Mitochondrial Gene Sequence (COI) Reveals the Genetic Structure and Demographic History of Lymantria dispar (Lepidoptera: Erebidae: Lymantriinae) in and around China
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. DNA Extraction
2.3. Primer Design, PCR Amplification and Sequencing
2.4. Gypsy Moth Mitochondrial COI Sequence Data
2.5. Analysis of Genetic Structure in Populations of Gypsy Moths
2.6. Phylogenetic Analyses among Populations of Gypsy Moths
3. Results
3.1. Assessing the Utility of Newly-Designed Primer Pairs
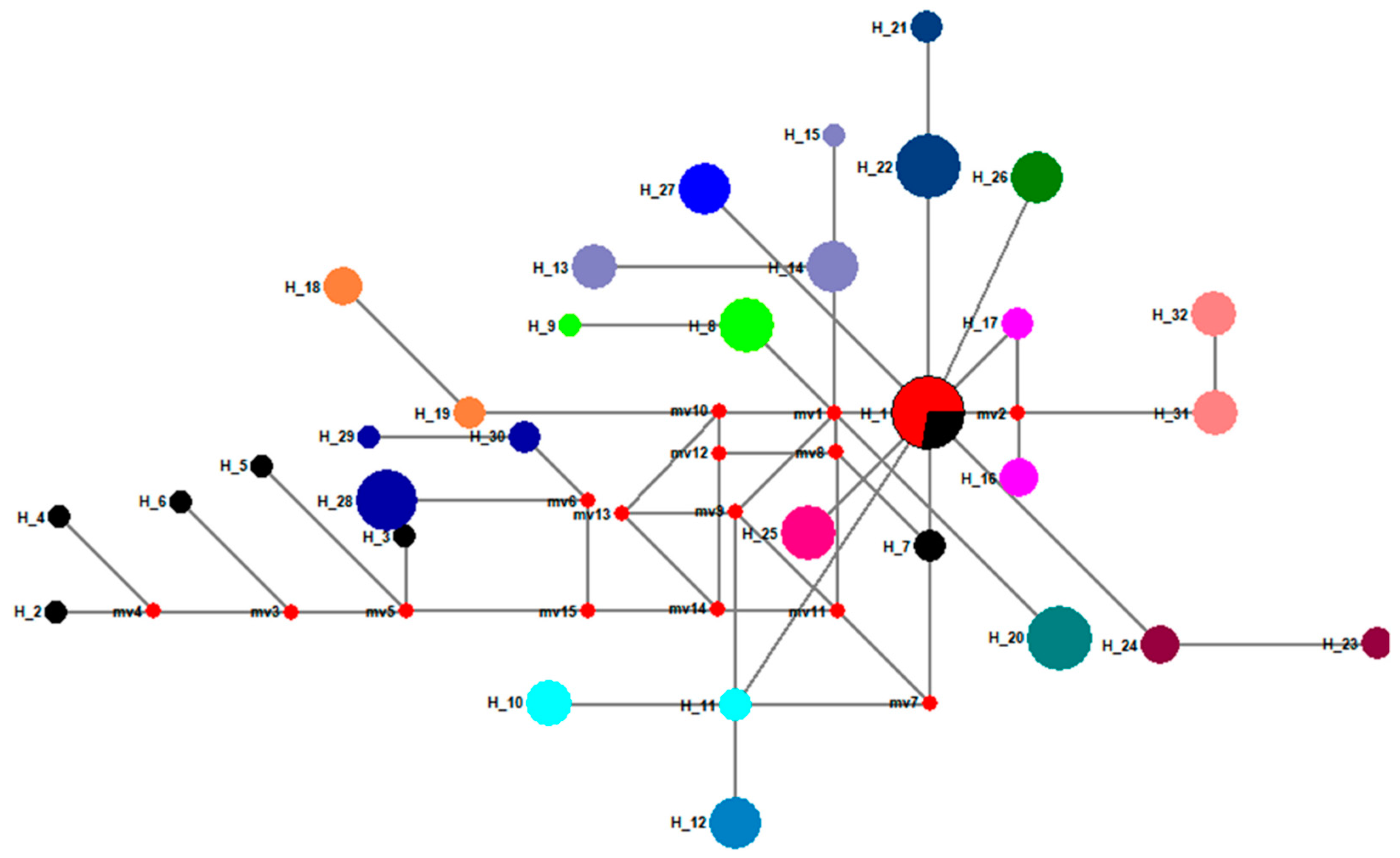
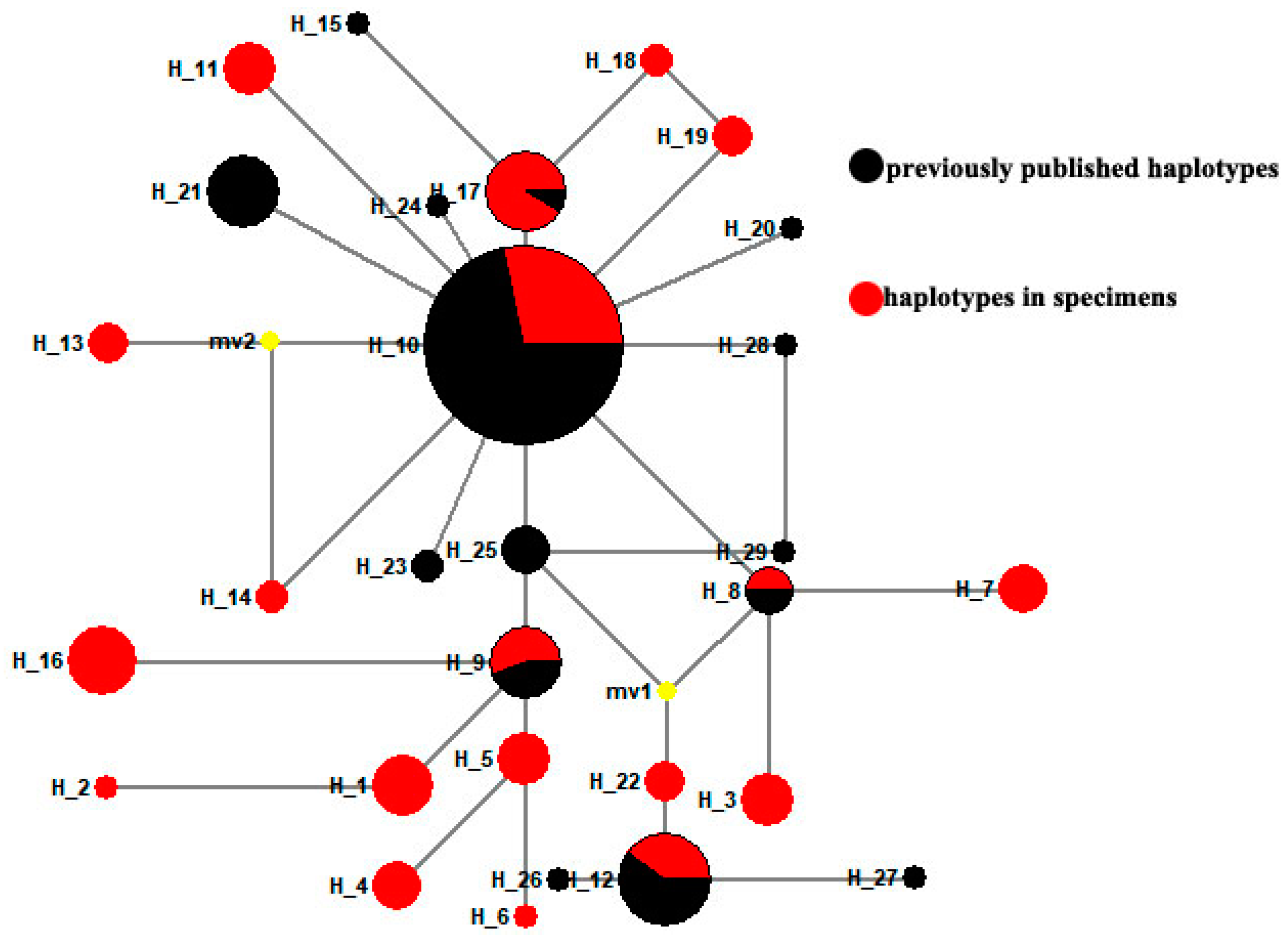
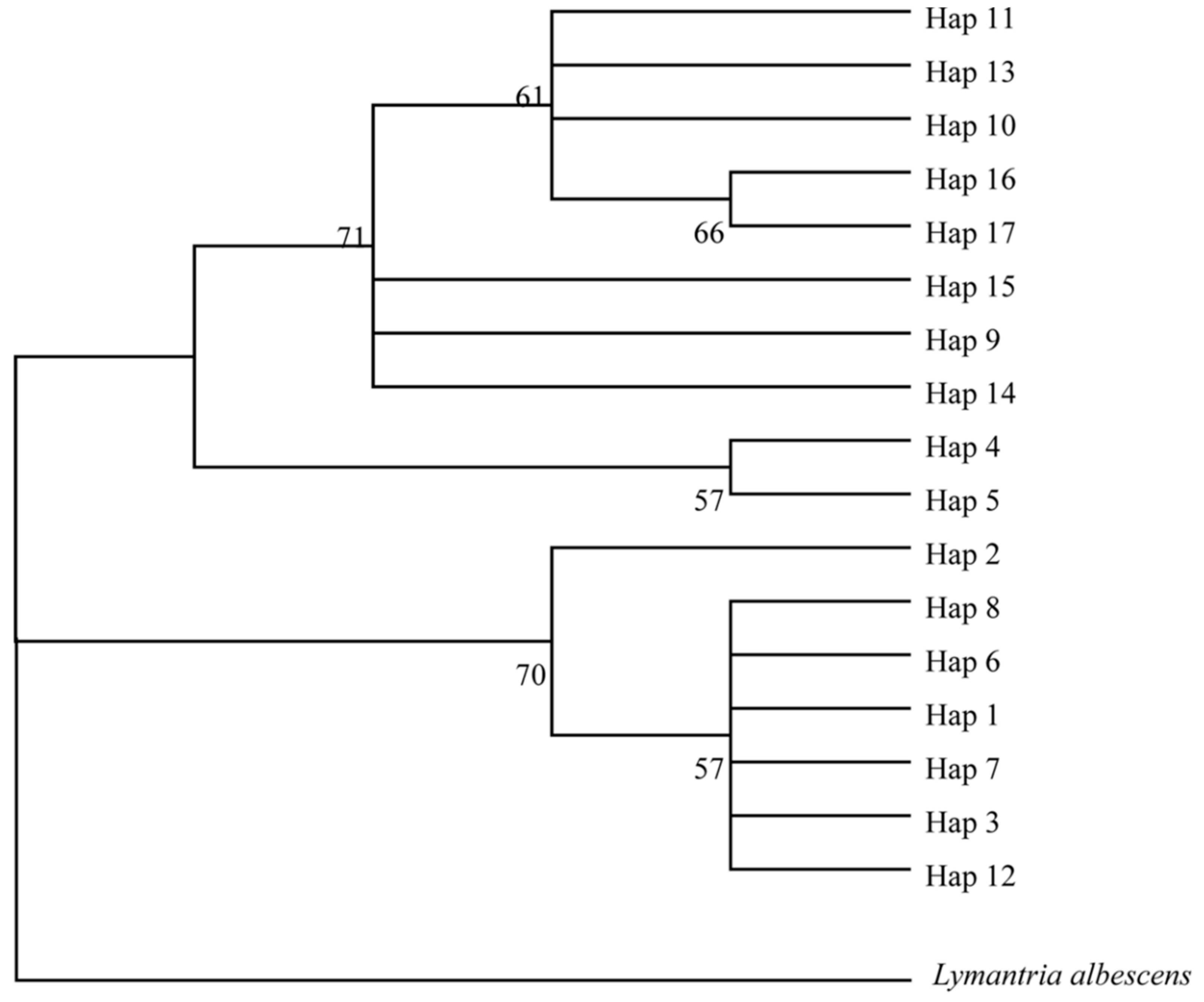
3.2. New Haplotypes from Old Gypsy Moth Specimens
3.3. Genetic Differentiation in Gypsy Moth Populations across Different Years
3.4. Variation in Population Structure for Three Geographical Populations over Different Years
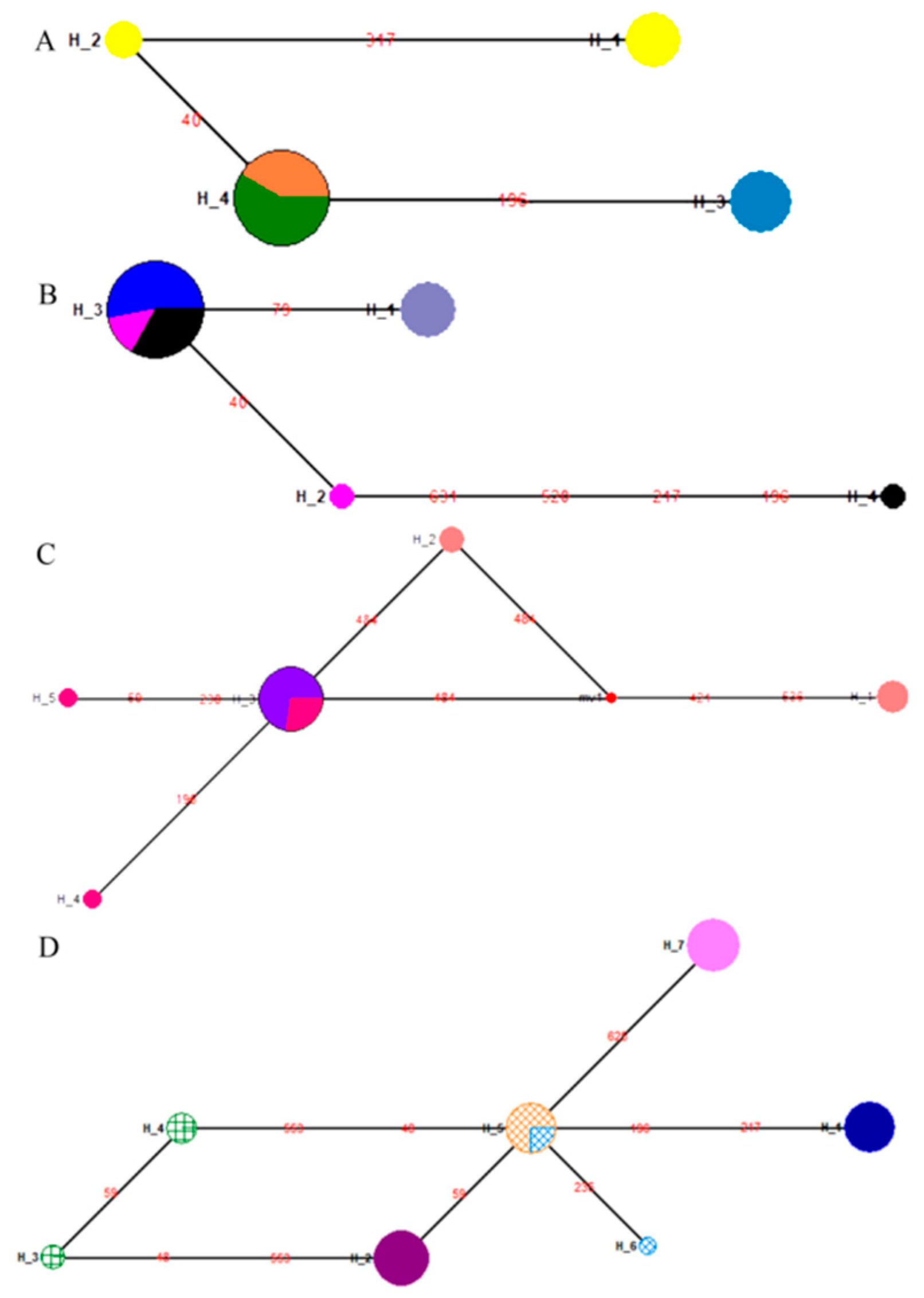
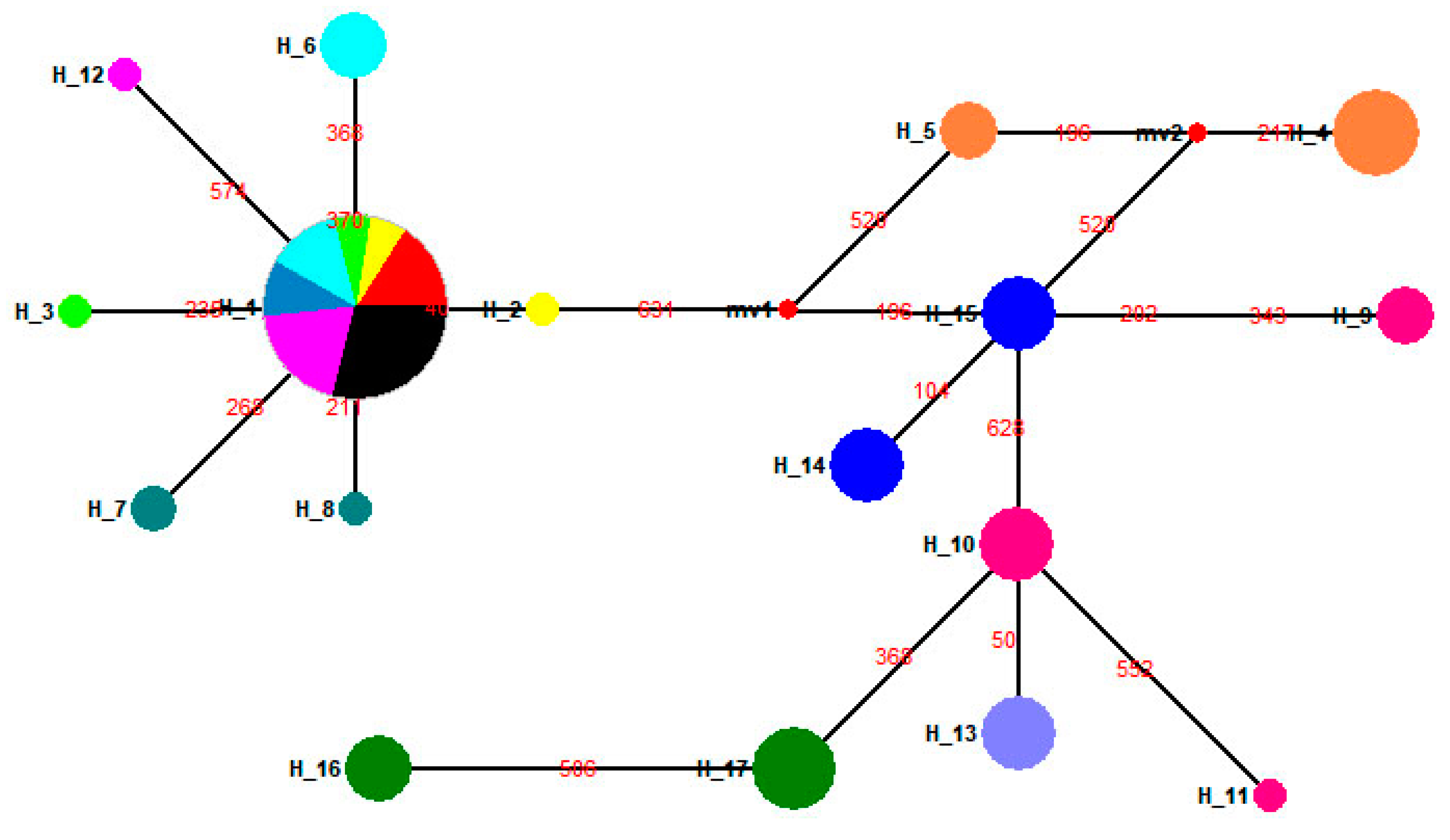
3.5. Population Structure of the Four Geographical Populations in the 1950s
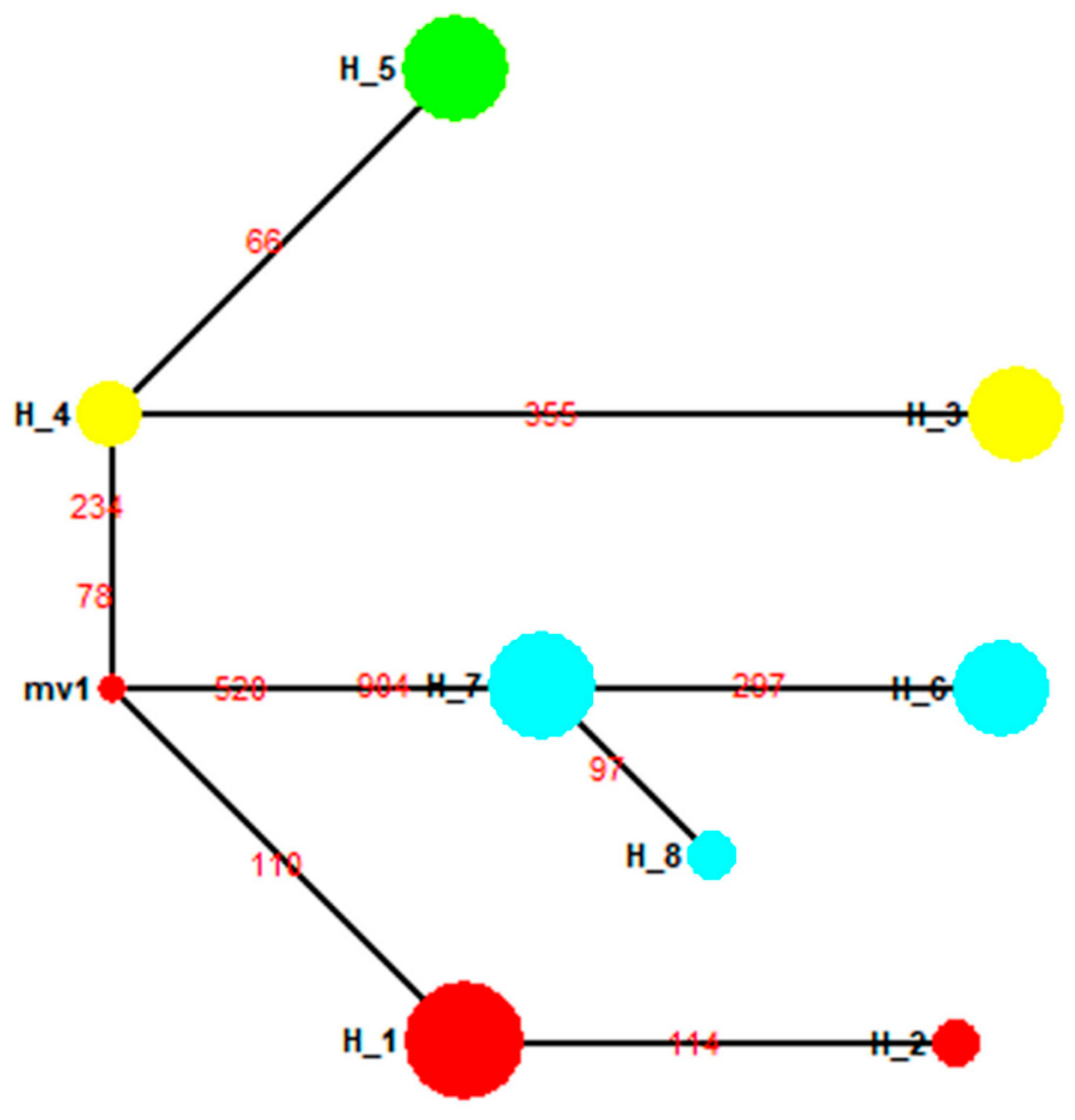
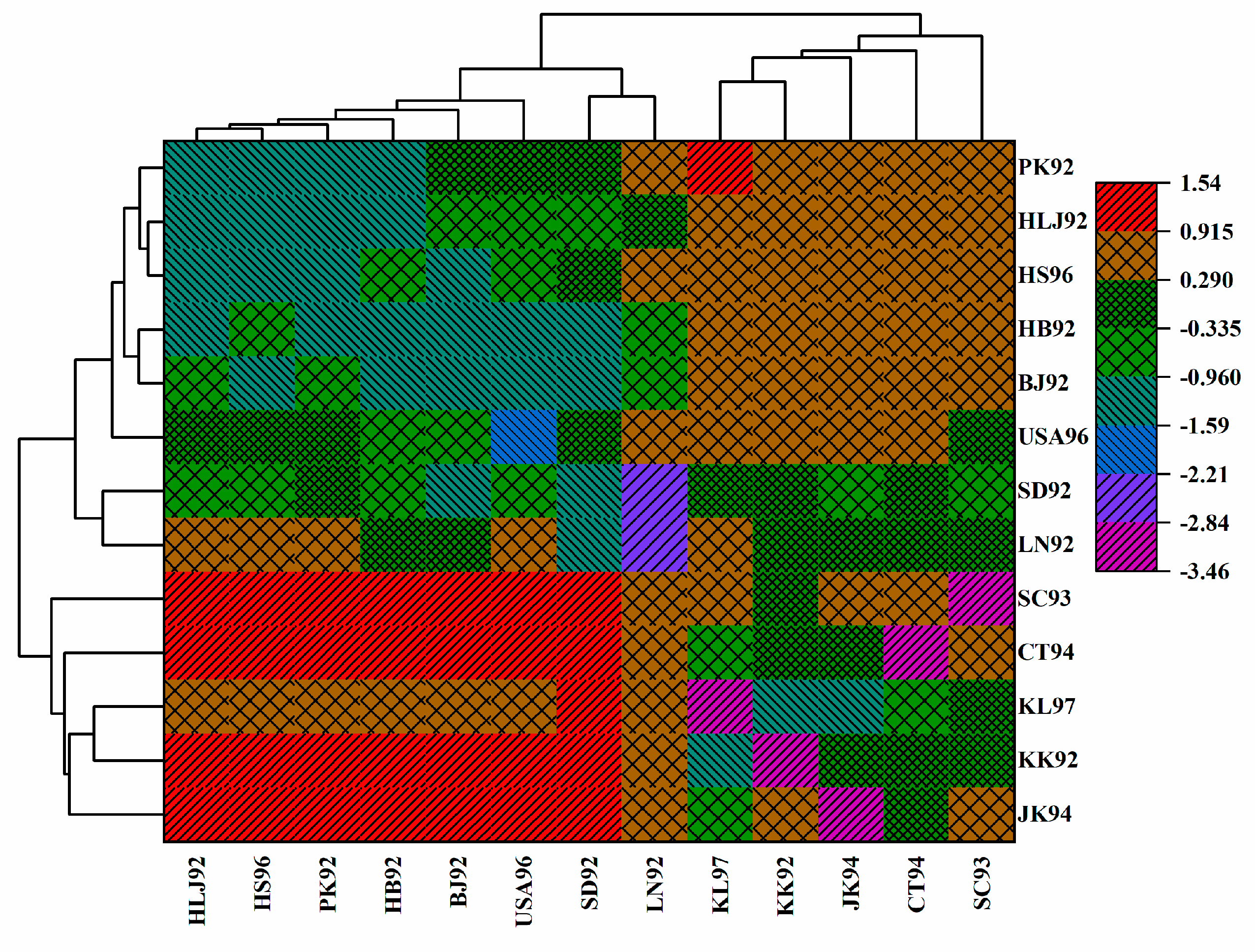
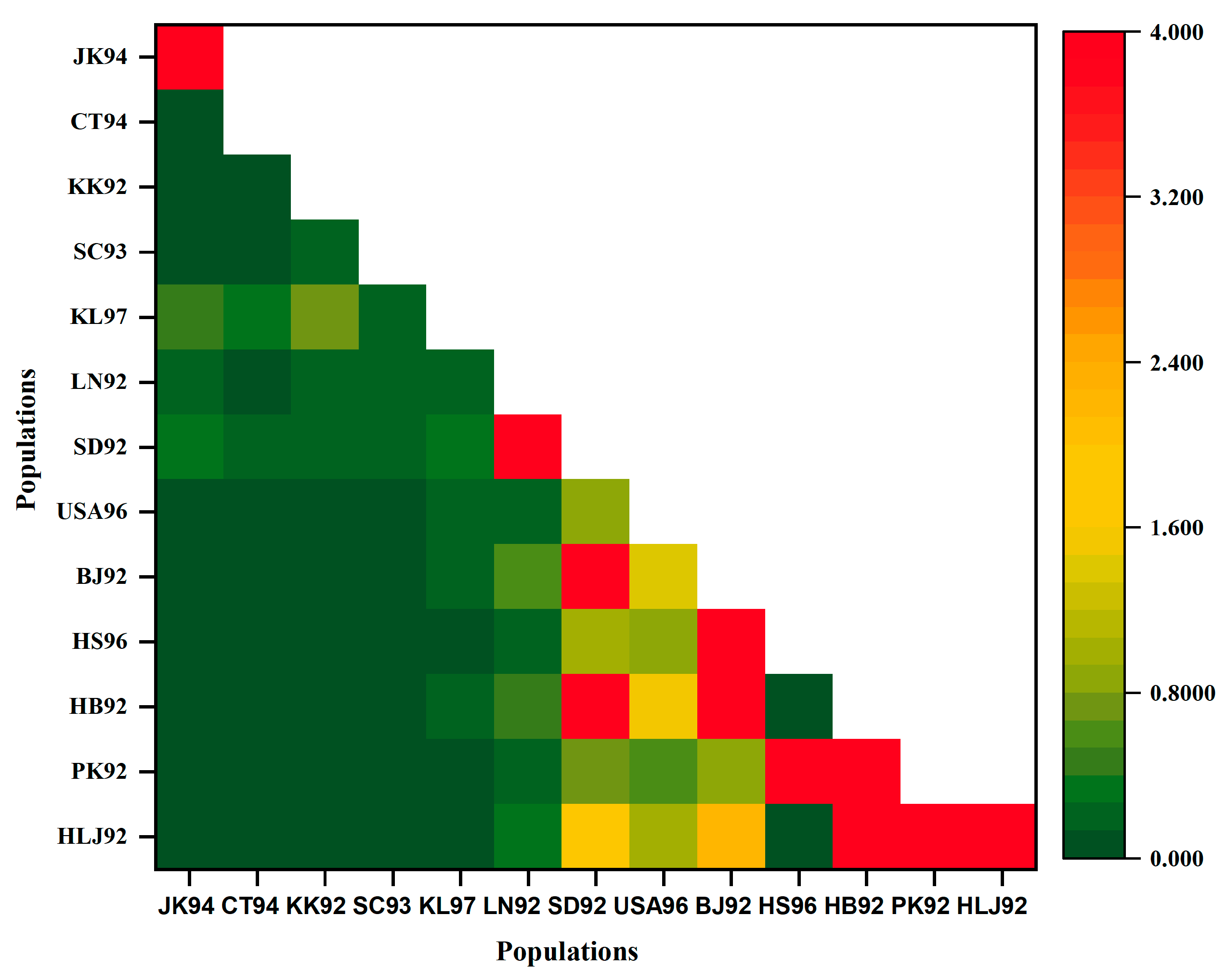
3.6. Population Structure for Geographical Populations in the 1990s
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ferguson, D.C. Fascicle 22.2 Noctuoidea, Lymantriidae. In The Moths of America North of Mexico; Dominick, R.B., Dominick, T., Ferguson, D.C., Franclemont, J.G., Hodges, R.W., Munroe, E.G., Classey, E.W., Eds.; Wedge Entomological Research Foundation: Faringdon, UK, 1978; pp. 1–110. [Google Scholar]
- Pogue, M.; Schaefer, P.W. Review of selected species of Lymantria hubner (1819) (Lepidoptera: Noctuidae: Lymantriinae) from subtropical and temperate regions of Asia, including the descriptions of three new species, some potentially invasive to North America. Int. J. Mech. Control 2007, 15, 31–35. [Google Scholar]
- Zahiri, R.; Holloway, J.D.; Kitching, I.J.; Lafontaine, J.D.; Mutanen, M.; Wahlberg, N. Molecular phylogenetics of Erebidae (Lepidoptera, Noctuoidea). Syst. Entomol. 2011, 37, 102–124. [Google Scholar] [CrossRef]
- Chen, N.Z. Bioecology of gypsy moth. In Gypsy Moth Quarantine and Control; China Forestry Publishing House: Beijing, China, 2013; pp. 20–28. [Google Scholar]
- Montgomery, M.E.; Wallner, W.E. The gypsy moth a westward migrant. In Dynamics of Forest Insect Populations, Patterns, Causes, Implications; Berryman, A.A., Ed.; Plenum Press: New York, NY, USA, 1988; Volume 4, pp. 354–378. [Google Scholar]
- Pimentel, D.; Zuniga, R.; Morrison, D. Update of the environmental and economic costs associated with alien-invasive species in the United States. Ecol. Econ. 2005, 52, 273–288. [Google Scholar] [CrossRef]
