Biophys. Acta BBA Proteins Proteom. 2016, 1864, 1372–1401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farrugia, M.A.; Puglielli, L. Nε-lysine acetylation in the endoplasmic reticulum—A novel cellular mechanism that regulates proteostasis and autophagy. J. Cell Sci. 2018, 131, jcs221747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, I.; Conrad, R.J.; Verdin, E.; Ott, M. Lysine Acetylation Goes Global: From Epigenetics to Metabolism and Therapeutics. Chem. Rev. 2018, 118, 1216–1252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narita, T.; Weinert, B.T.; Choudhary, C. Functions and mechanisms of non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 156–174. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Haberland, M.; Montgomery, R.L.; Olson, E.N. The many roles of histone deacetylases in development and physiology: Implications for disease and therapy. Nat. Rev. Genet. 2009, 10, 32–42. [Google Scholar] [CrossRef]
- Bassett, S.A.; Barnett, M.P.G. The Role of Dietary Histone Deacetylases (HDACs) Inhibitors in Health and Disease. Nutrients 2014, 6, 4273–4301. [Google Scholar] [CrossRef] [Green Version]
- Damjanovski, S.; Sachs, L.M.; Shi, Y.B. Multiple stage-dependent roles for histone deacetylases during amphibian embryogenesis: Implications for the involvement of extracellular matrix remodeling. Int. J. Dev. Biol. 2004, 44, 769–776. [Google Scholar]
- Schneider, A.; Chatterjee, S.; Bousiges, O.; Selvi, B.R.; Swaminathan, A.; Cassel, R.; Blanc, F.; Kundu, T.K.; Boutillier, A.-L. Acetyltransferases (HATs) as Targets for Neurological Therapeutics. Neurotherapeutics 2013, 10, 568–588. [Google Scholar] [CrossRef]
- Dancy, B.M.; Cole, P.A. Protein lysine acetylation by p300/CBP. Chem. Rev. 2015, 115, 2419–2452. [Google Scholar] [CrossRef]
- Zucconi, B.E.; Makofske, J.L.; Meyers, D.J.; Hwang, Y.; Wu, M.; Kuroda, M.I.; Cole, P.A. Combination Targeting of the Bromodomain and Acetyltransferase Active Site of p300/CBP. Biochemistry 2019, 58, 2133–2143. [Google Scholar] [CrossRef] [PubMed]
