Microsatellite Marker Discovery in the Stingless Bee Uruçu-Amarela (Melipona rufiventris Group, Hymenoptera, Meliponini) for Population Genetic Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and DNA Extraction
2.2. Library Preparation and High-Throughput Sequencing
2.3. SSR Mining and Primer Design
2.4. PCR Amplification and Validation of Selected SSRs
2.5. Data Analysis
3. Results and Discussion
3.1. Sequence Assembly and SSR Mining
3.2. SSR Validation
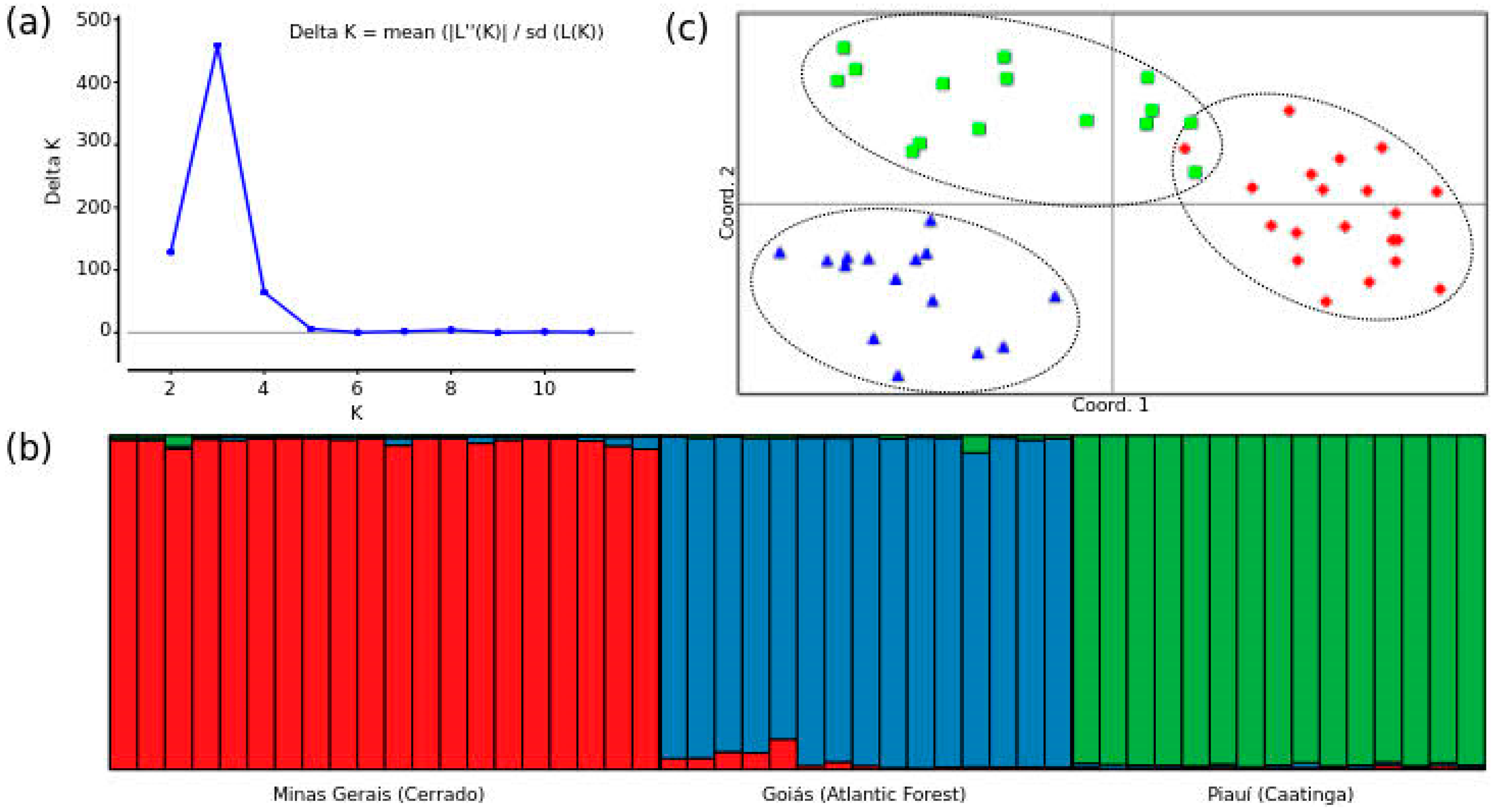
3.3. Genetic Divergence Among Populations in the ’Rufiventris Group’’
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Data Availability
References
- Silveira, F.A.; Melo, G.A.R.; Almeida, E.A.B. Abelhas Brasileiras: Sistemática e Identificação; Fundação Araucaria: Belo Horizonte, Brazil, 2002. [Google Scholar]
- Melo, G.A.R. Notas sobre meliponíneos neotropicais (Hymenoptera: Apidae), com a descrição de três novas espécies. In Apoidea Neotropica. Homenagem aos 90 Anos de Jesus Santiago Moure; Melo, G.A.R., Santos, I.A., Eds.; UNESC: Santa Catarina, Brazil, 2003; pp. 85–92. [Google Scholar]
- Moure, J.S.; Kerr, W.E. Sugestões para a modificação da sistemática do gênero Melipona (Hymenoptera: Apidae). Dusenia 1950, 1, 105–129. [Google Scholar]
- Moure, J.S. Notas sobre as espécies de Melipona descritas por Lepeletier em 1836 (Hymenoptera: Apidae). Rev. Bras. Biol. 1975, 35, 615–623. [Google Scholar]