- Xiao, G.R. Lymantridae. In Chinese Forest Insect; Xu, W.R., Ed.; China Forestry Publishing House: Beijing, China, 1992; p. 1086. [Google Scholar]
- Forbush, E.H.; Fernald, C.H. The Gypsy Moth; Wrightand Potter Press: Boston, MA, USA, 1896. [Google Scholar]
- Giese, R.L.; Schneider, M.L. Cartographic comparisons of Eurasian gypsy moth distribution (Lymantria dispar.; Lepidoptera: Lymantriidae). Entomol. News 1979, 90, 1–16. [Google Scholar]
- Reineke, A.; Karlovsky, P.; Zebitz, C.P.W. Amplified fragment length polymorphism analysis of different geographic populations of the gypsy moth, Lymantria dispar (Lepidoptera: Lymantriinae). Bull. Entomol. Res. 1999, 89, 79–88. [Google Scholar] [CrossRef]
- Zhou, X.; Chen, N.; Yang, D. Research advances in the population genetic relationships of Lymantria dispar Linnaeus based on molecular genetic markers. Plant Prot. 2009, 35, 1–5. [Google Scholar]
- Djoumad, A.; Nisole, A.; Zahiri, R.; Freschi, L.; Picq, S.; Gundersenrindal, D.E. Comparative analysis of mitochondrial genomes of geographic variants of the gypsy moth, Lymantria dispar, reveals a previously undescribed genotypic entity. Sci. Rep. 2017, 7, 14245. [Google Scholar] [CrossRef]
- Keena, M.A.; Grinberg, P.S.; Wallner, W.E. Inheritance of female flight in Lymantria dispar (Lepidoptera: Lymantriidae). Environ. Entomol. 2007, 36, 484. [Google Scholar] [CrossRef]
- Keena, M.A.; Côté, M.; Grinberg, P.S.; Wallner, W.E. World distribution of female flight and genetic variation in Lymantria dispar (Lepidoptera: Lymantriidae). Environ. Entomol. 2008, 37, 636–649. [Google Scholar] [CrossRef]
- Yang, F. The Study on Flight Ability among Different Geographic Populations of Asian Gypsy Moth, Lymantria dispar in China. Ph.D. Thesis, Beijing Forestry University, Beijing, China, 2013; pp. 55–59. [Google Scholar]
- Shi, J.; Chen, F.; Keena, M.A. Differences in wing morphometrics of Lymantria dispar (Lepidoptera: Erebidae) between populations that vary in female flight capability. Ann. Entomol. Soc. Am. 2015, 108, 528–535. [Google Scholar] [CrossRef]
- Chen, F.; Shi, J.; Keena, M. Evaluation of the effects of light intensity and time interval after the start of scotophase on the female flight propensity of Asian gypsy moth (Lepidoptera: Erebidae). Environ. Entomol. 2016, 45, 404–409. [Google Scholar] [CrossRef]
- Wallner, W.E.; Humble, L.M.; Levin, R.E.; Baranchikov, Y.N.; Cardé, R.T. Response of adult Lymantriid moths to illumination devices in the Russian Far East. J. Econ. Entomol. 1995, 88, 337–342. [Google Scholar] [CrossRef]