- Ramos, Y.F.M.; Hestand, M.S.; Verlaan, M.; Krabbendam, E.; Ariyurek, Y.; van Galen, M.; van Dam, H.; van Ommen, G.-J.B.; den Dunnen, J.T.; Zantema, A.; et al. Genome-wide assessment of differential roles for p300 and CBP in transcription regulation. Nucleic Acids Res. 2010, 38, 5396–5408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bordoli, L.; Netsch, M.; Lüthi, U.; Lutz, W.; Eckner, R. Plant orthologs of p300/CBP: Conservation of a core domain in metazoan p300/CBP acetyltransferase-related proteins. Nucleic Acids Res. 2001, 29, 589–597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dutto, I.; Scalera, C.; Prosperi, E. CREBBP and p300 lysine acetyl transferases in the DNA damage response. Cell. Mol. Life Sci. 2018, 75, 1325–1338. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Tang, Y.; Cole, P.A.; Marmorstein, R. Structure and chemistry of the p300/CBP and Rtt109 histone acetyltransferases: Implications for histone acetyltransferase evolution and function. Curr. Opin. Struct. Biol. 2008, 18, 741–747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akimaru, H.; Hou, D.-X.; Ishii, S. Drosophila CBP is required for dorsal–dependent twist gene expression. Nat. Genet. 1997, 17, 211–214. [Google Scholar] [CrossRef]
- Akimaru, H.; Chen, Y.; Dai, P.; Hou, D.-X.; Nonaka, M.; Smolik, S.M.; Armstrong, S.; Goodman, R.H.; Ishii, S. Drosophila CBP is a co-activator of cubitus interruptus in hedgehog signalling. Nature 1997, 386, 735–738. [Google Scholar] [CrossRef]
- Domínguez, M.; Brunner, M.; Hafen, E.; Basler, K. Sending and Receiving the Hedgehog Signal: Control by the Drosophila Gli Protein Cubitus interruptus. Science 1996, 272, 1621–1625. [Google Scholar] [CrossRef]
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. Publ. Protein Soc. 2019. [Google Scholar] [CrossRef]
- Lasko, L.M.; Jakob, C.G.; Edalji, R.P.; Qiu, W.; Montgomery, D.; Digiammarino, E.L.; Hansen, T.M.; Risi, R.M.; Frey, R.; Manaves, V.; et al. Discovery of a potent catalytic p300/CBP inhibitor that targets lineage-specific tumors. Nature 2017, 550, 128–132. [Google Scholar] [CrossRef]
- Goodman, R.H.; Smolik, S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000, 14, 1553–1577. [Google Scholar] [PubMed]