- Costa, R.G.; Tavares, M.G.; Dias, L.A.S.; Campos, L.A.O. Isoenzyme variation in Melipona rufiventris (Hymenoptera: Apidae, Meliponina) in Minas Gerais State, Brazil. Biochem. Genet. 2005, 43, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Barni, G.S.; Strapazzon, R.; Guerra, J.C.V.; Moretto, G. Mitochondrial genome differences between the stingless bees Melipona rufiventris and Melipona mondury (Apidae: Meliponini). Genet. Mol. Res. 2007, 6, 8–14. [Google Scholar]
- Pires, C.V. Filogenia de Espécies Melipona do Complexo rufiventris, com Base em Sequências de DNA Mitocondrial. Master’s Dissertation, Universidade Federal de Viçosa, Viçosa, Brazil, February 2010. [Google Scholar]
- Freitas, P.V.D.X.; Faquinello, P.; Costa, L.F.X.; Zanata, R.A.; Santos, Y.G.; Silva, C.C. Avaliação da área de voo da abelha Melipona rufiventris rufiventris por meio do teste de captura e recaptura. In Proceedings of the Congresso Estadual de Iniciação Científica do IF Goiano, Instituto Federal de Goiás, Goiania, Brazil, 21–24 September 2015; pp. 1–2. [Google Scholar]
- Beekman, M.; Ratnieks, F.L.W. Long range foraging in the honeybee. Funct. Ecol. 2000, 14, 490–496. [Google Scholar] [CrossRef]
- Lopes, D.M.; Silva, F.O.; Fernandes-Salomão, T.M.; Campos, L.A.O.; Tavares, M.G. Microsatellite loci for the stingless bee Melipona rufiventris (Hymenoptera: Apidae). Mol. Ecol. Resour. 2009, 9, 923–925. [Google Scholar] [CrossRef]
- ICMBIO. Instituto Chico Mendes de Biodiversidade Listas das Espécies da Fauna Brasileira Ameaçadas de Extinção vigentes (Portarias MMA nº 444/2014 e 445/2014). Available online: http://www.icmbio.gov.br/portal/faunabrasileira?id=6706:portarias-fauna-ameacada (accessed on 1 September 2015).