- Iwaizumi, R.; Arakawa, K. Report on female flight activity of the Asian gypsy moth, Lymantria dispar (Lepidoptera: Lymantriidae) and flight suppression with a yellow light source in Japan. Res. Bull. Plant Prot. Serv. Jpn. 2010, 46, 9–15. [Google Scholar]
- Dewaard, J.R.; Mitchell, A.; Keena, M.A.; Gopurenko, D.; Boykin, L.M.; Armstrong, K.F. Towards a global barcode library for Lymantria (Lepidoptera: Lymantriinae) tussock moths of biosecurity concern. PLoS ONE 2010, 5, e14280. [Google Scholar] [CrossRef]
- Bogdanowicz, S.M.; Mastro, V.C.; Prasher, D.C.; Harrison, R.G. Microsatellite DNA variation among Asian and North American gypsy moths (Lepidoptera: Lymantridae). Ann. Entomol. Soc. Am. 1997, 90, 768–775. [Google Scholar] [CrossRef]
- Bogdanowicz, S.M.; Schaefer, P.W.; Harrison, R.G. Mitochondrial DNA variation among worldwide populations of gypsy moths, Lymantria dispar. Mol. Phylogenet. Evol. 2000, 15, 487–495. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Shi, J.; Luo, Y.Q. Genetic characterization of the gypsy moth from China (Lepidoptera, Lymantriidae) using inter simple sequence repeats markers. PLoS ONE 2013, 8, e73017. [Google Scholar] [CrossRef]
- Chen, F.; Luo, Y.; Keena, M.A.; Wu, Y.; Wu, P.; Shi, J. DNA barcoding of gypsy moths from China (Lepidoptera: Erebidae) reveals new haplotypes and divergence patterns within gypsy moth subspecies. J. Econ. Entomol. 2015, 109, 366–374. [Google Scholar] [CrossRef]
- Kang, T.H.; Han, S.H.; Park, S.J. Development of seven microsatellite markers using next generation sequencing for the conservation on the Korean population of Dorcus hopei (E. Saunders, 1854) (Coleoptera, Lucanidae). Int. J. Mol. Sci. 2015, 16, 21330–21341. [Google Scholar] [CrossRef]
- Kang, T.H.; Han, S.H.; Park, S.J. Development of 12 microsatellite markers in Dorcus titanus castanicolor (Motschulsky, 1861) (Lucanidae, Coleoptera) from Korea using next-generation sequencing. Int. J. Mol. Sci. 2016, 17, 1621. [Google Scholar] [CrossRef]
- Qian, L. AFLP analysis of different geographic populations of the gypsy moth, Lymantria dispar (Lepidoptera: Lymantriidae). Sci. Silvae Sin. 2011, 47, 104–110. [Google Scholar]
- Zhang, H.; Chen, N.Z.; Li, Z.X. Analysis of RAPD markers and development of SCAR markers of six geographic populations of the gypsy moth, Lymantria dispar L. (Lepidoptera: Lymantriidae), from China. Acta Entomol. Sin. 2011, 54, 714–721. [Google Scholar]
- Wu, Y.; Molongoski, J.J.; Winograd, D.F.; Bogdanowicz, S.M.; Louyakis, A.S.; Lance, D.R. Genetic structure, admixture and invasion success in a Holarctic defoliator, the gypsy moth (Lymantria dispar, Lepidoptera: Erebidae). Mol. Ecol. 2015, 24, 1275–1291. [Google Scholar] [CrossRef]
