The Power of Electropenetrography in Enhancing Our Understanding of Host Plant-Vector Interactions
Abstract
1. Introduction
2. Materials and Methods
2.1. Plants
2.2. Insects
2.3. Electropenetrography Equipment and Settings
2.4. Electropenetrography Data Analysis
2.5. Ranking Resistance Levels in Mandarin Selections and Pummelo
2.6. Discriminant Analysis
2.7. Transitional Probabilities and Kinetograms
2.8. Leaf Metabolites
2.9. Metabolite Statistical Analysis
2.10. Diaphorina citri Oviposition and Survivorship on Mandarin Selections and Pummelo
3. Results
3.1. Diaphorina citri Probing Profiles on Mandarin Selections and Pummelo
3.2. Ranking Resistance Levels in Mandarin Selections and Pummelo
3.3. Transitional Probabilities and Kinetograms
3.4. Discriminant Analysis
3.5. Metabolite Profiles
3.6. Diaphorina citri Oviposition and Survivorship on Mandarin Selections and Pummelo
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CtoFrstG | The number of probes to first waveform xylem sap ingestion event |
| DurFirstE | Duration of first E event (mean)- may include phloem salivation (E1) only or include phloem ingestion (E2) if the first E2 event immediately follows the first E1. |
| DurFrstPrb | Duration of the first probe (mean) |
| DurG | Mean duration of the xylem ingestion waveform (G) |
| DurNnprbBfrFrstD | Cumulative duration of all non-probing events before first phloem contact waveform (D) event |
| DurNnprbBfrFrstE1 | Cumulative duration of all non-probing events before first phloem salivation event (E1) |
| DurNnprbBfrFrstG | Cumulative duration of all non-probing events before first xylem ingestion event (G) |
| DurScndPrb | Duration of the second probe |
| maxD | Longest waveform D event |
| maxE2 | Longest waveform E2 event |
| meanD | Average duration of waveform D events |
| MnDurC | Mean duration of pathway waveform (C) |
| MnDurE1 | Mean duration of E1 waveform events |
| NumLngD | Number of long (100+ seconds) phloem contact (D) events |
| NumLngE2 | Number of long (10+ minutes) E2 waveform events |
| NumLngG | Number of long (10+ minutes) G waveform events |
| PrcntPrbC | Of the cumulative time spent in a probe, the percentage of time spent in pathway (waveform C) |
| PrcntPrbD | Of the cumulative time spent in a probe, the percentage of time spent in phloem contact (waveform D) |
| PrcntPrbE2 | Of the cumulative time spent in a probe, the percentage of time spent in phloem ingestion (waveform E2) |
| PrcntE2SusE2 | Of all time spent ingesting phloem, the proportion of that time spent as sustained (>10 minutes) events. |
| TmFrmFrstPrbFrst D | Time from first probe to first D waveform event (mean) |
| TmFrmFrstPrbFrstE | Time from first probe to first E waveform event (mean) |
| TmFrstE2FrmFrstPrb | Time from first probe to first E2 waveform event (mean) |
| TmFrstE2StrtEPG | Time to first E2 waveform event from start of recording (mean) |
| TmFrstPrbFrmStrt | Time to first probe from start of recording (mean) Also equals the duration of first non-probing event. |
| TmFrstSusDFrstPrb | Time to first sustained D from first probe (mean) |
| TmFrstSusE2 | Time to first sustained waveform E2 event (mean) |
| TmFrstSusE2FrstPrb | Time from first probe to first sustained (>10 minutes) E2 waveform event |
| TmFrstSusGFrstPrb | Time from first probe to first sustained (>10 minutes) xylem ingestion (G) waveform event |
| TmLstE2EndRcrd | Time from the last E2 to end of recording. |
| TtlDurC | Total duration of C waveform events |
| TtlDurE | Total duration spent performing both E1 and E2 waveforms |
| TtlDurE1FllwdE2PlsE2 | Total duration of waveform E1 when followed by E2 plus the duration of all E2 events |
| TtlDurE2 | Total duration of E2 waveform events |
| TtlDurNnPhlPhs | Total duration of non-phloem phase, recording time less the time spent in E1 or E2. |
| TtlDurNP | Total duration of time spent not probing (mean) |