- Tavares, M.G.; Dias, L.A.S.; Borges, A.A.; Lopes, D.M.; Busse, A.H.P.; Costa, R.G.; Fernandes-Salomão, T.M.; Campos, L.A.O. Genetics divergence between populations of the stingless bee uruçu-amarela (Melipona rufiventris group, Hymenoptera, Meliponini): Is there a new Melipona species in the Brazilian state of Minas Gerais? Genet. Mol. Biol. 2007, 30, 667–675. [Google Scholar] [CrossRef]
- May-Itzá, W.J.; Quezada-Euán, J.J.G.; Medina, L.A.M.; Enriquez, E.; De La Rúa, P. Morphometric and genetic differentiation in isolated populations of the endangered Mesoamerican stingless bee Melipona yucatanica (Hymenoptera: Apoidea) suggest the existence of a two species complex. Conserv. Genet. 2010, 11, 2079–2084. [Google Scholar] [CrossRef]
- Lopes, D.M.; Campos, L.A.O.; Fernandes-Salomão, T.M.; Tavares, M.G. Comparative study on the use of specific and heterologous microsatellite primers in the stingless bees Melipona rufiventris and M. mondury (Hymenoptera, Apidae). Genet. Mol. Biol. 2010, 33, 390–393. [Google Scholar] [CrossRef]
- Beaumont, M.A.; Bruford, M.W. Microsatellites in conservation genetics. In Microsatellites: Evolution and Applications; Goldstein, D.B., Schlötterer, C., Eds.; Oxford University Press: New York, NY, USA, 1999; pp. 165–182. [Google Scholar]
- Estoup, A.; Cornuet, J.M. Microsatellite evolution: Inferences from population data. In Microsatellites: Evolution and Applications; Goldstein, D.B., Schlötterer, C., Eds.; Oxford University Press: New York, NY, USA, 1999; pp. 49–65. [Google Scholar]
- Ellegren, H. Microsatellites: Simple sequences with complex evolution. Nat. Rev. Genet. 2004, 5, 435–445. [Google Scholar] [CrossRef]
- Peters, J.M.; Queller, D.C.; Imperatriz-Fonseca, V.L.; Strassmann, J.E. Microsatellite loci for stingless bees. Mol. Ecol. 1998, 7, 784–787. [Google Scholar] [PubMed]
- Francini, I.B.; Sforça, D.A.; Sousa, A.C.B.; Campos, T.; Zucchi, M.I.; Souza, A.P.; Nunes-Silva, C.G.; Carvalho-Zilse, G.A. Microsatellite loci for an endemic stingless bee Melipona seminigra merrillae (Apidae, Meliponini) from Amazon. Conserv. Genet. Resour. 2009, 1, 487–490. [Google Scholar] [CrossRef]
- Francini, I.B.; Sousa, A.C.B.; Sforça, D.A.; Costa-Pinto, M.F.F.; Campos, T.; Nunes-Silva, C.G.; Zucchi, M.I.; Souza, A.P.; Carvalho-Zilse, G.A. Isolation and characterization of microsatellite loci in the stingless bee Melipona interrupta manaosensis (Apidae: Meliponini). Conserv. Genet. Resour. 2010, 2, 27–30. [Google Scholar] [CrossRef]