- Kang, T.H.; Sang, H.H.; Lee, H.S. Genetic structure and demographic history of Lymantria dispar (Linnaeus, 1758) (Lepidoptera: Erebidae) in its area of origin and adjacent areas. Ecol. Evol. 2017, 7, 9162–9178. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.S.; Kang, T.H.; Jeong, J.W.; Ryu, D.P.; Lee, H.S. Taxonomic review of the genus Lymantria (Lepidoptera: Erebidae: Lymantriinae) in Korea. Entomol. Res. 2015, 45, 225–234. [Google Scholar] [CrossRef]
- Wandeler, P.; Hoeck, P.E.A.; Keller, L.F. Back to the future: Museum specimens in population genetics. Trends Ecol. Evol. 2007, 22, 634–642. [Google Scholar] [CrossRef] [PubMed]
- Ewers, R.M.; Didham, R.K. Confounding factors in the detection of species responses to habitat fragmentation. Biol. Rev. 2006, 81, 117–142. [Google Scholar] [CrossRef]
- Lister, A.M. Natural history collections as sources of long-term data sets. Trends Ecol. Evol. 2011, 26, 150–154. [Google Scholar]
- Holmes, M.W.; Hammond, T.T.; Wogan, G.O.; Walsh, R.E.; LaBarbera, K.; Wommack, E.A. Natural history collections as windows on evolutionary processes. Mol. Ecol. 2016, 25, 864–881. [Google Scholar] [CrossRef]
- Krehenwinkel, H.; Pekar, S. An analysis of factors affecting genotyping success from museum specimens reveals an increase of genetic and morphological variation during a historical range expansion of a European spider. PLoS ONE 2015, 10, e0136337. [Google Scholar] [CrossRef]
- Mikheyev, A.S.; Tin, M.M.; Arora, J.; Seeley, T.D. Museum samples reveal rapid evolution by wild honey bees exposed to a novel parasite. Nat. Commun. 2015, 6, 7991. [Google Scholar] [CrossRef]
- Hajibabaei, M.; Smith, M.A.; Janzen, D.H.; Rodriguez, J.J.; Whitfield, J.B.; Hebert, P.D.N. A minimalist barcode can identify a specimen whose DNA is degraded. Mol. Ecol. Notes 2006, 6, 959–964. [Google Scholar] [CrossRef]
- Rowley, D.L.; Coddington, J.A.; Gates, M.W.; Norrbom, R.A.; Ochoa, R.; Vandenberg, N.J. Vouchering DNA-barcoded specimens: Test of a nondestructive extraction protocol for terrestrial arthropods. Mol. Ecol. Notes 2007, 7, 915–924. [Google Scholar] [CrossRef]
- Meusnier, I.; Singer, G.A.; Landry, J.F.; Hickey, D.A.; Hebert, P.D.; Hajibabaei, M. A universal DNA mini-barcode for biodiversity analysis. BMC Genom. 2008, 9, 214. [Google Scholar] [CrossRef]
- Zimmermann, J.; Hajibabaei, M.; Blackburn, D.C.; Hanken, J.; Cantin, E.; Posfai, J. DNA damage in preserved specimens and tissue samples: A molecular assessment. Front. Zool. 2008, 5, 18. [Google Scholar] [CrossRef]
- Hausmann, A.; Herbert, P.D.; Mitchell, A.; Rougerie, R.; Sommerer, M.; Edwards, T. Revision of the Australian Oenochroma vinaria Guenée, 1858 species-complex (Lepidoptera: Geometridae, Oenochrominae): DNA barcoding reveals cryptic diversity and assesses status of type specimen without dissection. Zootaxa 2009, 2239, 1–21. [Google Scholar]