| TtlPrbTm | Total probing time |
References
- Da Graca, J.V.; Douhan, G.W.; Halbert, S.E.; Keremane, M.L.; Lee, R.F.; Vidalakis, G.; Zhao, H. Huanglongbing: An overview of a complex pathosystem ravaging the world’s citrus. J. Integr. Plant Biol. 2016, 58, 373–387. [Google Scholar] [CrossRef] [PubMed]
- Sétamou, M.; da Graça, J.V.; Sandoval, J.L. Suitability of native North American Rutaceae to serve as host plants for the Asian citrus psyllid (Hemiptera: Liviidae). J. Appl. Entomol. 2016, 140, 645–654. [Google Scholar] [CrossRef]
- Manjunath, K.L.; Halbert, S.E.; Ramadugu, C.; Webb, S.; Lee, R.F. Detection of ‘Candidatus Liberibacter asiaticus’ in Diaphorina citri and its importance in the management of citrus huanglongbing in Florida. Phytopathology 2008, 98, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef]
- Ebert, T.A.; Backus, E.A.; Shugart, H.J.; Rogers, M.E. Behavioral plasticity in probing by Diaphorina citri (Hemiptera, Liviidae): Ingestion from phloem versus xylem is influenced by leaf age and surface. J. Insect Behav. 2018, 31, 119–137. [Google Scholar] [CrossRef]
- Luo, X.; Yen, A.L.; Powell, K.S.; Wu, F.; Wang, Y.; Zeng, L.; Yang, Y.; Cen, Y. Feeding behavior of Diaphorina citri (Hemiptera: Liviidae) and its acquisition of ‘Candidatus Liberibacter asiaticus’, on huanglongbing-infected Citrus reticulata leaves of several maturity stages. Fla. Entomol. 2015, 98, 186–192. [Google Scholar] [CrossRef]
- Cen, Y.; Yang, C.; Holford, P.; Beattie, G.A.C.; Spooner-Hart, R.N.; Liang, G.; Deng, X. Feeding behaviour of the Asiatic citrus psyllid, Diaphorina citri, on healthy and huanglongbing-infected citrus. Entomol. Exp. Appl. 2012, 143, 13–22. [Google Scholar] [CrossRef]
- Backus, E.A.; Bennett, W.H. The AC-DC correlation monitor: New EPG design with flexible input resistors to detect both R and emf components for any piercing-sucking hemipteran. J. Insect Physiol. 2009, 55, 869–884. [Google Scholar] [CrossRef]
- Walker, G.P. A beginner’s guide to electronic monitoring of homopteran probing behavior. In Principles and Applications of Electronic Monitoring and Other Techniques in the Study of Homopteran Feeding Behavior; Walker, G.P., Backus, E.A., Eds.; Thomas Say Publications in Entomology: Annapolis, MD, USA, 2000; pp. 14–40. [Google Scholar]
- Backus, E.A.; Cervantes, F.A.; Guedes, R.N.C.; Li, A.Y.; Wayadande, A.C. AC–DC electropenetrography for in-depth studies of feeding and oviposition behaviors. Ann. Entomol. Soc. Am. 2019, 112, 236–248. [Google Scholar] [CrossRef]
- Hu, H.; Li, J.; Delatte, T.; Vervoort, J.; Gao, L.; Verstappen, F.; Xiong, W.; Gan, J.; Jongsma, M.A.; Wang, C. Modification of chrysanthemum odour and taste with chrysanthemol synthase induces strong dual resistance against cotton aphids. Plant Biotechnol. J. 2018, 16, 1434–1445. [Google Scholar] [CrossRef]
- Zhang, Z.; Cui, B.; Zhang, Y. Electrical penetration graphs indicate that tricin is a key secondary metabolite of rice, inhibiting phloem feeding of brown planthopper, Nilaparvata lugens. Entomol. Exp. Appl. 2015, 156, 14–27. [Google Scholar] [CrossRef]
- Barrios-San Martín, J.; Quiroz, A.; Verdugo, J.A.; Parra, L.; Hormazabal, E.; Astudillo, L.A.; Rojas-Herrera, M.; Ramírez, C.C. Host selection and probing behavior of the poplar aphid Chaitophorus leucomelas (Sternorrhyncha: Aphididae) on two poplar hybrids with contrasting susceptibility to aphids. J. Econ. Entomol. 2014, 107, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Koch, K.; Palmer, N.; Stamm, M.; Bradshaw, J.D.; Blankenship, E.; Baird, L.M.; Sarath, G.; Heng-Moss, T. Characterization of greenbug feeding behavior and aphid (Hemiptera: Aphididae) host preference in relation to resistant and susceptible tetraploid switchgrass populations. BioEnergy Res. 2015, 8, 165–174. [Google Scholar] [CrossRef]