- Yao, T.P.; Oh, S.P.; Fuchs, M.; Zhou, N.D.; Ch’ng, L.E.; Newsome, D.; Bronson, R.T.; Li, E.; Livingston, D.M.; Eckner, R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 1998, 93, 361–372. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Nicolas, A.; Belles, X. CREB-binding protein contributes to the regulation of endocrine and developmental pathways in insect hemimetabolan pre-metamorphosis. Biochim. Biophys. Acta BBA Gen. Subj. 2016, 1860, 508–515. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; George, S.; Palli, S.R. Multiple functions of CREB-binding protein during postembryonic development: Identification of target genes. BMC Genom. 2017, 18, 996. [Google Scholar] [CrossRef]
- Roy, A.; Palli, S.R. Epigenetic modifications acetylation and deacetylation play important roles in juvenile hormone action. BMC Genom. 2018, 19, 934. [Google Scholar] [CrossRef]
- Legeai, F.; Shigenobu, S.; Gauthier, J.-P.; Colbourne, J.; Rispe, C.; Collin, O.; Richards, S.; Wilson, A.C.C.; Murphy, T.; Tagu, D. AphidBase: A centralized bioinformatic resource for annotation of the pea aphid genome. Insect Mol. Biol. 2010, 19, 5–12. [Google Scholar] [CrossRef] [Green Version]
- Skaljac, M.; Vogel, H.; Wielsch, N.; Mihajlovic, S.; Vilcinskas, A. Transmission of a Protease-Secreting Bacterial Symbiont among Pea Aphids via Host Plants. Front. Physiol. 2019, 10, 438. [Google Scholar] [CrossRef]
- The International Aphid Genomics Consortium. Genome Sequence of the Pea Aphid Acyrthosiphon pisum. PLoS Biol. 2010, 8, e1000313. [Google Scholar]
- Van Emden, H.F.; Harrington, R. Aphids as Crop Pests, 2nd ed.; CABI: Wallingford, UK, 2017. [Google Scholar]
- Kirfel, P.; Skaljac, M.; Grotmann, J.; Kessel, T.; Seip, M.; Michaelis, K.; Vilcinskas, A. Inhibition of histone acetylation and deacetylation enzymes affects longevity, development, and fecundity in the pea aphid (Acyrthosiphon pisum). Arch. Insect Biochem. Physiol. 2019, 103, e21614. [Google Scholar] [CrossRef] [Green Version]
- Rider, S.D.; Srinivasan, D.G.; Hilgarth, R.S. Chromatin-remodelling proteins of the pea aphid, Acyrthosiphon pisum (Harris): Aphid chromatin genes. Insect Mol. Biol. 2010, 19, 201–214. [Google Scholar] [CrossRef]
- Baudach, A.F.; Mukherjee, K. Epigenetic Control of Polyphenism in Aphids. In Biology and Ecology of Aphids; Taylor & Francis Group: Abingdon-on-Thames, UK, 2016; pp. 98–108. [Google Scholar]