- Espeland, M.; Irestedt, M.; Johanson, K.A.; Akerlund, M.; Bergh, J.E.; Källersjö, M. Dichlorvos exposure impedes extraction and amplification of DNA from insects in museum collections. Front. Zool. 2010, 7, 2. [Google Scholar] [CrossRef]
- Nagy, Z.T.; Breman, F.C.; Dall’Asta, U. DNA barcoding of museum specimens of Lymantriidae preserved in the Royal Museum for Central Africa. Entomol. Roman. 2010, 15, 11–16. [Google Scholar]
- Welch, A.J.; Wiley, A.E.; James, H.F.; Ostrom, P.H.; Stafford, S.T., Jr.; Fleischer, R.C. Ancient DNA reveals genetic stability despite demographic decline: 3000 years of population history in the endemic Hawaiian petrel. Mol. Biol. Evol. 2012, 29, 3729–3740. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.A.; Beentjes, K.K.; Helsdingen, P.; Ijland, S. Which specimens from a museum collection will yield DNA barcodes? A time series study of spiders in alcohol. Zookeys 2013, 365, 245–261. [Google Scholar] [CrossRef]
- Hernández-Triana, L.M.; Prosser, S.; Rodríguez-Perez, M.A.; Chaverri, L.G.; Hebert, P.D.; Gregory, T.R. Recovery of DNA barcodes from blackfly museum specimens (Diptera: Simuliidae) using primer sets that target a variety of sequence lengths. Mol. Ecol. Resour. 2014, 214, 508–518. [Google Scholar] [CrossRef]
- Malmström, H.; Linderholm, A.; Skoglund, P.; Storå, J.; Sjödin, P.; Gilbert, M.T. Ancient mitochondrial DNA from the Northern fringe of the neolithic farming expansion in Europe sheds light on the dispersion process. Philos. Trans. R. Soc. Biol. Sci. 2015, 370, 20130373. [Google Scholar] [CrossRef]
- Price, B.W.; Henry, C.S.; Hall, A.C.; Mochizuki, A.; Duelli, P.; Brooks, S.J. Singing from the grave: DNA from a 180-year old type specimen confirms the identity of Chrysoperla carnea (Stephens). PLoS ONE 2015, 10, e0121127. [Google Scholar] [CrossRef] [PubMed]
- White, L.C.; Mitchell, K.J.; Austin, J.J. Ancient mitochondrial genomes reveal the demographic history and phylogeography of the extinct, enigmatic Thylacine (Thylacinus cynocephalus). J. Biogeogr. 2018, 45, 1–13. [Google Scholar] [CrossRef]
- Alzohairy, A.M. Bioedit: An important software for molecular biology. GERF Bull. Biosci. 2011, 2, 60–61. [Google Scholar]
- Whitlock, M.C.; Mccauley, D.E. Indirect measures of gene flow and migration: FST not equal to 1/(4nm + 1). Heredity 1999, 82, 117–125. [Google Scholar] [CrossRef]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol. Bioinform. Online 2005, 1, 47–50. [Google Scholar] [CrossRef]