- Lopes, D.M.; Silva, F.O.; Fernandes-Salomão, T.M.; Campos, L.A.O.; Tavares, M.G. Scientific note on the characterization of microsatellite locus for Melipona mondury (Hymenoptera: Apidae). Apidologie 2010, 41, 138–140. [Google Scholar] [CrossRef]
- Souza, I.G.B.; Paterson, I.; McBride, M.C.; Souza, B.A.; Pereira, F.M.; Lopes, M.T.R.; Bentzen, P.; Diniz, F.M. Isolation and characterization of 23 microsatellite loci in the stingless bee Melipona subnitida using next-generation sequencing. Conserv. Genet. Resour. 2015, 7, 239–241. [Google Scholar] [CrossRef]
- Silva, G.R.; Souza, I.G.B.; Pereira, F.M.; Souza, B.A.; Lopes, M.T.R.; Bentzen, P.; Diniz, F.M. Genome-wide discovery and characterization of microsatellite markers from Melipona fasciculata (Hymenoptera: Apidae), cross-amplification and a snapshot assessment of the genetic diversity in two stingless bee populations. Eur. J. Entomol. 2018, 115, 614–619. [Google Scholar] [CrossRef]
- Selkoe, K.A.; Toonen, R.J. Microsatellites for ecologists: A practical guide to using and evaluating microsatellite marker. Ecol. Lett. 2006, 9, 615–629. [Google Scholar] [CrossRef]
- Abdelkrim, J.; Robersten, B.C.; Stanton, J.A.; Gemmell, N.J. Fast, cost-effective development of species-specific microsatellite markers by genome sequencing. Biotechniques 2009, 46, 185–191. [Google Scholar] [CrossRef]
- Faircloth, B. Msatcommander: Detection of microsatellite repeat arrays and automated, locus-specific primer design. Mol. Ecol. Resour. 2008, 8, 92–94. [Google Scholar] [CrossRef]
- van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. Micro-Checker: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Goudet, J. FSTAT Version 2.9.3.2: A computer program to calculate F-statistics. J. Hered. 1995, 86, 485–486. [Google Scholar] [CrossRef]
- Kalinowski, S.T.; Taper, M.L.; Marshal, T.C. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef] [PubMed]
- Raymond, M.; Rousset, F. An exact test for population differentiation. Evolution 1995, 49, 1280–1283. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Earl, D.A.; Vonholdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 65: Genetic analysis in Excel Population genetic software for teaching and research: An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Graur, D. Gene diversity in Hymenoptera. Evolution 1985, 39, 190–199. [Google Scholar] [CrossRef]