- Grantham, M.; Brisson, J.A.; Tagu, D.; Le Trionnaire, G. Integrative Genomic Approaches to Studying Epigenetic Mechanisms of Phenotypic Plasticity in the Aphid. In Short Views on Insect Genomics and Proteomics; Entomology in Focus; Springer International Publishing AG: Cham, Switzerland, 2015; Volume 3, p. np. [Google Scholar]
- Srinivasan, D.G.; Brisson, J.A. Aphids: A Model for Polyphenism and Epigenetics. Genet. Res. Int. 2012, 2012, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, M.; Gatehouse, J.A.; Fitches, E.C. A Systematic Study of RNAi Effects and dsRNA Stability in Tribolium castaneum and Acyrthosiphon pisum, Following Injection and Ingestion of Analogous dsRNAs. Int. J. Mol. Sci. 2018, 19, 1079. [Google Scholar] [CrossRef] [Green Version]
- Sapountzis, P.; Duport, G.; Balmand, S.; Gaget, K.; Jaubert-Possamai, S.; Febvay, G.; Charles, H.; Rahbé, Y.; Colella, S.; Calevro, F. New insight into the RNA interference response against cathepsin-L gene in the pea aphid, Acyrthosiphon pisum: Molting or gut phenotypes specifically induced by injection or feeding treatments. Insect Biochem. Mol. Biol. 2014, 51, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Will, T.; Schmidtberg, H.; Skaljac, M.; Vilcinskas, A. Heat shock protein 83 plays pleiotropic roles in embryogenesis, longevity, and fecundity of the pea aphid Acyrthosiphon pisum. Dev. Genes Evol. 2017, 227, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Will, T.; Vilcinskas, A. The structural sheath protein of aphids is required for phloem feeding. Insect Biochem. Mol. Biol. 2015, 57, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Cagliari, D.; dos Santos, E.A.; Dias, N.; Smagghe, G.; Zotti, M.J. Nontransformative Strategies for RNAi in Crop Protection. Modul. Gene Expr. Abridging RNAi CRISPR-Cas9 Technol. 2019. [Google Scholar] [CrossRef] [Green Version]
- Joga, M.R.; Zotti, M.J.; Smagghe, G.; Christiaens, O. RNAi Efficiency, Systemic Properties, and Novel Delivery Methods for Pest Insect Control: What We Know So Far. Front. Physiol. 2016, 7, 657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knorr, E.; Fishilevich, E.; Tenbusch, L.; Frey, M.L.F.; Rangasamy, M.; Billion, A.; Worden, S.E.; Gandra, P.; Arora, K.; Lo, W.; et al. Gene silencing in Tribolium castaneum as a tool for the targeted identification of candidate RNAi targets in crop pests. Sci. Rep. 2018, 8, 2061. [Google Scholar] [CrossRef]