- Mutti, N.S.; Louis, J.; Pappan, L.K.; Pappan, K.; Begum, K.; Chen, M.-S.; Park, Y.; Dittmer, N.; Marshall, J.; Reese, J.C.; et al. A protein from the salivary glands of the pea aphid, Acyrthosiphon pisum, is essential in feeding on a host plant. Proc. Natl. Acad. Sci. USA 2008, 105, 9965–9969. [Google Scholar] [CrossRef]
- Cornara, D.; Garzo, E.; Morente, M.; Moreno, A.; Alba-Tercedor, J.; Fereres, A. EPG combined with micro-CT and video recording reveals new insights on the feeding behavior of Philaenus spumarius. PLoS ONE 2018, 13, e0199154. [Google Scholar] [CrossRef]
- Ebert, T.A.; Rogers, M.E. Effect of substrate voltage on EPG recordings of ingestion and probing behavior in Diaphorina citri (Hemiptera: Liviidae). Fla. Entomol. 2016, 99, 528–534. [Google Scholar] [CrossRef]
- Tholt, G.; Samu, F.; Kiss, B. Feeding behaviour of a virus-vector leafhopper on host and non-host plants characterised by electrical penetration graphs. Entomol. Exp. Appl. 2015, 155, 123–136. [Google Scholar] [CrossRef]
- Pompon, J.; Quiring, D.; Giordanengo, P.; Pelletier, Y. Role of xylem consumption on osmoregulation in Macrosiphum euphorbiae (Thomas). J. Insect Physiol. 2010, 56, 610–615. [Google Scholar] [CrossRef]
- Serrano, M.S.; Backus, E.A.; Cardona, C. Comparison of AC electronic monitoring and field data for estimating tolerance to Empoasca kraemeri (Homoptera: Cicadellidae) in common bean genotypes. J. Econ. Entomol. 2000, 93, 1796–1809. [Google Scholar] [CrossRef]
- Arredondo de Ibarra, M.J. Abundance and Population Dynamics of the Asian Citrus Psyllid, Diaphorina citri, Kuwayama (Hemiptera: Psyllidae), as Affected by Flush Shoots in Different Host Plants. Master’s Thesis, Texas A&M University, College Station, TX, USA, 2009. [Google Scholar]
- Backus, E.A.; Bennett, W.H. Electrical Penetration Graph System. U.S. Patent US8004292B1, 22 October 2008. [Google Scholar]
- Bonani, J.P.; Fereres, A.; Garzo, E.; Miranda, M.P.; Appezzato-Da-Gloria, B.; Lopes, J.R.S. Characterization of electrical penetration graphs of the Asian citrus psyllid, Diaphorina citri, in sweet orange seedlings. Entomol. Exp. Appl. 2010, 134, 35–49. [Google Scholar] [CrossRef]
- Shugart, H.J. Probing Behavior and Host Preference in the Asian Citrus Psyllid, Diaphorina citri (Hemiptera: Liviidae) Dissertation. Ph.D. Thesis, University of Florida, Gainesville, FL, USA, 2019; p. 151. [Google Scholar]
- Ebert, T.A.; Backus, E.A.; Cid, M.; Fereres, A.; Rogers, M.E. A new SAS program for behavioral analysis of electrical penetration graph data. Comput. Electron. Agric. 2015, 116, 80–87. [Google Scholar] [CrossRef]
- Killiny, N.; Hijaz, F.; Ebert, T.A.; Rogers, M.E. A plant bacterial pathogen manipulates its insect vector’s energy metabolism. Appl. Environ. Microbiol. 2017, 83, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Killiny, N.; Valim, M.F.; Jones, S.E.; Omar, A.A.; Hijaz, F.; Gmitter, F.G., Jr.; Grosser, J.W. Metabolically speaking: Possible reasons behind the tolerance of ‘Sugar Belle’ mandarin hybrid to huanglongbing. Plant Physiol. Biochem. 2017, 116, 36–47. [Google Scholar] [CrossRef] [PubMed]
- Cifuentes-Arenas, J.C.; de Goes, A.; de Miranda, M.P.; Beattie, G.A.C.; Lopes, S.A. Citrus flush shoot ontogeny modulates biotic potential of Diaphorina citri. PLoS ONE 2018, 13, e0190563. [Google Scholar] [CrossRef] [PubMed]
- Patt, J.M.; Setamou, M. Responses of the Asian citrus psyllid to volatiles emitted by the flushing shoots of its rutaceous host plants. Environ. Entomol. 2010, 39, 618–624. [Google Scholar] [CrossRef] [PubMed]
- Hijaz, F.; Nehela, Y.; Killiny, N. Possible role of plant volatiles in tolerance against huanglongbing in citrus. Plant Signal. Behav. 2016, 11, e1138193. [Google Scholar] [CrossRef]
- Ford, E.S. Anatomy and histology of the eureka lemon. Bot. Gaz. 1942, 104, 288–305. [Google Scholar] [CrossRef]