- Bandelt, H.J.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. Dnasp v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. Mega 7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Feng, G.E. Challenges facing entomologists in a changing global climate. Chin. J. Appl. Entomol. 2011, 48, 1117–1122. [Google Scholar]
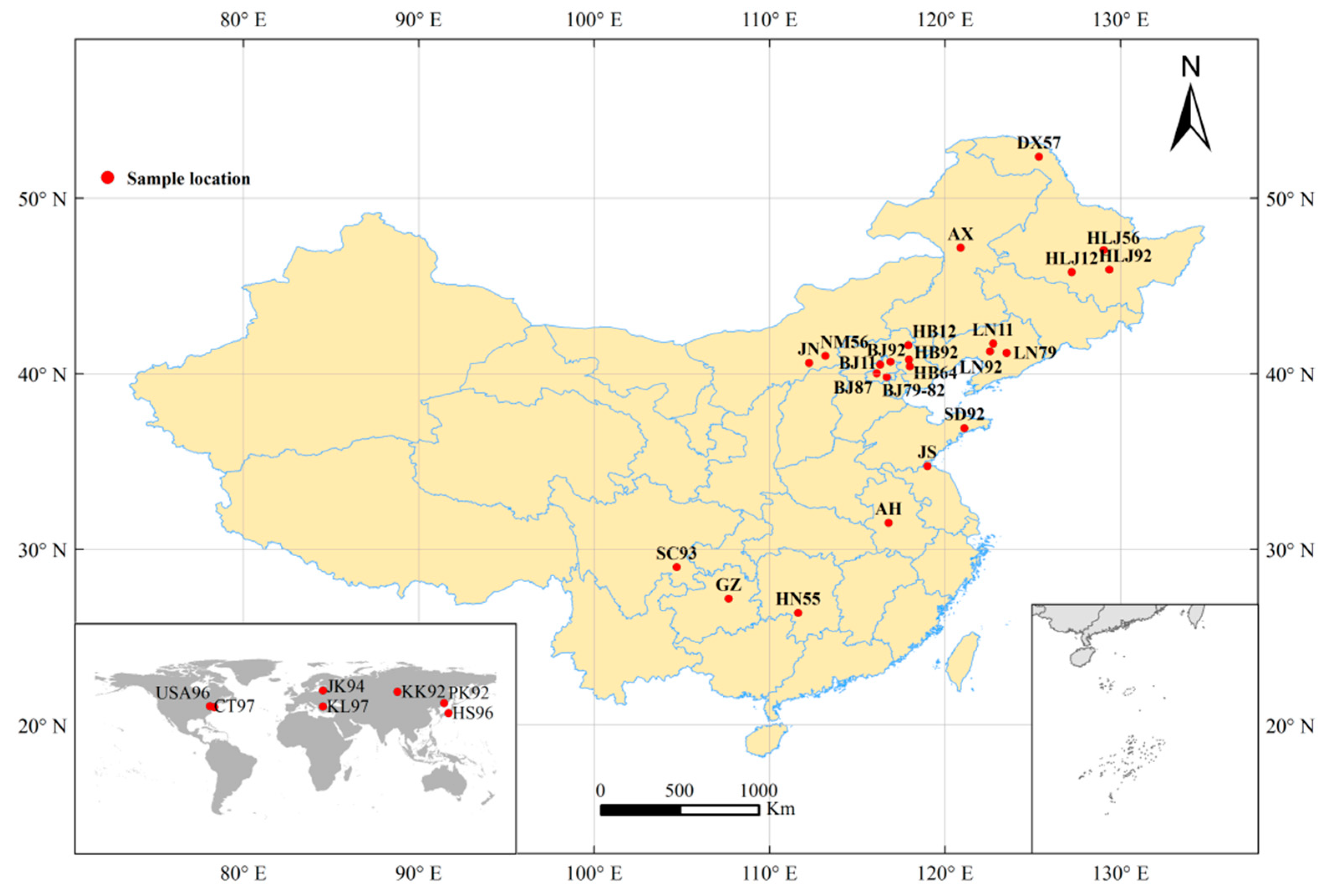

| Population Code | Collection Locality | Collection Year | Stage | Type of Preservation | Number of Samples |
|---|---|---|---|---|---|
| HN55 | Hunan | 1955 | Adult | Dry | 7 |
| HLJ56 | Heilongjiang | 1956 | Adult | Dry | 6 |
| NM56 | Inner Mongolia | 1956 | Larva | Formalin | 5 |
| DX57 | Greater Hinggan | 1957 | Adult | Dry | 10 |
| HB64 | Hebei | 1964 | Adult | Dry | 5 |
| HLJ79 | Heilongjiang | 1979 | Adult | Dry | 5 |
| LN79 | Liaoning | 1979 | Larva | Formalin | 5 |
| BJ79 | Beijing | 1979 | Adult | Dry | 8 |
| BJ81 | Beijing | 1981 | Adult | Dry | 10 |
| BJ82 | Beijing | 1982 | Adult | Dry | 5 |
| BJ87 | Beijing | 1987 | Adult | Dry | 6 |
| HLJ92 | Heilongjiang | 1992 | Larva | Formalin | 5 |
| SC93 | Sichuan | 1993 | Adult | Dry | 10 |
| USA96 | USA, Newark, DE | 1996 | Adult | Dry | 8 |
| LN08 | Liaoning | 2008 | Larva | Ethanol | 8 |
| Haplotype Code | Population Code | Country | Prefecture/Province/City/Region | Collection Year | Number of Samples |
|---|---|---|---|---|---|
| H1(4) | SHCIQ | China | Yichun, Heilongjiang | 2008 | 1 |
| LdaRM | Russia | Primorski, far east | 2016 | 1 | |
| LdaTJ | China | Tianjin | 2016 | 1 | |
| LN08 | China | Anshan, Liaoning | 2008 | 8 | |
| H2(1) | LddNJ | USA | New Jersey | 2016 | 1 |
| H3(1) | LddKG | Greece | Kavála, Macedonia | 2016 | 1 |
| H4(1) | LddLJ | Lithuania | Juodkrante, Kuzsin Nezijos | 2016 | 1 |
| H5(1) | LddKZ | Kazakhstan | Chuy Valley | 2016 | 1 |
| H6(1) | LddRB | Russia | Krasnoyarsk, Siberia | 2016 | 1 |
| H7(2) | LdjJN | Japan | Honshu | 2016 | 1 |
| LdjID | Japan | Iwate district | 2016 | 1 | |
| H8(1) | HN55 | China | Dong an, Hunan | 1955 | 6 |
| H9(1) | HN55 | China | Dong an, Hunan | 1955 | 1 |
| H10(1) | HLJ56 | China | Yichun, dailing, Heilongjiang | 1956 | 4 |
| H11(1) | HLJ56 | China | Yichun, dailing, Heilongjiang | 1956 | 2 |
| H12(1) | NM56 | China | Caihaer, Inner Mongolia | 1956 | 5 |
| H13(1) | DX57 | China | Daxinganling | 1957 | 4 |
| H14(1) | DX57 | China | Daxinganling | 1957 | 5 |
| H15(1) | DX57 | China | Daxinganling | 1957 | 1 |
| H16(1) | HB64 | China | Xinglong, Hebei | 1964 | 3 |
| H17(1) | HB64 | China | Xinglong, Hebei | 1964 | 2 |
| H18(1) | HLJ79 | China | Yichun, dailing, Heilongjiang | 1979 | 3 |
| H19(1) | HLJ79 | China | Yichun, dailing, Heilongjiang | 1979 | 2 |
| H20(1) | BJ79 | China | Mentougou, Beijing | 1979 | 8 |
| H21(1) | BJ81 | China | Mentougou, Beijing | 1981 | 2 |