- Mamta, B.; Rajam, M.V. RNAi technology: A new platform for crop pest control. Physiol. Mol. Biol. Plants 2017, 23, 487–501. [Google Scholar] [CrossRef]
- Niu, J.; Taning, C.N.T.; Christiaens, O.; Smagghe, G.; Wang, J.-J. Chapter One—Rethink RNAi in Insect Pest Control: Challenges and Perspectives. In Advances in Insect Physiology; Protection, C., Smagghe, G., Eds.; Academic Press: Cambridge, MA, USA, 2018; Volume 55, pp. 1–17. [Google Scholar]
- Vogel, E.; Santos, D.; Mingels, L.; Verdonckt, T.-W.; Broeck, J.V. RNA Interference in Insects: Protecting Beneficials and Controlling Pests. Front. Physiol. 2019, 9, 9. [Google Scholar] [CrossRef] [Green Version]
- Will, T.; Vilcinskas, A. Aphid-proof plants: Biotechnology-based approaches for aphid control. Adv. Biochem. Eng. Biotechnol. 2013, 136, 179–203. [Google Scholar] [PubMed]
- Luna-Ramirez, K.; Skaljac, M.; Grotmann, J.; Kirfel, P.; Vilcinskas, A. Orally Delivered Scorpion Antimicrobial Peptides Exhibit Activity against Pea Aphid (Acyrthosiphon pisum) and Its Bacterial Symbionts. Toxins 2017, 9, 261. [Google Scholar] [CrossRef] [PubMed]
- Skaljac, M.; Kirfel, P.; Grotmann, J.; Vilcinskas, A. Fitness costs of infection with Serratia symbiotica are associated with greater susceptibility to insecticides in the pea aphid Acyrthosiphon pisum. Pest Manag. Sci. 2018, 74, 1829–1836. [Google Scholar] [CrossRef] [PubMed]
- Ayoubi, A.; Talebi, A.A.; Fathipour, Y.; Mehrabadi, M. Coinfection of the secondary symbionts, Hamiltonella defensa and Arsenophonus sp. contribute to the performance of the major aphid pest, Aphis gossypii (Hemiptera: Aphididae). Insect Sci. 2020, 27, 86–98. [Google Scholar] [CrossRef]
- Miura, T.; Braendle, C.; Shingleton, A.; Sisk, G.; Kambhampati, S.; Stern, D.L. A comparison of parthenogenetic and sexual embryogenesis of the pea aphid Acyrthosiphon pisum (Hemiptera: Aphidoidea). J. Exp. Zoolog. B Mol. Dev. Evol. 2003, 295, 59–81. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Chan, H.M.; La Thangue, N.B. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 2001, 114, 2363–2373. [Google Scholar] [PubMed]