- Al-Mousawi, A.H.; Richardson, P.E.; Burton, R.L. Ultrastructural studies of greenbug (Hemiptera: Aphididae) feeding damage to susceptible and resistant wheat cultivars. Ann. Entomol. Soc. Am. 1983, 76, 964–971. [Google Scholar] [CrossRef]
- McAllen, J.W.; Adams, J.B. The significance of pectinase in plant penetration by aphids. Can. J. Zool. 1961, 39, 305–310. [Google Scholar] [CrossRef]
- Shugart, H.J.; Rogers, M.E. Composition of Leaf Fibrous Ring Mediates Diaphorina Citri Preference of Citrus Sinensis over Citrus Aurantium; American Phytopathological Society: Tampa, FL, USA, 2016; Volume 106, pp. 134–135. [Google Scholar]
- Ammar, E.-D.; Richardson, M.L.; Abdo, Z.; Hall, D.G.; Shatters, R.G., Jr. Differences in stylet sheath occurrence and the fibrous ring (sclerenchyma) between xCitroncirus plants relatively resistant or susceptible to adults of the Asian citrus psyllid Diaphorina citri (Hemiptera: Liviidae). PLoS ONE 2014, 9, e110919. [Google Scholar] [CrossRef]
- George, J.; Ammar, E.D.; Hall, D.G.; Lapointe, S.L. Sclerenchymatous ring as a barrier to phloem feeding by Asian citrus psyllid: Evidence from electrical penetration graph and visualization of stylet pathways. PLoS ONE 2017, 12, e0173520. [Google Scholar] [CrossRef] [PubMed]
- Sandanayaka, W.R.M.; Moreno, A.; Tooman, L.K.; Page-Weir, N.E.M.; Fereres, A. Stylet penetration activities linked to the acquisition and inoculation of Candidatus Liberibacter solanacearum by its vector tomato potato psyllid. Entomol. Exp. Appl. 2014, 151, 170–181. [Google Scholar] [CrossRef]
- Yu, X.; Killiny, N. The secreted salivary proteome of Asian citrus psyllid Diaphorina citri. Physiol. Entomol. 2018, 43, 324–333. [Google Scholar] [CrossRef]
- Douglas, A.E. Phloem-sap feeding by animals: Problems and solutions. J. Exp. Bot. 2006, 57, 747–754. [Google Scholar] [CrossRef] [PubMed]
- Hijaz, F.; Killiny, N. Composition of citrus phloem sap and honeydew produced by the phloem sap feeder, the Asian citrus psyllid, Diaphorina citri (Homoptera: Psyllidae). J. Citrus Pathol. 2014, 1, 157–158. [Google Scholar]
- Hijaz, F.; Killiny, N. Collection and chemical composition of phloem sap from Citrus sinensis L. Osbeck (sweet orange). PLoS ONE 2014, 9, e101830. [Google Scholar] [CrossRef]
- Marutani-Hert, M.; Hunter, W.B.; Morgan, J.K. Associated bacteria of Asian Citrus Psyllid (Hemiptera: Psyllidae: Diaphorina citri). Southwest. Entomol. 2011, 36, 323–330. [Google Scholar] [CrossRef]
- Jing, X.; Wong, A.C.; Chaston, J.M.; Colvin, J.; McKenzie, C.L.; Douglas, A.E. The bacterial communities in plant phloem-sap-feeding insects. Mol. Ecol. 2014, 23, 1433–1444. [Google Scholar] [CrossRef]
- Pompon, J.; Quiring, D.; Goyer, C.; Giordanengo, P.; Pelletier, Y. A phloem-sap feeder mixes phloem and xylem sap to regulate osmotic potential. J. Insect Physiol. 2011, 57, 1317–1322. [Google Scholar] [CrossRef]
- Killiny, N. Metabolite signature of the phloem sap of fourteen citrus varieties with different degrees of tolerance to Candidatus Liberibacter asiaticus. Physiol. Mol. Plant Pathol. 2017, 97, 20–29. [Google Scholar] [CrossRef]
- Borgoni, P.C.; Vendramim, J.D.; Lourencao, A.L.; Machado, M.A. Resistance of Citrus and Related Genera to Diaphorina citri Kuwayama (Hemiptera: Liviidae). Neotrop. Entomol. 2014, 43, 465–469. [Google Scholar] [CrossRef] [PubMed]
- Alves, G.R.; Beloti, V.H.; Faggioni-Floriano, K.M.; de Carvalho, S.A.; Moral, R.A.; Demitrio, C.G.B.; Parra, J.R.P.; Yamamoto, P.T. Does the scion or rootstock of Citrus sp. affect the feeding and biology of Diaphorina citri Kuwayama (Hemiptera: Liviidae)? Arthropod-Plant Int. 2017, 12, 77–84. [Google Scholar] [CrossRef]
- Tsagkarakis, A.E.; Rogers, M.E. Suitability of ‘Cleopatra’ mandarin as a host plant for Diaphorina citri (Hemiptera: Psyllidae). Fla. Entomol. 2010, 93, 451–453. [Google Scholar] [CrossRef]