- Singh, R.S.; Rhomberg, L.R. A comprehensive study of genic variation in natural populations of Drosophila melanogaster. Genetics 1987, 115, 313–322. [Google Scholar]
- Cornuet, J.M.; Luikart, G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 1996, 144, 2001–2014. [Google Scholar]
- Potts, S.G.; Biesmeijer, J.C.; Kremen, C.; Neumann, P.; Schweiger, O.; Kunin, W.E. Global pollinator declines: Trends, impacts and drivers. Trends Ecol. Evol. 2010, 25, 345–353. [Google Scholar] [CrossRef]
- Wright, S. Evolution and the Genetics of Populations, Variability within and among Natural Populations (Vol. 4.); The University of Chicago Press: Chicago, IL, USA, 1978. [Google Scholar]
- Quezada-Euan, J.J.G.; Paxton, R.J.; Palmer, K.A.; May-Itza, W.J.; Tay, W.T.; Oldroyd, B.P. Morphological and molecular characters reveal differentiation in a Neotropical social bee, Melipona beecheii (Apidae: Meliponini). Apidologie 2007, 38, 247–258. [Google Scholar] [CrossRef]
- Nascimento, M.A. Variabilidade genética de Melipona quadrifasciata (Hymenoptera: Apidae) no estado de Minas Gerais com marcadores ISSR. Master’s Dissertation, Universidade Federal de Viçosa, Viçosa, Brazil, February 2008. [Google Scholar]
- Beebee, T.J.C.; Rowe, G. Conservation genetics. In Introduction to Molecular Ecology; Beebee, T.J.C., Rowe, G., Eds.; Oxford University Press: New York, NY, USA, 2004; pp. 199–222. [Google Scholar]
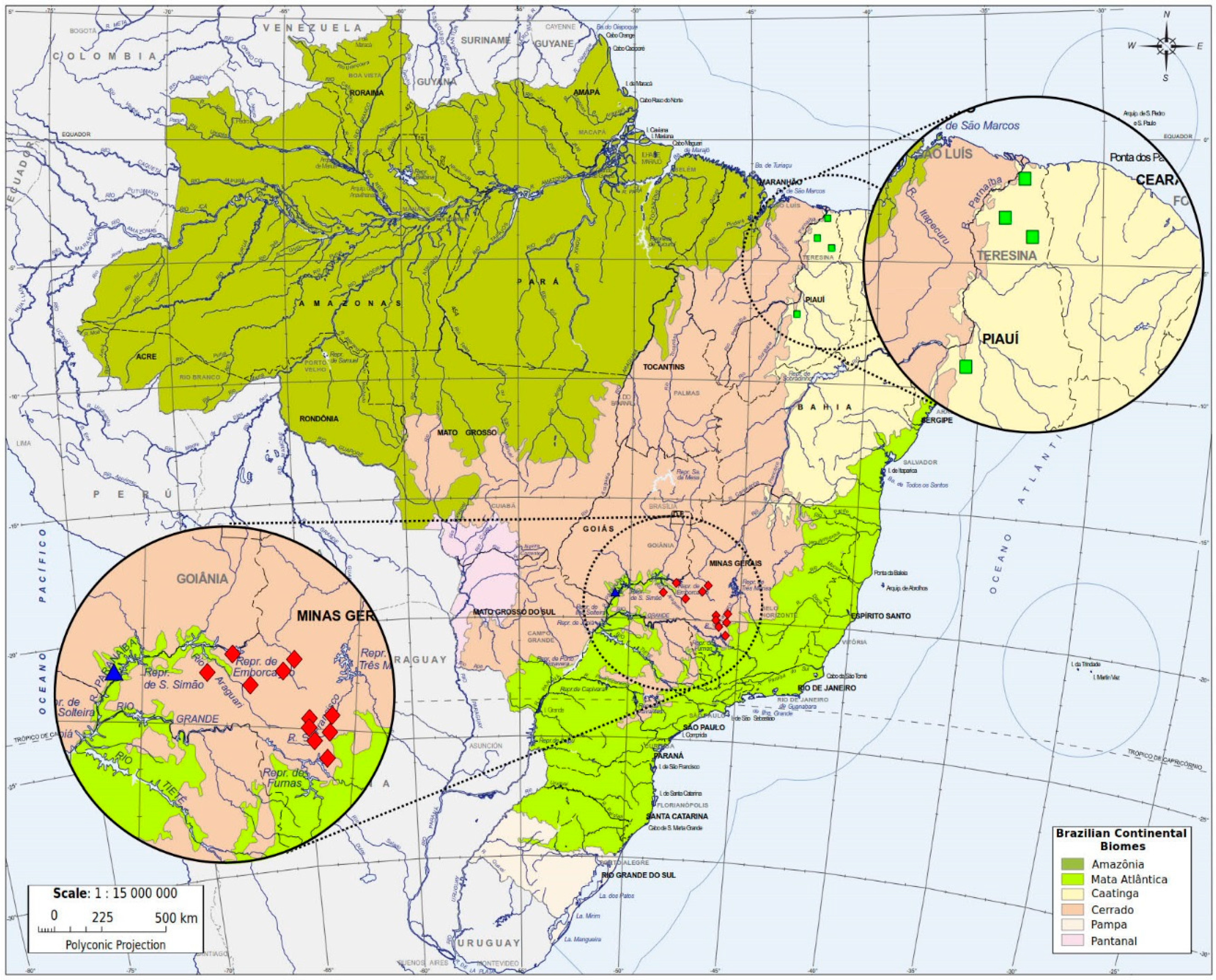
| Localities | State | Biome | Latitude/Longitude | Number of Colonies (Worker Bees) |
|---|---|---|---|---|
| Guimarânia | Minas Gerais | Cerrado | −18°50′/−46°47′ | 4 (4) |
| Araguari | Minas Gerais | Cerrado | −18°38′/−48°11′ | 3 (3) |
| Patos de Minas | Minas Gerais | Cerrado | −18°34′/−46°31′ | 1 (1) |
| Uberlândia | Minas Gerais | Cerrado | −18°55′/−48°16′ | 1 (1) |
| Águas do Paraíso | Minas Gerais | Cerrado | −20°19′/−45°27′ | 1 (1) |
| Iguatama | Minas Gerais | Cerrado | −20°10′/−45°42′ | 2 (2) |
| Tapiraí | Minas Gerais | Cerrado | −19°53′/−46°01′ | 1 (1) |
| Bambuí | Minas Gerais | Cerrado | −20°00′/−45°58′ | 1 (1) |
| Arcos | Minas Gerais | Cerrado | −20°16′/−45°32′ | 4 (4) |
| Lagoa da Prata | Minas Gerais | Cerrado | −20°01′/−45°32′ | 1 (1) |
| Córrego Danta | Minas Gerais | Cerrado | −19°49′/−45°54′ | 1 (1) |