- Kumar, J.P.; Jamal, T.; Doetsch, A.; Turner, F.R.; Duffy, J.B. CREB binding protein functions during successive stages of eye development in Drosophila. Genetics 2004, 168, 877–893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lilja, T.; Aihara, H.; Stabell, M.; Nibu, Y.; Mannervik, M. The acetyltransferase activity of Drosophila CBP is dispensable for regulation of the Dpp pathway in the early embryo. Dev. Biol. 2007, 305, 650–658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simola, D.F.; Graham, R.J.; Brady, C.M.; Enzmann, B.L.; Desplan, C.; Ray, A.; Zwiebel, L.J.; Bonasio, R.; Reinberg, D.; Liebig, J.; et al. Epigenetic (re)programming of caste-specific behavior in the ant Camponotus floridanus. Science 2016, 351, aac6633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Wu, C.; Sheng, Q.; Jiang, C.; Chen, Q.; Lv, Z.; Yao, J.; Nie, Z. Lysine acetylation stabilizes SP2 protein in the silkworm Bombyx mori. J. Insect Physiol. 2016, 91–92, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, D.; Okan, N.A.; Bales, E.; Nascimento, L.; Cole, P.A.; Medrano, E.E. Down-regulation of p300/CBP histone acetyltransferase activates a senescence checkpoint in human melanocytes. Cancer Res. 2002, 62, 6231–6239. [Google Scholar]
- Bedford, D.C.; Brindle, P.K. Is histone acetylation the most important physiological function for CBP and p300? Aging 2012, 4, 247–255. [Google Scholar] [CrossRef] [Green Version]
- Yan, G.; Eller, M.S.; Elm, C.; Larocca, C.A.; Ryu, B.; Panova, I.P.; Dancy, B.M.; Bowers, E.M.; Meyers, D.; Lareau, L.; et al. Selective Inhibition of p300 HAT Blocks Cell Cycle Progression, Induces Cellular Senescence, and Inhibits the DNA Damage Response in Melanoma Cells. J. Investig. Dermatol. 2013, 133, 2444–2452. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Poplawski, M.; Yen, K.; Cheng, H.; Bloss, E.; Zhu, X.; Patel, H.; Mobbs, C.V. Role of CBP and SATB-1 in aging, dietary restriction, and insulin-like signaling. PLoS Biol. 2009, 7, e1000245. [Google Scholar] [CrossRef] [Green Version]