- Hopkins, D.P.; Cameron, D.D.; Butlin, R.K. The chemical signatures underlying host plant discrimination by aphids. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Ejsmond, M.; Provenza, F. Is doping of cognitive performance an anti-herbivore adaptation? Alkaloids inhibiting acetylcholinesterase as a case. Ecosphere 2018, 9, 1–14. [Google Scholar] [CrossRef]
- Yousaf, H.K.; Shan, T.; Chen, X.; Ma, K.; Shi, X.; Desneux, N.; Biondi, A.; Gao, X. Impact of the secondary plant metabolite Cucurbitacin B on the demographical traits of the melon aphid, Aphis gossypii. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Wittstock, U.; Gershenzon, J. Constitutive plant toxins and their role in defense against herbivores and pathogens. Curr. Opin. Plant Biol. 2002, 5, 1–8. [Google Scholar] [CrossRef]
- Albrecht, U.; Fiehn, O.; Bowman, K.D. Metabolic variations in different citrus rootstock cultivars associated with different responses to Huanglongbing. Plant Physiol. Biochem. 2016, 107, 33–44. [Google Scholar] [CrossRef]
- Asai, T.; Matsukawa, T.; Kajiyama, S. Metabolomic analysis of primary metabolites in citrus leaf during defense responses. J. Biosci. Bioeng. 2017, 123, 376–381. [Google Scholar] [CrossRef]
- Chen, Q.; Lu, X.; Guo, X.; Liu, J.; Liu, Y.; Guo, Q.; Tang, Z. The specific responses to mechanical wound in leaves and roots of Catharanthus roseus seedlings by metabolomics. J. Plant Interact. 2018, 13, 450–460. [Google Scholar] [CrossRef]
- Obata, T.; Fernie, A.R. The use of metabolomics to dissect plant responses to abiotic stresses. Cell. Mol. Life Sci. 2012, 69, 3225–3243. [Google Scholar] [CrossRef] [PubMed]
- Hijaz, F.; El-Shesheny, I.; Killiny, N. Herbivory by the insect Diaphorina citri induces greater change in the citrus plant volatile profile than does infection by the bacterium, Candidatus Liberibacter asiaticus. Plant Signal. Behav. 2013, 8, e25677. [Google Scholar] [CrossRef] [PubMed]
- Portillo-Estrada, M.; Niinemets, Ü. Massive release of volatile organic compounds due to leaf midrib wounding in Populus tremula. Plant Ecol. 2018, 219, 1021–1028. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.M. Plant Resistance to Insects—A Fundamental Approach; John Wiley & Sons: New York, NY, USA, 1989; p. 286. [Google Scholar]
- Ma, Y.; Wang, P.; Wang, M.; Sun, M.; Gu, Z.; Yang, R. GABA mediates phenolic compounds accumulation and the antioxidant system enhancement in germinated hulless barley under NaCl stress. Food Chem. 2019, 270, 593–601. [Google Scholar] [CrossRef]
- Hijaz, F.; Killiny, N. Exogenous GABA is quickly metabolized to succinic acid and fed into the plant TCA cycle. Plant Signal. Behav. 2019, 14, e1573096. [Google Scholar] [CrossRef]
- Bown, A.W.; Macgregor, K.B.; Shelp, B.J. Gamma-aminobutyrate: Defense against invertebrate pests? Trends Plant Sci. 2006, 11, 424–427. [Google Scholar] [CrossRef]
- Ramputh, A.I.; Bown, A.W. Rapid gamma-aminobutyric acid synthesis and the inhibition of growth and development of oblique-banded leafroller larvae. Plant Physiol. 1996, 111, 1349–1352. [Google Scholar] [CrossRef]
- Macgregor, K.B.; Shelp, B.J.; Peiris, S.; Bown, A.W. Overexpression of glutamate decarboxylase in transgenic tobacco plants deters feeding by phytophagous insect larvae. J. Chem. Ecol. 2003, 29, 2177–2182. [Google Scholar] [CrossRef]
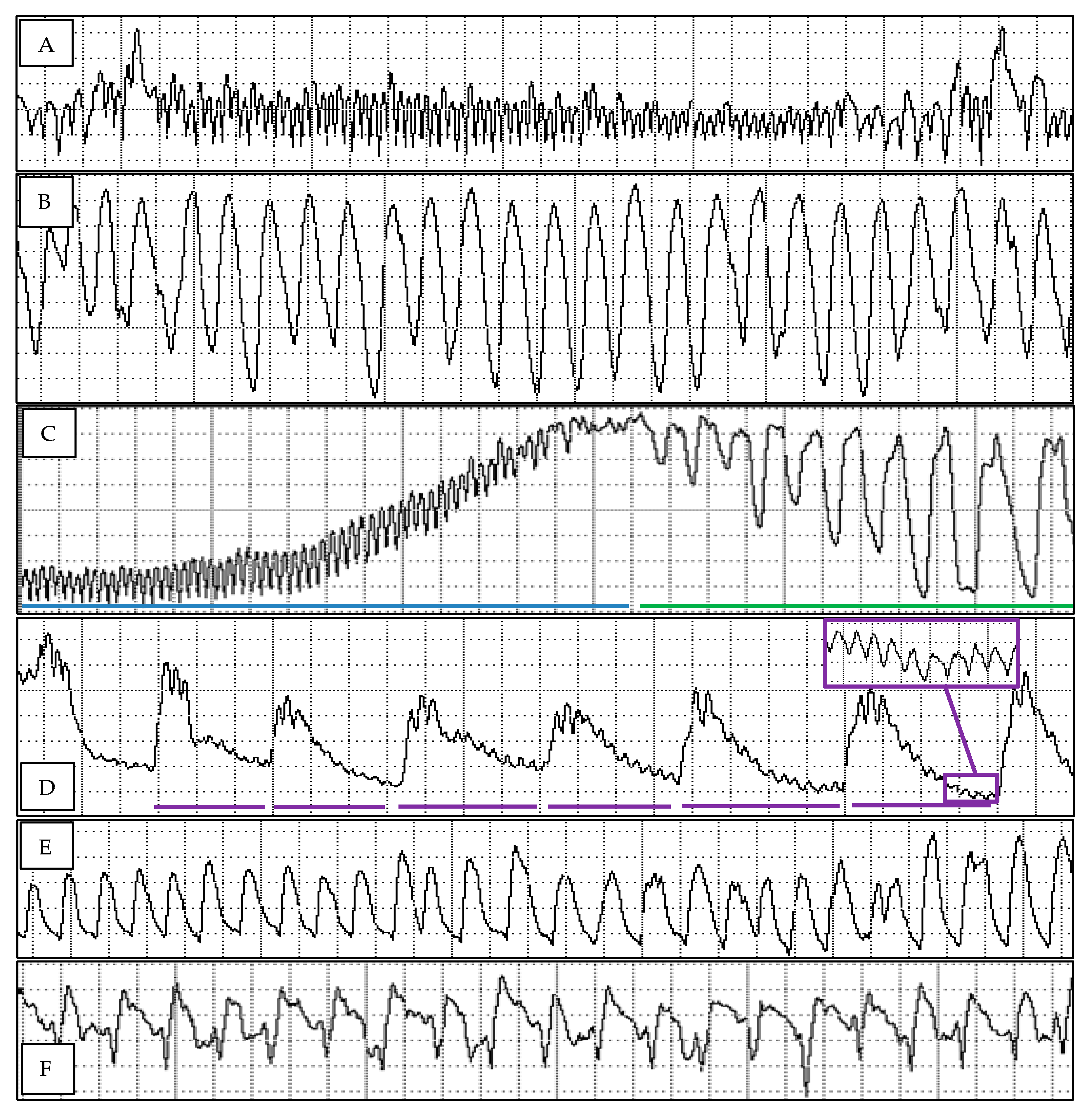
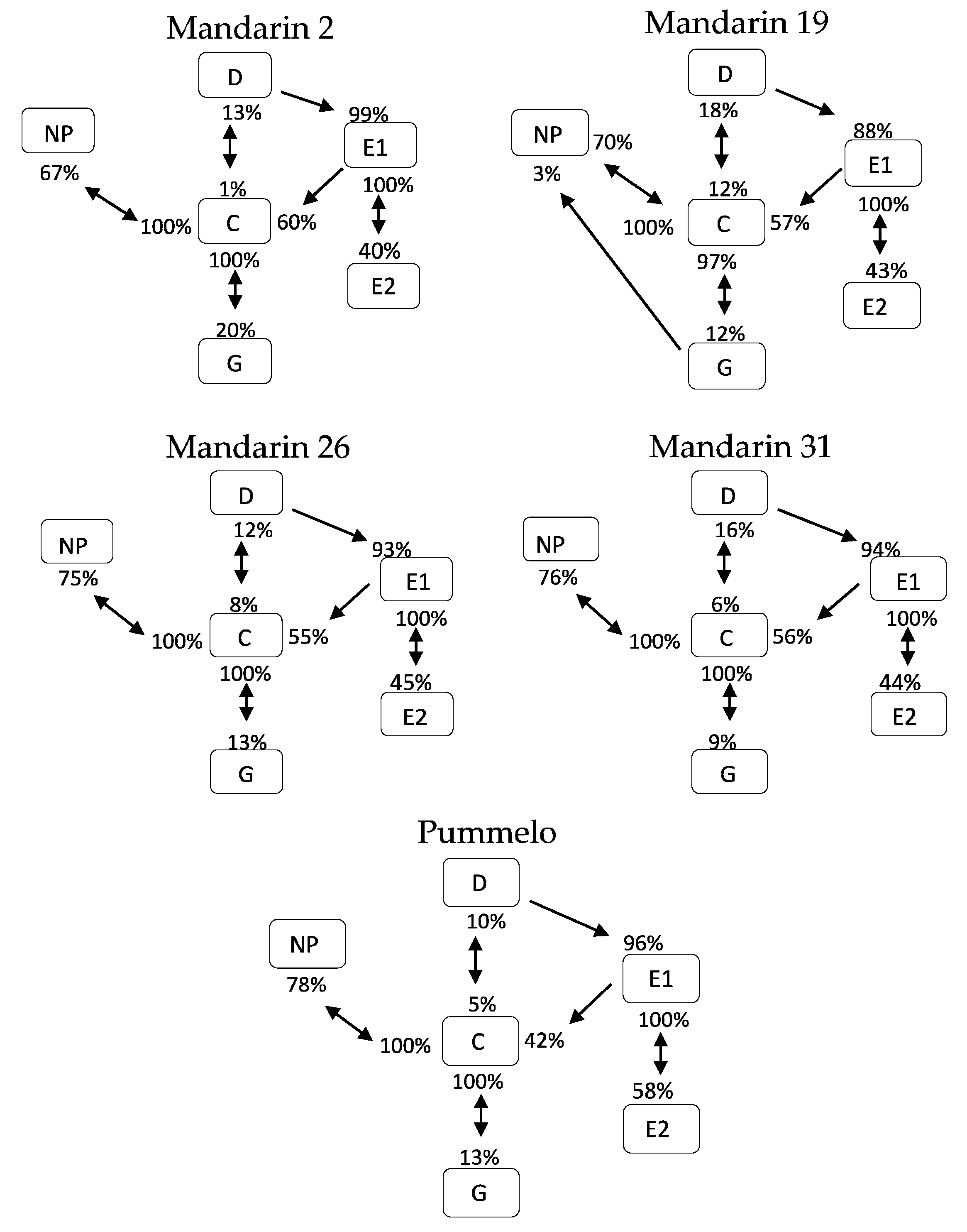
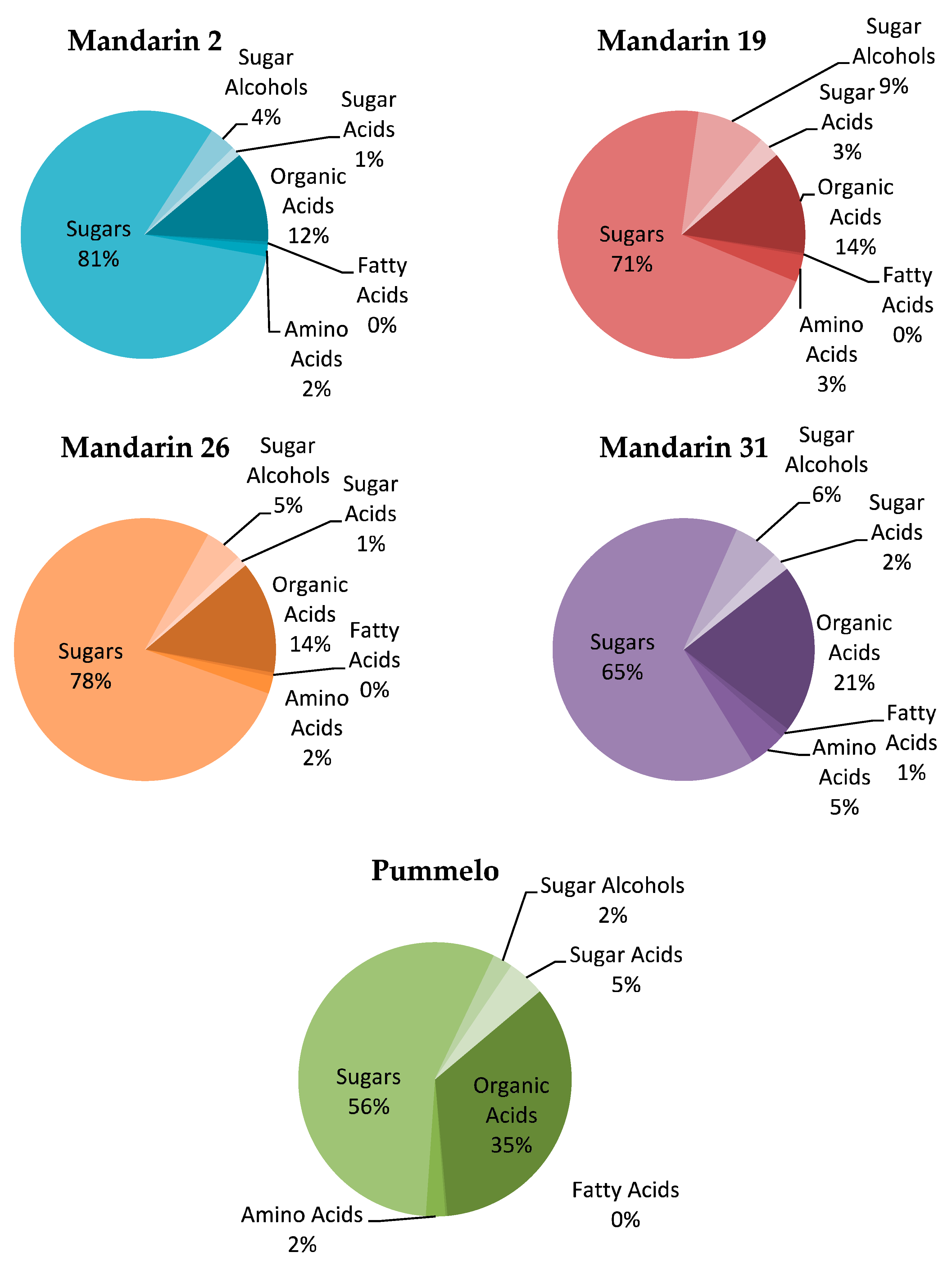
| Variable | Unit | Mandarin 2 | Mandarin 19 | Mandarin 26 | Mandarin 31 | Pummelo | p-Value | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DurFrstPrb | hr | 0.8 ± 0.295 | c | 1.9 ± 0.295 | a | 1 ± 0.295 | bc | 0.2 ± 0.295 | d | 1.1 ± 0.295 | b | 0.0001 |
| DurScndPrb | hr | 1.7 ± 0.288 | a | 1.6 ± 0.288 | a | 0.8 ± 0.288 | c | 0.1 ± 0.288 | d | 1.3 ± 0.288 | b | 0.0001 |
| TtlPrbTm | hr | 17.8 ± 0.406 | a | 17.6 ± 0.406 | a | 16.2 ± 0.406 | b | 14.8 ± 0.406 | c | 16.6 ± 0.406 | b | 0.0001 |
| TtlDurNP | hr | 4.7 ± 0.411 | c | 5.1 ± 0.411 | c | 7 ± 0.411 | a | 7.9 ± 0.411 | a | 6.4 ± 0.411 | b | 0.0001 |