| Caçu | Goiás | Atlantic Forest | −18°33′/−51°07′ | 5 (15) |
| Murici dos Portelas | Piauí | Caatinga | −03°19′/−42°05′ | 3 (4) |
| Campo Maior | Piauí | Caatinga | −04°49′/−42°10′ | 2 (5) |
| Castelo do Piauí | Piauí | Caatinga | −05°19′/−41°33′ | 1 (3) |
| Guadalupe | Piauí | Caatinga | −06°47′/−43°34′ | 1 (3) |
| Total | - | - | - | 32 (50) |
| Locus | Primer Sequence (5′–3′) | Motifs Repeats | PCR Profile | Ta (°C) | Size Range (bp) | NA | GenBank Accession No. |
|---|---|---|---|---|---|---|---|
| Mruf 1 | F: CAGTCGCCCAAGTAAATACG R: CTTATGAAACGAACCACAAGCC | (AGCC)8 | PCRSTD1 | 54 | 142–166 | 6 | MK133898 |
| Mruf 4 | F: GTTACGTTGGCAGGAGAGC R: AACTTGATTATTAGCGCGTGA | (TGCT)8 | PCRSTD1 | 52 | 146−166 | 6 | MK133901 |
| Mruf 5 | F: AGTGAAATCCGAGAGTGGGTT R: TCTCCACCGTCTTTTGTTTCTT | (AGAA)9 | PCRSTD1 | 51 | 154−170 | 4 | MK133902 |
| Mruf 6 | F: GTGCCTCGTTACCACCTTCTC R: TTAAAAGTGCGACGGGGA | (CT)9N13(GTCT)6 | PCRSTD1 | 54 | 106−110 | 2 | MK133903 |
| Mruf 8 | F: CATCGTCCTCCCGTGAATATAG R: TGCTTTTCCTTCCACGACC | (GCTG)10 | PCRSTD1 | 50 | 102−122 | 5 | MK133905 |
| Mruf 9 | F: TATACTTACGAGAGCGCACGAG R: TATTTTCTACGGTCCCACTTCG | (AACG)9 | PCRSTD1 | 55 | 118−140 | 7 | MK133906 |
| Mruf 11 | F: TGTGACGTTTTGGACGTAATTC R: CGCTTCCTTTGATCTCTCGAT | (TTTA)13 | PCRSTD2 | 48 | 112−124 | 4 | MK133908 |
| Mruf 13 | F: GCTAGGGGACCTTCTTCTTCTT R: GTGATAAGGCGGAGTGTAATC | (TTC)12 | PCRSTD1 | 50 | 100−106 | 2 | MK133910 |
| Mruf 15 | F: AAGTGGGGAGCTAATAAGGGAG R: TGCAGAGGAGCAGTACAGAGAG | (TTC)12 | PCRSTD3 | 51 | 157−172 | 4 | MK133912 |
| Mruf 17 | F: GTCGAGGACGACTACACAACAA R: CTCACCGCACACAGGGTT | (ACG)15 | PCRSTD1 | 52 | 161−173 | 3 | MK133914 |
| Mruf 18 | F: AAGCGGACAAGCAGATCACT R: ACTGTATGTCGTTCCTCGTCCT | (AAT)9 | PCRSTD1 | 52 | 108−126 | 6 | MK133915 |
| Mruf 19 | F: CACTGTCTTGTATTTAGACGCAATC R: GGTCGGGGACTTTAGTGTTTTA | (TTG)14 | PCRSTD1 | 55 | 125−134 | 2 | MK133916 |
| Mruf 20 | F: CGGGTAGTATTAAGGGAATTGA R: TGTGTCAGGAAGAAAAGCAA | (ATT)9 | PCRSTD1 | 54 | 184−193 | 2 | MK133917 |
| Mruf 21 | F: CTACCGAGAGTAGCGACGACAT R: TCAGTTCTCAATGTTGCAGGC | (ACG)11 | PCRSTD1 | 54 | 150−162 | 2 | MK133918 |
| Mruf 22 | F: CGACTTCGCGTGGTGCTAC R: AGAGGTTTCGGCGGCTTC | (ACG)9 | PCRSTD1 | 54 | 125−131 | 2 | MK133919 |
| Mruf 25 | F: AACAAGAGCAAAGTAACGACGA R: GAAGGAACAAGTCGAAACCAAC | (ACG)7N11(GAA)13 | PCRSTD3 | 51 | 137–155 | 6 | MK133922 |