| H22(1) | BJ81 | China | Mentougou, Beijing | 1981 | 8 |
| H23(1) | BJ82 | China | Mentougou, Beijing | 1982 | 2 |
| H24(1) | BJ82 | China | Mentougou, Beijing | 1982 | 3 |
| H25(1) | BJ87 | China | Xishan, Beijing | 1987 | 6 |
| H26(1) | LN79 | China | Anshan, Liaoning | 1979 | 5 |
| H27(1) | HLJ92 | China | Heihekou, Heilongjiang | 1992 | 5 |
| H28(1) | SC93 | China | Yibin, Sichuan | 1993 | 7 |
| H29(1) | SC93 | China | Yibin, Sichuan | 1993 | 1 |
| H30(1) | SC93 | China | Yibin, Sichuan | 1993 | 2 |
| H31(1) | USA96 | USA | Newark, DE | 1996 | 4 |
| H32(1) | USA96 | USA | Newark, DE | 1996 | 4 |
| Haplotype Code | Population Code | Prefecture/Province/City/Region | Collection Year | Number of Samples |
|---|---|---|---|---|
| H1(1) | HN55 | Dong an, Hunan | 1955 | 6 |
| H2(1) | HN55 | Dong an, Hunan | 1955 | 1 |
| H3(1) | NM56 | Caihaer, Inner Mongolia | 1956 | 5 |
| H4(1) | DX57 | Daxinganling | 1957 | 4 |
| H5(1) | DX57 | Daxinganling | 1957 | 5 |
| H6(1) | DX57 | Daxinganling | 1957 | 1 |
| H7(1) | HLJ56 | Yichun, dailing, Heilongjiang | 1956 | 4 |
| H8(3) | HLJ56 | Yichun, dailing, Heilongjiang | 1956 | 2 |
| LN92 | Sitaizi, Liaoning | 1992 | 1 | |
| AX | Arxan, Inner Mongolia | 2011 | 1 | |
| H9(3) | AX | Arxan, Inner Mongolia | 2011 | 3 |
| HLJ79 | Yichun, dailing, Heilongjiang | 1979 | 5 | |
| HB12 | Longhua, Hebei | 2012 | 1 | |
| H10(13) | HLJ92 | Qitaihe, Heilongjiang | 1992 | 5 |
| HLJ12 | Heihekou, Heilongjiang | 2012 | 7 | |
| LN92 | Anshan, Liaoning | 1992 | 2 | |
| LN08 | Anshan, Liaoning | 2008 | 8 | |
| LN11 | Sandeli, Liaoning | 2011 | 5 | |
| HB92 | Chengde, Hebei | 1992 | 3 | |
| HB12 | Longhua, Hebei | 2012 | 8 | |
| BJ87 | Xishan, Beijing | 1987 | 6 | |
| BJ92 | Labagou, Beijing | 1992 | 2 | |
| AH11 | Liu an, Anhui | 2011 | 5 | |
| JN | Jining, Inner Mongolia | 2011 | 5 | |
| JS | Lianyunguang, Jiangsu | 2011 | 10 | |
| AX | Arxan, Inner Mongolia | 2011 | 1 | |
| H11(1) | LN79 | Anshan, Liaoning | 1979 | 5 |
| H12(1) | LN11 | Sandeli, Liaoning | 2011 | 1 |
| SC93 | Yibin, Sichuan | 1993 | 6 | |
| GZ | Xifeng, Guizhou | 2011 | 8 | |
| H13(1) | HB64 | Xinglong, Hebei | 1964 | 3 |
| H14(1) | HB64 | Xinglong, Hebei | 1964 | 2 |
| H15(1) | HB12 | Longhua, Hebei | 2012 | 1 |
| H16(1) | BJ79 | Mentougou, Beijing | 1979 | 8 |
| H17(1) | BJ81 | Mentougou, Beijing | 1981 | 10 |
| AX | Arxan, Inner Mongolia | 2011 | 1 | |
| H18(1) | BJ82 | Mentougou, Beijing | 1982 | 2 |
| H19(1) | BJ82 | Mentougou, Beijing | 1982 | 3 |
| H20(1) | BJ92 | Labagou, Beijing | 1992 | 1 |
| H21(1) | BJ11 | Yanzikou, Beijing | 2011 | 9 |
| H22(1) | SC93 | Yibin, Sichuan | 1993 | 3 |
| H23(1) | SD92 | Laixi, Shandong | 1992 | 2 |