| TmFrstPrbFrmStrt | hr | 0.2 ± 0.102 | c | 0.4 ± 0.099 | a | 0.3 ± 0.107 | ab | 0.6 ± 0.104 | a | 0.2 ± 0.104 | c | 0.0493 |
| DurNnprbBfrFrstE1 | hr | 2.9 ± 0.389 | b | 2.1 ± 0.389 | c | 3.6 ± 0.389 | a | 3.8 ± 0.389 | a | 2.6 ± 0.389 | b | 0.0001 |
| TtlDurC | hr | 11 ± 0.408 | a | 8.6 ± 0.408 | d | 10.1 ± 0.408 | b | 9.4 ± 0.408 | c | 7.9 ± 0.408 | e | 0.0001 |
| MnDurC | hr | 0.4 ± 0.058 | a | 0.4 ± 0.057 | a | 0.4 ± 0.061 | a | 0.3 ± 0.059 | b | 0.2 ± 0.059 | c | 0.0450 |
| PrcntPrbC | % | 62 ± 0.210 | b | 51 ± 0.20 | c | 62 ± 0.22 | b | 66 ± 0.21 | a | 48 ± 0.21 | c | 0.0502 |
| DurG | hr | 3.0 ± 0.326 | a | 2.0 ± 0.326 | b | 2.2 ± 0.342 | bc | 1.4 ± 0.342 | d | 2.4 ± 0.342 | b | 0.0159 |
| CtoFrstG | sec | 2.6 ± 0.97 | e | 3.2 ± 0.97 | d | 4.8 ± 1.02 | c | 6.8 ± 1.02 | b | 7.5 ± 1.02 | a | 0.0016 |
| DurNnprbBfrFrstG | hr | 0.7 ± 0.253 | c | 0.8 ± 0.253 | c | 1.2 ± 0.265 | b | 2.2 ± 0.265 | a | 1 ± 0.265 | b | 0.0013 |
| NumLngG | # | 4.6 ± 0.38 | a | 3.3 ± 0.38 | c | 3.0 ± 0.40 | d | 3.4 ± 0.40 | c | 3.7 ± 0.40 | b | 0.0362 |
| TmFrstSusGFrstPrb | hr | 1.5 ± 0.327 | d | 2.7 ± 0.327 | bc | 2.9 ± 0.327 | b | 3.5 ± 0.327 | a | 2.5 ± 0.327 | c | 0.0001 |
| meanD | sec | 66 ± 9.01 | c | 74 ± 7.62 | b | 63 ± 8.01 | c | 90 ± 8.01 | a | 43 ± 7.80 | d | 0.0019 |
| DurNnprbBfrFrstD | hr | 2.9 ± 0.316 | a | 2 ± 0.316 | b | 2.9 ± 0.316 | a | 2.7 ± 0.316 | a | 1.8 ± 0.316 | b | 0.0001 |
| TmFrmFrstPrbFrstD | hr | 7.1 ± 0.704 | b | 6.2 ± 0.704 | c | 7.8 ± 0.704 | a | 5.9 ± 0.704 | d | 6.4 ± 0.704 | c | 0.0001 |
| NumLngD | # | 1.1 ± 0.75 | b | 2.1 ± 0.64 | a | 0.3 ± 0.67 | c | 2.4 ± 0.67 | a | 0.2 ± 0.65 | c | 0.0497 |
| TmFrstSusDFrstPrb | hr | 14.1 ± 1.099 | b | 15.4 ± 1.099 | a | 6.7 ± 1.099 | c | 13.7 ± 1.099 | b | 15.5 ± 1.099 | a | 0.0001 |
| maxD | sec | 97.6 ± 22.99 | b | 129.9 ± 19.43 | a | 91.5 ± 20.43 | b | 142 ± 20.43 | a | 61 ± 19.91 | c | 0.0426 |
| PrcntPrbD | % | 6.2 ± 0.20 | c | 8.5 ± 0.19 | b | 6.4 ± 0.21 | c | 10.3 ± 0.20 | a | 8.10 ± 0.20 | b | 0.0503 |
| MnDurE1 | sec | 76.4 ± 9.68 | a | 50.2 ± 8.19 | b | 69 ± 8.61 | a | 45.6 ± 8.61 | c | 45.9 ± 8.39 | c | 0.0463 |
| TmFrmFrstPrbFrstE | hr | 12.3 ± 0.775 | a | 7.5 ± 0.775 | c | 8.6 ± 0.775 | b | 7.5 ± 0.775 | c | 7.2 ± 0.775 | c | 0.0001 |
| DurFirstE | hr | 1.1 ± 0.448 | b | 1.9 ± 0.379 | a | 0.7 ± 0.398 | c | 0.7 ± 0.398 | c | 1.9 ± 0.388 | a | 0.0501 |
| NumLngE2 | # | 1.3 ± 0.46 | d | 2.7 ± 0.45 | b | 2.2 ± 0.49 | c | 2.4 ± 0.47 | bc | 3.6 ± 0.47 | a | 0.0175 |
| TtlDurE2 | hr | 6 ± 0.392 | b | 10.4 ± 0.392 | a | 4.8 ± 0.392 | c | 4.7 ± 0.392 | c | 6.8 ± 0.392 | b | 0.0001 |
| TmFrstSusE2FrstPrb | hr | 15 ± 0.8 | a | 8.8 ± 0.8 | c | 11.2 ± 0.8 | b | 11.2 ± 0.8 | b | 7.4 ± 0.8 | d | 0.0001 |
| TmFrstE2StrtEPG | hr | 13.8 ± 0.8 | a | 9.1 ± 0.8 | c | 10.9 ± 0.8 | b | 10.8 ± 0.8 | b | 7.4 ± 0.8 | d | 0.0001 |
| TmFrstE2FrmFrstPrb | hr | 13.6 ± 0.793 | a | 8.7 ± 0.793 | c | 10.7 ± 0.793 | b | 10.2 ± 0.793 | b | 7.2 ± 0.793 | d | 0.0001 |
| TmLstE2EndRcrd | hr | 5.6 ± 0.965 | b | 2.6 ± 0.965 | c | 5.9 ± 0.965 | b | 8.3 ± 0.965 | a | 1.9 ± 0.965 | c | 0.0001 |
| maxE2 | hr | 3.5 ± 0.547 | b | 4.8 ± 0.509 | a | 2.8 ± 0.493 | c | 2.3 ± 0.478 | c | 3 ± 0.452 | bc | 0.0081 |
| PrcntPrbE2 | % | 29 ± 0.41 | b | 57 ± 0.38 | a | 19.5 ± 0.37 | c | 22.5 ± 0.36 | c | 37 ± 0.34 | b | 0.0149 |
| PrcntE2SusE2 | % | 44 ± 0.35 | b | 69 ± 0.29 | a | 72 ± 0.33 | a | 50 ± 0.27 | b | 72 ± 0.27 | a | 0.0184 |
| TtlDurE | hr | 5.3 ± 0.441 | c | 7.6 ± 0.441 | a | 4.2 ± 0.441 | d | 4.3 ± 0.441 | d | 6.6 ± 0.441 | b | 0.0001 |
| TtlDurE1FllwdE2PlsE2 | hr | 6.1 ± 0.395 | b | 10.5 ± 0.395 | a | 4.9 ± 0.395 | c | 4.7 ± 0.395 | c | 6.9 ± 0.395 | b | 0.0001 |
| TtlDurNnPhlPhs | hr | 17.3 ± 0.432 | b | 15.3 ± 0.432 | d | 19.1 ± 0.432 | a | 18.4 ± 0.432 | a | 16.4 ± 0.432 | c | 0.0001 |
| TmFrstSusE2 | hr | 15.1 ± 0.79 | a | 9.1 ± 0.79 | c | 11.4 ± 0.79 | b | 11.6 ± 0.79 | b | 7.5 ± 0.79 | d | 0.0001 |