| Locus | Minas Gerais (n = 20) | Goiás (n = 15) | Piauí (n = 15) | FST | RST | DEST | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AR | HO/HE | PIC | pHWE | Null | AR | HO/HE | PIC | pHWE | Null | AR | HO/HE | PIC | pHWE | Null | ||||
| Mruf 1 | 1.0 | 0.000/0.000 | 0.000 | - | 0.000 | 3.5 | 0.286/0.664 | 0.569 | 0.001 * | 0.216 | 4.6 | 0.357/0.762 | 0.689 | 0.001 * | 0.218 | 0.288 ¶ | 0.279 ¶ | 0.333 ¶ |
| Mruf 4 | 2.9 | 0.500/0.472 | 0.410 | 0.664 | −0.027 | 3.6 | 0.786/0.643 | 0.562 | 0.473 | −0.102 | 3.0 | 0.429/0.593 | 0.501 | 0.224 | 0.091 | 0.329 ¶ | 0.676 ¶ | 0.622 ¶ |
| Mruf 5 | 2.0 | 0.077/0.212 | 0.183 | 0.118 | 0.106 | 3.0 | 0.615/0.625 | 0.532 | 0.025 | −0.009 | 3.0 | 0.429/0.569 | 0.485 | 0.355 | 0.077 | 0.287 ¶ | 0.428 ¶ | 0.357 ¶ |
| Mruf 6 | 2,0 | 0.917/0.518 | 0.373 | 0.014 | −0.281 | 1.0 | 0.000/0.000 | 0.000 | − | 0.000 | 2.0 | 1.000/0.520 | 0.375 | 0.001 * | −0.333 | 0.270 ¶ | 0.270 ¶ | 0.229 ¶ |
| Mruf 8 | 2,0 | 0.400/0.337 | 0.269 | 1.000 | −0.061 | 2.6 | 0.385/0.335 | 0.290 | 1.000 | −0.047 | 3.8 | 0.500/0.540 | 0.482 | 0.166 | 0.011 | 0.433 ¶ | 0.363 ¶ | 0.508 ¶ |
| Mruf 9 | 3.9 | 0.294/0.709 | 0.630 | 0.000 * | 0.234 | 4.8 | 0.800/0.766 | 0.701 | 0.351 | −0.035 | 3.9 | 0.556/0.732 | 0.631 | 0.036 | 0.080 | 0.058 | 0.160 | 0.193 ¶ |
| Mruf 11 | 4.0 | 1.000/0.779 | 0.696 | 0.010 | −0.147 | 3.8 | 0.786/0.717 | 0.633 | 0.000 * | −0.056 | 4.0 | 0.667/0.752 | 0.657 | 0.050 | 0.025 | 0.030 | −0.014 | 0.083 |
| Mruf 13 | 2.0 | 0.933/0.515 | 0.374 | 0.002 * | −0.291 | 2.0 | 0.818/0.506 | 0.367 | 0.066 | −0.226 | 2.0 | 1.000/0.524 | 0.375 | 0.003 * | −0.333 | 0.002 | 0.002 | 0.002 |
| Mruf 15 | 3.1 | 0.714/0.521 | 0.433 | 0.430 | −0.141 | 2.7 | 0.308/0.283 | 0.255 | 1.000 | −0.028 | 2.8 | 0.400/0.416 | 0.347 | 0.223 | −0.004 | 0.053 | 0.085 | 0.035 ¶ |
| Mruf 17 | 1.0 | 0.000/0.000 | 0.000 | - | 0.000 | 1.0 | 0.000/0.000 | 0.000 | - | 0.000 | 2.0 | 0.462/0.492 | 0.361 | 1.000 | 0.008 | 0.850 ¶ | 0.771 ¶ | 0.847 ¶ |
| Mruf 18 | 2.4 | 0.053/0.437 | 0.354 | 0.000 * | 0.261 | 4.0 | 0.833/0.775 | 0.695 | 0.005 | −0.052 | 2.7 | 0.917/0.562 | 0.432 | 0.013 | −0.246 | 0.309 ¶ | 0.333 ¶ | 0.613 ¶ |
| Mruf 19 | 2.0 | 0.850/0.501 | 0.369 | 0.002 * | −0.243 | 2.0 | 1.000/0.517 | 0.375 | 0.000 * | −0.333 | 2.0 | 1.000/0.517 | 0.375 | 0.000 * | −0.333 | 0.005 | 0.005 | 0.005 |