- Laughton, A.M.; Fan, M.H.; Gerardo, N.M. The Combined Effects of Bacterial Symbionts and Aging on Life History Traits in the Pea Aphid, Acyrthosiphon pisum. Appl. Environ. Microbiol. 2014, 80, 470–477. [Google Scholar] [CrossRef] [Green Version]
- Iyer, N.G.; Özdag, H.; Caldas, C. p300/CBP and cancer. Oncogene 2004, 23, 4225–4231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Victor, M.; Bei, Y.; Gay, F.; Calvo, D.; Mello, C.; Shi, Y. HAT activity is essential for CBP-1-dependent transcription and differentiation in Caenorhabditis elegans. EMBO Rep. 2002, 3, 50–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdellatef, E.; Will, T.; Koch, A.; Imani, J.; Vilcinskas, A.; Kogel, K.-H. Silencing the expression of the salivary sheath protein causes transgenerational feeding suppression in the aphid Sitobion avenae. Plant Biotechnol. J. 2015, 13, 849–857. [Google Scholar] [CrossRef] [PubMed]
- Peserico, A.; Simone, C. Physical and Functional HAT/HDAC Interplay Regulates Protein Acetylation Balance. J. Biomed. Biotechnol. 2011, 2011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christiaens, O.; Swevers, L.; Smagghe, G. DsRNA degradation in the pea aphid (Acyrthosiphon pisum) associated with lack of response in RNAi feeding and injection assay. Peptides 2014, 53, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Christiaens, O.; Smagghe, G. The challenge of RNAi-mediated control of hemipterans. Curr. Opin. Insect Sci. 2014, 6, 15–21. [Google Scholar] [CrossRef]
- Baumann, A.; Lehmann, R.; Beckert, A.; Vilcinskas, A.; Franta, Z. Selection and Evaluation of Tissue Specific Reference Genes in Lucilia sericata during an Immune Challenge. PLoS ONE 2015, 10, e0135093. [Google Scholar] [CrossRef]
- Fukaya, T.; Tomari, Y. MicroRNAs Mediate Gene Silencing via Multiple Different Pathways in Drosophila. Mol. Cell 2012, 48, 825–836. [Google Scholar] [CrossRef] [Green Version]
- Iwasaki, S.; Kawamata, T.; Tomari, Y. Drosophila Argonaute1 and Argonaute2 Employ Distinct Mechanisms for Translational Repression. Mol. Cell 2009, 34, 58–67. [Google Scholar] [CrossRef] [Green Version]
- Iwakawa, H.; Tomari, Y. The Functions of MicroRNAs: mRNA Decay and Translational Repression. Trends Cell Biol. 2015, 25, 651–665. [Google Scholar] [CrossRef]