| Variable | Mandarin 2 | Mandarin 19 | Mandarin 26 | Mandarin 31 | Pummelo | |
|---|---|---|---|---|---|---|
| 1 | TtlDurNP | 1 | 2 | 4 | 5 | 3 |
| 2 | TmFrstPrbFrmStrt | 1 | 4 | 3 | 5 | 1 |
| 3 | DurNnprbBfrFrstE1 | 3 | 1 | 4 | 5 | 2 |
| 4 | TtlDurC | 5 | 2 | 4 | 3 | 1 |
| 5 | PrcntPrbC | 4 | 2 | 3 | 5 | 1 |
| 6 | DurG | 5 | 2 | 3 | 1 | 4 |
| 7 | NumLngD | 3 | 4 | 2 | 5 | 1 |
| 8 | maxD | 3 | 4 | 2 | 5 | 1 |
| 9 | meanD | 3 | 4 | 2 | 5 | 1 |
| 10 | DurNnprbBfrFrstD | 5 | 2 | 4 | 3 | 1 |
| 11 | TmFrmFrstPrbFrstD | 4 | 2 | 5 | 1 | 3 |
| 12 | PrcntPrbD | 1 | 4 | 2 | 5 | 3 |
| 13 | TtlDurNnPhlPhs | 3 | 1 | 5 | 4 | 2 |
| 14 | TmFrstSusE2 | 5 | 2 | 3 | 4 | 1 |
| 15 | TmFrmFrstPrbFrstE | 5 | 2 | 4 | 3 | 1 |
| 16 | TmFrstSusE2FrstPrb | 5 | 2 | 4 | 3 | 1 |
| 17 | TmFrstE2StrtEPG | 5 | 2 | 4 | 3 | 1 |
| 18 | TmFrstE2FrmFrstPrb | 5 | 2 | 4 | 3 | 1 |
| 19 | TmLstE2EndRcrd | 3 | 2 | 4 | 5 | 1 |
| 20 | maxE2 | 2 | 1 | 4 | 5 | 3 |
| 21 | PrcntPrbE2 | 3 | 1 | 5 | 4 | 2 |
| 22 | PrcntE2SusE2 | 5 | 3 | 1 | 4 | 2 |
| 23 | NumLngE2 | 5 | 2 | 4 | 3 | 1 |
| 24 | TtlDurE2 | 3 | 1 | 4 | 5 | 2 |
| Column Sum | 87 | 54 | 84 | 94 | 40 | |
| Rank | 4 | 2 | 3 | 5 | 1 | |
| Most Resis. | Most Susc. |
| Mandarin 2 | Mandarin 19 | Mandarin 26 | Mandarin 31 | Pummelo | |
|---|---|---|---|---|---|
| Mandarin 2 | 0 | 2.43925 | 1.0376 | 4.44439 | 3.27002 |
| Mandarin 19 | 0.0037 | 0 | 2.35387 | 5.59795 | 5.97433 |
| Mandarin 26 | 0.2727 | 0.0069 | 0 | 1.85684 | 1.89187 |
| Mandarin 31 | 0.0001 | 0.0001 | 0.038 | 0 | 5.55242 |
| Pummelo | 0.0004 | 0.0001 | 0.0345 | 0.0001 | 0 |
| Metabolite | Mandarin 2 | Mandarin 19 | Mandarin 26 | Mandarin 31 | Pummelo | p-Value | |
|---|---|---|---|---|---|---|---|
| 1 | Ferulic Acid 338 | 7.06 ± 6.81 | 26.87 ± 32.38 | 45.78 ± 40.40 | 33.77 ± 37.14 | 7.76 ± 8.95 | 0.096 |
| 2 | Gluconic Acid | 11.26 ± 14.76 | 7.33 ± 7.70 | 31.29 ± 30.03 | 4.06 ± 3.21 | 3.90 ± 6.62 | 0.7348 |
| 3 | Citric Acid | 3989 ± 5276 | 2316 ± 4308 | 5901 ± 4018 | 3287 ± 6973 | 8546 ± 11,588 | 0.23 |
| 4 | Quinic Acid | 9.75 ± 0.84 a | 9.09 ± 0.84 a | 4.17 ± 0.84 b | 9.08 ± 0.84 a | 8.48 ± 0.84 a | 0.0203 |
| 5 | Malic Acid | 729.46 ± 1459.26 | 119.33 ± 165.17 | 275.41 ± 280.76 | 1,465.81 ± 869.77 | 422.74 ± 249.38 | 0.0864 |
| 6 | Synephrine | 228.52 ± 365.65 | 274.02 ± 357.05 | 246.44 ± 337.61 | 141.95 ± 218.21 | 0 | 0.3256 |
| 7 | Succinic Acid | 4.24 ± 2.46 ab | 1.18 ± 2.46 b | 4.49 ± 2.46 ab | 3.43 ± 2.46 ab | 12.99 ± 2.46 a | 0.0297 |
| 8 | Fumaric Acid | 0.84 ± 1.10 | 2.41 ± 4.02 | 2.98 ± 3.39 | 1.22 ± 0.94 | 1.22 ± 1.24 | 0.834 |
| 9 | Phosphoric Acid | 13.76 ± 19.06 | 0 | 0 | 0 | 66.54 ± 17.25 | 0.2055 |
| 10 | Maleic Acid | 21.86 ± 22.15 | 20.79 ± 24.03 | 110.88 ± 144.76 | 76.40 ± 59.14 | 129.57 ± 149.87 | 0.3013 |
| 11 | Lactic Acid | 4.00 ± 7.89 | 8.90 ± 13.51 | 26.46 ± 53.07 | 10.24 ± 15.38 | 5.85 ± 5.52 | 0.7579 |
| 12 | Oxalic Acid | 4.14 ± 2.66 | 2.46 ± 3.04 | 3.92 ± 3.3 | 52.28 ± 88.07 | 27.03 ± 50.68 | 0.3798 |
| 13 | Palmitic Acid | 171.63 ± 146.79 | 62.77 ± 131.28 | 204.87 ± 250.98 | 172.55 ± 209.70 | 45.40 ± 33.12 | 0.4819 |
| 14 | Oleic Acid | 1.11 ± 0.72 bc | 2.44 ± 0.72 a | 1.85 ± 0.72 ab | 0.4 ± 0.72 abc | 1.47 ± 0.8 c | 0.0072 |
| 15 | Stearic Acid | 4.27 ± 2.47 | 2.82 ± 4.78 | 18.59 ± 21.38 | 120.75 ± 256.19 | 19.72 ± 26.09 | 0.327 |
| 16 | L-Alanine | 24.96 ± 25.13 | 22.81 ± 20.35 | 2.84 ± 2.01 | 22.08 ± 36.15 | 43.70 ± 77.58 | 0.3133 |
| 17 | Glutamic Acid | 8.13 ± 11.47 | 0.97 ± 1.11 | 23.93 ± 41.10 | 22.07 ± 36.68 | 77.57 ± 95.77 | 0.6388 |
| 18 | L-Aspartic Acid | 18.05 ± 31.34 | 5.03 ± 7.42 | 19.66 ± 17.27 | 8.77 ± 9.81 | 20.48 ± 22.38 | 0.8808 |
| 19 | γ- Aminobutyric Acid | 360.04 ± 664.31 | 590.50 ± 715.28 | 725.34 ± 717.59 | 974.47 ± 529.83 | 79.91 ± 93.21 | 0.6335 |
| 20 | L-Threonine | 12.69 ± 9.53 | 10.16 ± 13.50 | 41.98 ± 57.13 | 18.63 ± 21.13 | 35.38 ± 35.41 | 0.6296 |