| H24(1) | SD92 | Laixi, Shandong | 1992 | 1 |
| H25(1) | AH11 | Liu an, Anhui | 2011 | 4 |
| H26(1) | GZ | Xifeng, Guizhou | 2011 | 1 |
| H27(1) | GZ | Xifeng, Guizhou | 2011 | 1 |
| H28(1) | JN | Jining, Inner Mongolia | 2011 | 1 |
| H29(1) | AX | Arxan, Inner Mongolia | 2011 | 1 |
| Haplotype Code | Population Code | Prefecture/Province/City/Region | Collection Year | Number of Samples |
|---|---|---|---|---|
| H1(7) | HLJ92 | Qitaihe, Heilongjiang | 1992 | 5 |
| LN92 | Sitaizi, Liaoning | 1992 | 2 | |
| BJ92 | Labagou, Beijing | 1992 | 2 | |
| USA96 | Newark, DE, USA | 1996 | 4 | |
| HB92 | Chengde, Hebei | 1992 | 3 | |
| HS96 | Honshu, Japan | 1996 | 6 | |
| PK92 | Primorsky Krai, Russia | 1992 | 9 | |
| H2(1) | BJ92 | Labagou, Beijing | 1992 | 1 |
| H3(1) | LN92 | Sitaizi, Liaoning | 1992 | 1 |
| H4(1) | SC93 | Yibin, Sichuan | 1993 | 7 |
| H5(1) | SC93 | Yibin, Sichuan | 1993 | 3 |
| H6(1) | USA96 | Newark, DE, USA | 1996 | 6 |
| H7(1) | SD92 | Laixi, Shandong | 1992 | 2 |
| H8(1) | SD92 | Laixi, Shandong | 1992 | 1 |
| H9(1) | KL97 | Kavala, Greece | 1997 | 3 |
| H10(1) | KL97 | Kavala, Greece | 1997 | 5 |
| H11(1) | KL97 | Kavala, Greece | 1997 | 1 |
| H12(1) | HS96 | Honshu, Japan | 1996 | 1 |
| H13(1) | JK94 | Juodkrante, Lithuania | 1994 | 5 |
| H14(1) | KK92 | Krasnoyarsk Krai, Russia | 1992 | 5 |
| H15(1) | KK92 | Krasnoyarsk Krai, Russia | 1992 | 5 |
| H16(1) | CT94 | Connecticut, United States | 1994 | 4 |
| H17(1) | CT94 | Connecticut, United States | 1994 | 6 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Zhang, S.; Wang, H.; Wang, M.; Li, G. Mitochondrial Gene Sequence (COI) Reveals the Genetic Structure and Demographic History of Lymantria dispar (Lepidoptera: Erebidae: Lymantriinae) in and around China. Insects 2019, 10, 146. https://doi.org/10.3390/insects10050146
Xu Y, Zhang S, Wang H, Wang M, Li G. Mitochondrial Gene Sequence (COI) Reveals the Genetic Structure and Demographic History of Lymantria dispar (Lepidoptera: Erebidae: Lymantriinae) in and around China. Insects. 2019; 10(5):146. https://doi.org/10.3390/insects10050146
Chicago/Turabian StyleXu, Yao, Sufang Zhang, Hongbin Wang, Mei Wang, and Guohong Li. 2019. "Mitochondrial Gene Sequence (COI) Reveals the Genetic Structure and Demographic History of Lymantria dispar (Lepidoptera: Erebidae: Lymantriinae) in and around China" Insects 10, no. 5: 146. https://doi.org/10.3390/insects10050146
APA StyleXu, Y., Zhang, S., Wang, H., Wang, M., & Li, G. (2019). Mitochondrial Gene Sequence (COI) Reveals the Genetic Structure and Demographic History of Lymantria dispar (Lepidoptera: Erebidae: Lymantriinae) in and around China. Insects, 10(5), 146. https://doi.org/10.3390/insects10050146