- Asgari, S. MicroRNA functions in insects. Insect Biochem. Mol. Biol. 2013, 43, 388–397. [Google Scholar] [CrossRef] [PubMed]
- Holmes, K.; Williams, C.M.; Chapman, E.A.; Cross, M.J. Detection of siRNA induced mRNA silencing by RT-qPCR: Considerations for experimental design. BMC Res. Notes 2010, 3, 53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pitino, M.; Coleman, A.D.; Maffei, M.E.; Ridout, C.J.; Hogenhout, S.A. Silencing of Aphid Genes by dsRNA Feeding from Plants. PLoS ONE 2011, 6, e25709. [Google Scholar] [CrossRef] [PubMed]
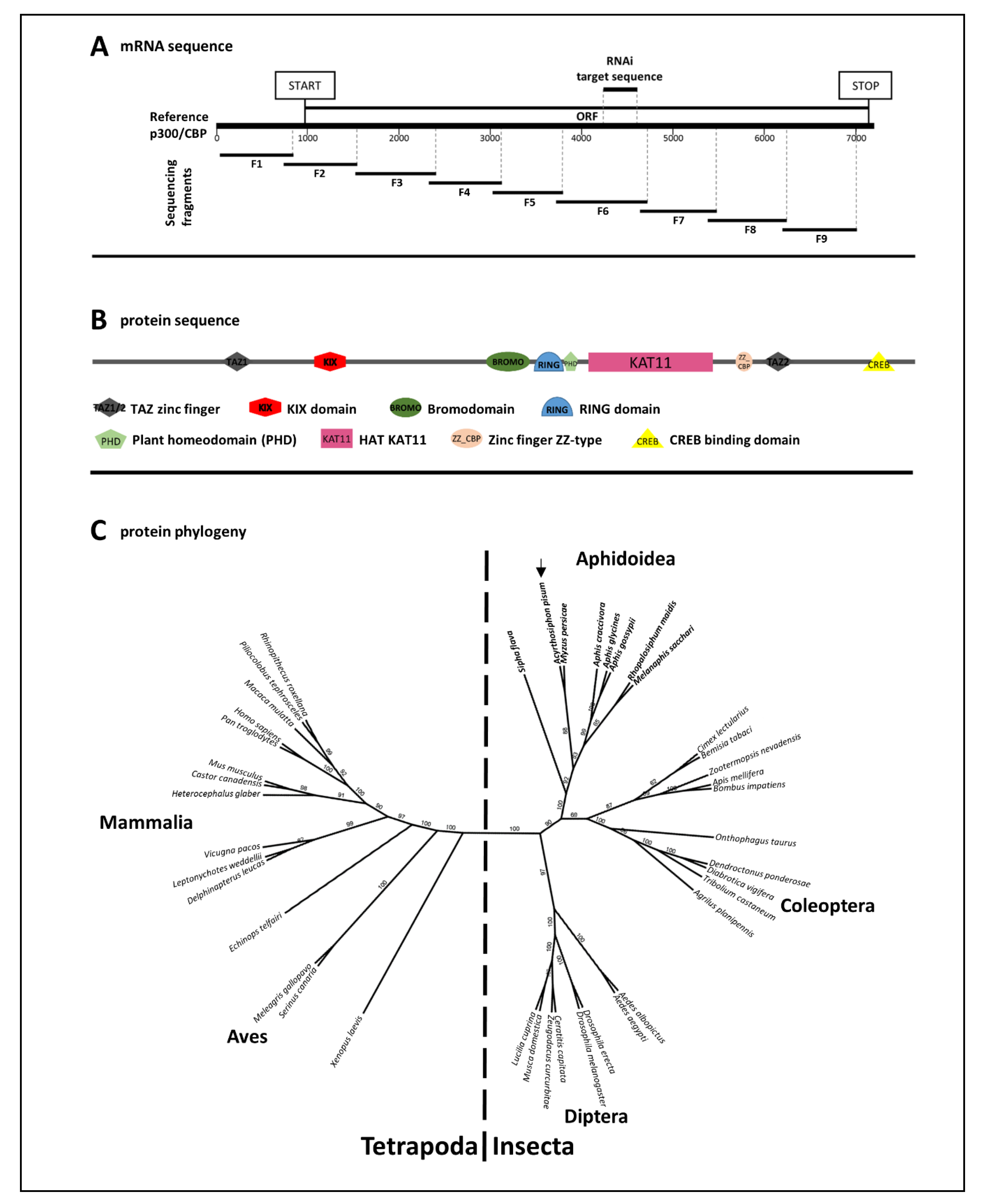
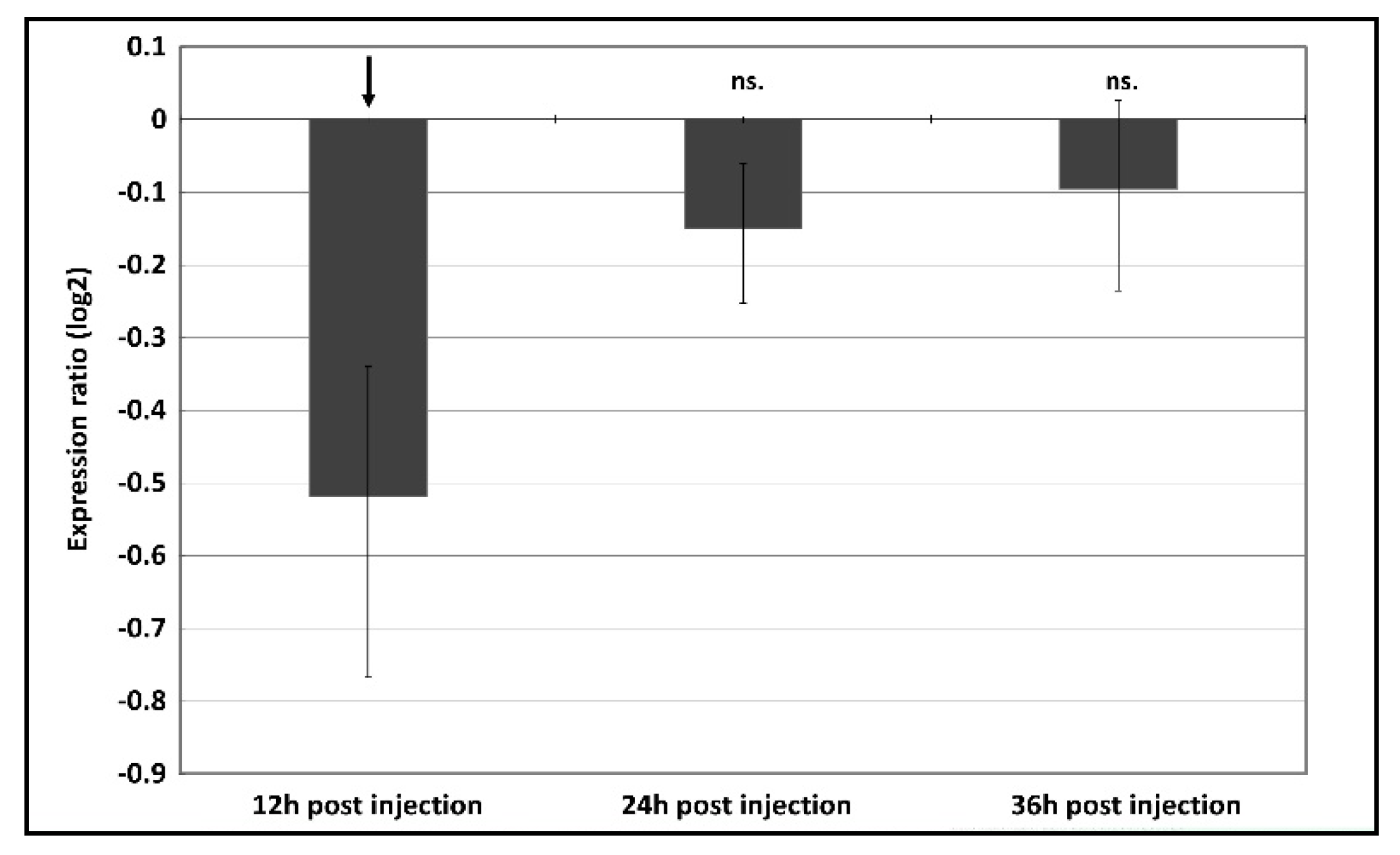

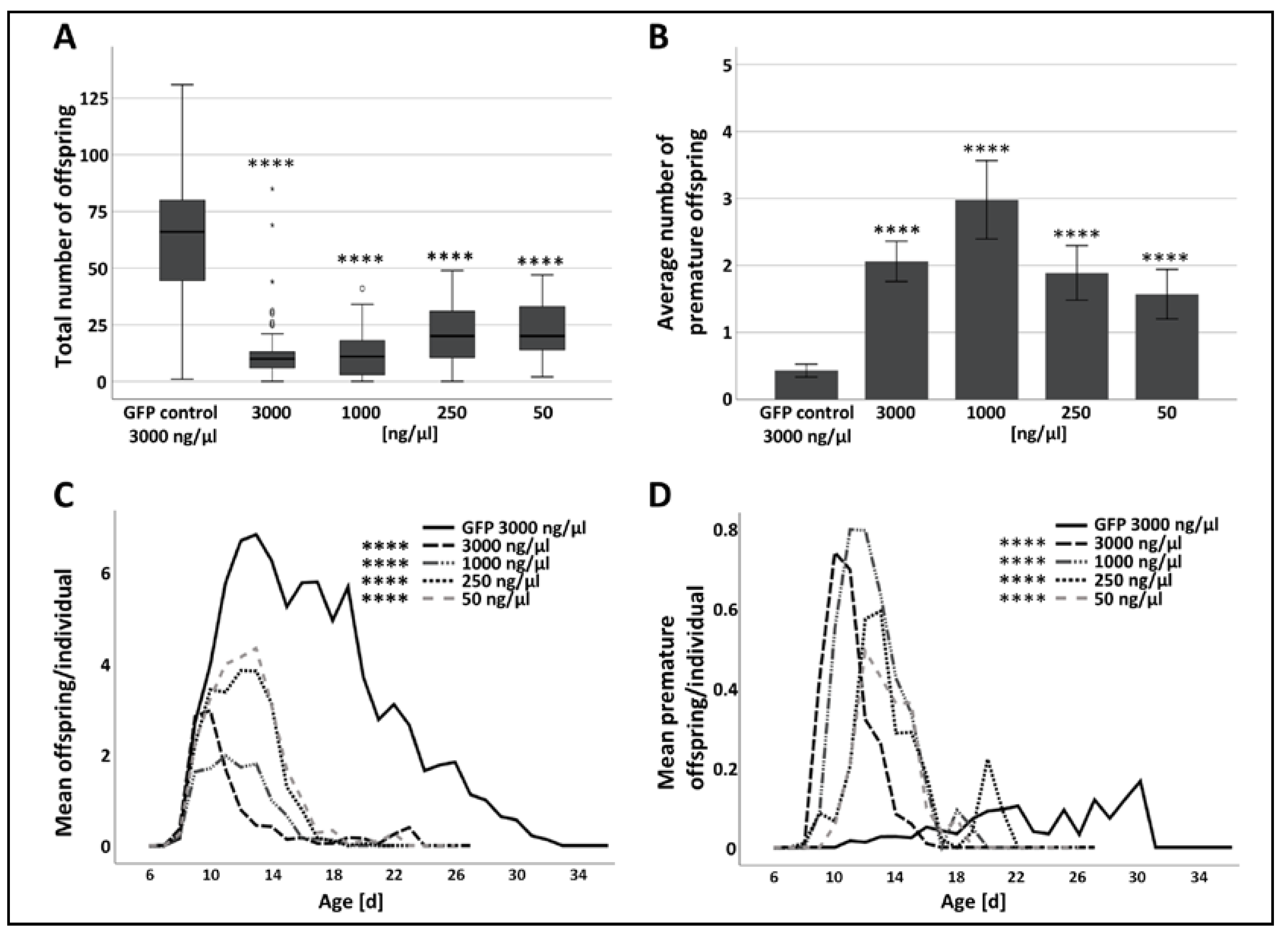
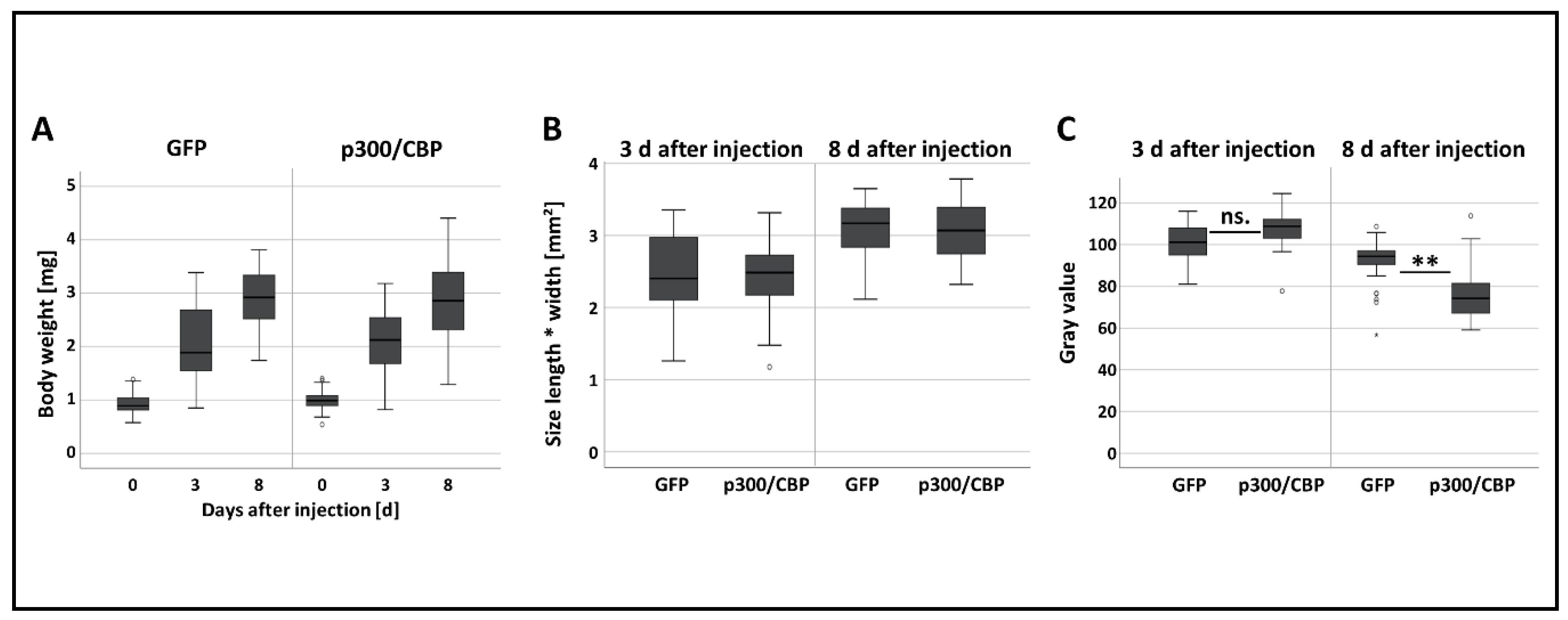
| Treatment | Post-Injection Survival Frequency [%] | |||
|---|---|---|---|---|
| After 5 Days | After 10 Days | After 15 Days | After 20 Days | |
| GFP 3000 ng/µL | 79 | 65 | 45 | 20 |
| p300/CBP 3000 ng/µL | 79 | 51 | 13 | 1 |
| p300/CBP 1000 ng/µL | 85 | 57 | 8 | 0 |
| p300/CBP 250 ng/µL | 88 | 60 | 11 | 0 |
| p300/CBP 50 ng/µL | 85 | 61 | 12 | 3 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kirfel, P.; Vilcinskas, A.; Skaljac, M. Lysine Acetyltransferase p300/CBP Plays an Important Role in Reproduction, Embryogenesis and Longevity of the Pea Aphid Acyrthosiphon pisum. Insects 2020, 11, 265. https://doi.org/10.3390/insects11050265
Kirfel P, Vilcinskas A, Skaljac M. Lysine Acetyltransferase p300/CBP Plays an Important Role in Reproduction, Embryogenesis and Longevity of the Pea Aphid Acyrthosiphon pisum. Insects. 2020; 11(5):265. https://doi.org/10.3390/insects11050265
Chicago/Turabian StyleKirfel, Phillipp, Andreas Vilcinskas, and Marisa Skaljac. 2020. "Lysine Acetyltransferase p300/CBP Plays an Important Role in Reproduction, Embryogenesis and Longevity of the Pea Aphid Acyrthosiphon pisum" Insects 11, no. 5: 265. https://doi.org/10.3390/insects11050265
APA StyleKirfel, P., Vilcinskas, A., & Skaljac, M. (2020). Lysine Acetyltransferase p300/CBP Plays an Important Role in Reproduction, Embryogenesis and Longevity of the Pea Aphid Acyrthosiphon pisum. Insects, 11(5), 265. https://doi.org/10.3390/insects11050265