| Mruf 20 | 2.0 | 0.500/0.386 | 0.305 | 0.526 | −0.091 | 2.0 | 0.308/0.271 | 0.226 | 1.000 | −0.038 | 1.0 | 0.000/0.000 | 0.000 | − | 0.000 | 0.106 | 0.106 | 0.033 |
| Mruf 21 | 2.0 | 0.375/0.315 | 0.258 | 1.000 | −0.054 | 2.0 | 0.636/0.455 | 0.340 | 0.481 | −0.141 | 2.0 | 0.250/0.233 | 0.195 | 1.000 | −0.026 | 0.019 | 0.019 | 0.012 |
| Mruf 22 | 2.0 | 0.800/0.513 | 0.375 | 0.022 | −0.200 | 2.0 | 0.692/0.471 | 0.350 | 0.208 | −0.165 | 2.0 | 0.333/0.287 | 0.239 | 1.000 | −0.044 | 0.112 ¶ | 0.112 ¶ | 0.083 ¶ |
| Mruf 25 | 3.5 | 0.813/0.579 | 0.498 | 0.206 | −0.162 | 3.0 | 0.500/0.633 | 0.511 | 0.322 | 0.059 | 3.0 | 0.700/0.532 | 0.442 | 0.503 | −0.130 | 0.278 ¶ | 0.649 ¶ | 0.491 ¶ |
| Mean | 2.4 | 0.514/0.425 | 0.345 | - | 2.7 | 0.547/0.479 | 0.400 | - | 2.7 | 0.563/0.502 | 0.412 | - | 0.252 ¶ | 0.317 ¶ | 0.284 ¶ | |||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Negreiros, A.B.; Silva, G.R.; Oliveira, F.A.S.; Resende, H.C.; Fernandes-Salomão, T.M.; Maggioni, R.; Pereira, F.M.; Souza, B.A.; Lopes, M.T.R.; Diniz, F.M. Microsatellite Marker Discovery in the Stingless Bee Uruçu-Amarela (Melipona rufiventris Group, Hymenoptera, Meliponini) for Population Genetic Analysis. Insects 2019, 10, 450. https://doi.org/10.3390/insects10120450
Negreiros AB, Silva GR, Oliveira FAS, Resende HC, Fernandes-Salomão TM, Maggioni R, Pereira FM, Souza BA, Lopes MTR, Diniz FM. Microsatellite Marker Discovery in the Stingless Bee Uruçu-Amarela (Melipona rufiventris Group, Hymenoptera, Meliponini) for Population Genetic Analysis. Insects. 2019; 10(12):450. https://doi.org/10.3390/insects10120450
Chicago/Turabian StyleNegreiros, Aline B., Geice R. Silva, Francisca A. S. Oliveira, Helder C. Resende, Tânia M. Fernandes-Salomão, Rodrigo Maggioni, Fabia M. Pereira, Bruno A. Souza, Maria T. R. Lopes, and Fábio M. Diniz. 2019. "Microsatellite Marker Discovery in the Stingless Bee Uruçu-Amarela (Melipona rufiventris Group, Hymenoptera, Meliponini) for Population Genetic Analysis" Insects 10, no. 12: 450. https://doi.org/10.3390/insects10120450
APA StyleNegreiros, A. B., Silva, G. R., Oliveira, F. A. S., Resende, H. C., Fernandes-Salomão, T. M., Maggioni, R., Pereira, F. M., Souza, B. A., Lopes, M. T. R., & Diniz, F. M. (2019). Microsatellite Marker Discovery in the Stingless Bee Uruçu-Amarela (Melipona rufiventris Group, Hymenoptera, Meliponini) for Population Genetic Analysis. Insects, 10(12), 450. https://doi.org/10.3390/insects10120450