| 21 | Serine | 24.21 ± 36.44 b | 18.02 ± 36.44 b | 76.78 ± 36.44 ab | 26.26 ± 36.44 b | 187.76 ± 36.44 a | 0.0173 |
| 22 | L-Isoleucine | 5.28 ± 3.35 | 2.45 ± 3.85 | 2.15 ± 4.21 | 1.29 ± 0.54 | 6.75 ± 6.69 | 0.3506 |
| 23 | L-Proline | 155.95 ± 111.91 | 13.57 ± 9.52 | 40.55 ± 26.16 | 10.80 ± 8.45 | 128.26 ± 206.64 | 0.0669 |
| 24 | Glycine | 0 | 1.93 ± 2.38 | 20.81 ± 17.85 | 25.00 ± 35.82 | 18.09 ± 17.96 | 0.2206 |
| 25 | L-Valine | 2.62 ± 2.28 | 2.82 ± 4.78 | 18.59 ± 21.38 | 4.02 ± 7.02 | 1.10 ± 1.06 | 0.324 |
| 26 | 2-Aminopropanol | 37.54 ± 38.67 | 52.81 ± 36.33 | 51.34 ± 41.49 | 15.73 ± 4.03 | 11.93 ± 11.12 | 0.297 |
| 27 | Xylose 1 | 3.37 ± 2.35 b | 4.15 ± 2.35 b | 6.30 ± 2.35 b | 3.08 ± 2.35 b | 16.78 ± 2.35 a | 0.0473 |
| 28 | Xylose 2 | 48.96 ± 58.79 | 13.06 ± 17.08 | 32.76 ± 58.84 | 36.09 ± 48.58 | 45.15 ± 43.79 | 0.8393 |
| 29 | Arabinose | 1.50 ± 1.04 | 7.56 ± 5.72 | 4.46 ± 3.34 | 5.25 ± 6.92 | 3.77 ± 2.01 | 0.5822 |
| 30 | Erythrose | 156.54 ± 78.21 | 239.49 ± 64.14 | 254.54 ± 100.14 | 260.12 ± 119.96 | 181.15 ± 5.04 | 0.2415 |
| 31 | Fructose | 414.94 ± 471.91 | 648.13 ± 864.54 | 2827 ± 4297 | 1183 ± 1404 | 1162 ± 1412 | 0.3747 |
| 32 | Mannose | 104.01 ± 177.97 | 128.47 ± 149.63 | 140.10 ± 125.65 | 192.22 ± 103.78 | 59.57 ± 71.99 | 0.433 |
| 33 | Glucose | 7001 ± 12025 | 1569 ± 2011 | 5770 ± 12,120 | 1037 ± 1240 | 2398 ± 4143 | 0.9915 |
| 34 | Glucopyranose | 11.19 ± 16.33 | 32.25 ± 35.77 | 19.94 ± 21.38 | 18.18 ± 15.79 | 3.29 ± 3.95 | 0.3198 |
| 35 | Threose | 8.13 ± 11.20 | 12.64 ± 15.14 | 17.22 ± 10.86 | 19.24 ± 13.40 | 8.61 ± 10.88 | 0.6122 |
| 36 | Pyranoside 204/338 | 1.18 ± 0.44 | 5.16 ± 6.12 | 1.15 ± 1.39 | 0.16 ± 0.06 | 0.24 ± 0.45 | 0.8232 |
| 37 | α-galactoside | 5.06 ± 1.93 ab | 2.95 ± 1.93 ab | 10.13 ± 1.93 a | 2.50 ± 1.93 ab | 1.71 ± 1.93 b | 0.039 |
| 38 | Deoxy-galactoside | 393.40 ± 869.20 | 74.87 ± 102.35 | 59.01 ± 43.88 | 19.72 ± 25.23 | 8.58 ± 15.46 | 0.2983 |
| 39 | Glucoheptose | 932 ± 1318 | 1350 ± 1316 | 1300 ± 2053 | 1404 ± 2826 | 356.07 ± 566.44 | 0.2401 |
| 40 | Sucrose | 24,586 ± 20,814 | 10,144 ± 9511 | 26,319 ± 56,240 | 11,322 ± 15,132 | 10,305 ± 10,812 | 0.1514 |
| 41 | Maltose | 66.46 ± 50.97 | 332.82 ± 376.90 | 31.92 ± 26.38 | 69.59 ± 54.02 | 229.71 ± 157.41 | 0.7644 |
| 42 | Unk Disaccharide 361 | 218.88 ± 222.90 | 94.45 ± 152.11 | 188.48 ± 340.13 | 283.68 ± 261.95 | 67.99 ± 111.31 | 0.1241 |
| 43 | Unk Disaccharide | 87.59 ± 69.97 | 11.82 ± 12.26 | 19.89 ± 20.037 | 53.94 ± 50.23 | 69.23 ± 86.47 | 0.5592 |
| 44 | Inositol-2- Phosphate | 15.89 ± 21.34 | 10.65 ± 7.55 | 18.13 ± 23.89 | 11.55 ± 14.80 | 1.79 ± 1.86 | 0.946 |
| 45 | Scyllo- Inositol | 218.96 ± 167.70 | 193.23 ± 156.54 | 326.86 ± 411.38 | 281.57 ± 189.51 | 197.42 ± 256.13 | 0.7775 |
| 46 | Myo- Inositol | 351.59 ± 546.51 | 2.46 ± 3.04 | 3.92 ± 3.30 | 93.55 ± 184.35 | 205.76 ± 227.12 | 0.1965 |
| 47 | Phytol 143 | 13.94 ± 18.45 | 0 | 45.89 ± 56.93 | 9.72 ± 3.68 | 0.02 ± 0.04 | 0.3732 |
| 48 | Glycerol | 1.97 ± 0.67 b | 4.9 ± 0.67 a | 5.06 ± 0.67 a | 4.32 ± 0.67 ab | 4.46 ± 0.67 ab | 0.0267 |
| 49 | Xylitol | 87.91 ± 181.95 | 252.24 ± 251.94 | 382.21 ± 346.67 | 110.18 ± 137.42 | 117.65 ± 150.62 | 0.318 |
| 50 | Glucitol | 11.87 ± 16.82 | 5.53 ± 6.40 | 7.27 ± 10.61 | 3.35 ± 3.11 | 0.96 ± 1.57 | 0.6759 |
| 51 | Mannitol | 34.73 ± 38.52 | 37.67 ± 55.35 | 48.29 ± 39.73 | 17.44 ± 21.41 | 97.69 ± 108.58 | 0.3695 |
| 52 | Chiro-Inositol | 730.71 ± 818.83 | 1337 ± 1033 | 1334 ± 2109 | 803.14 ± 720.79 | 0 | 0.4593 |
| 53 | Sugar Alcohol 217/319 | 6.17 ±4.01 | 5.78 ± 6.52 | 11.96 ± 10.08 | 11.03 ± 8.31 | 3.31 ± 5.84 | 0.4336 |
| 54 | Ribonic Acid | 93.48 ± 112.08 | 34.67 ± 43.88 | 64.91 ± 78.58 | 140.54 ± 121.14 | 71.06 ± 78.88 | 0.2806 |
| 55 | Saccharic Acid | 14.12 ± 24.50 | 5.88 ± 4.38 | 20.83 ± 23.03 | 12.21 ± 21.97 | 8.61 ± 5.06 | 0.6895 |
| 56 | Sugar Acid 204/333 | 17.10 ± 14.16 b | 19.12 ± 14.16 b | 73.84 ± 14.16 a | 9.22 ± 14.16 b | 14.37 ± 14.16 b | 0.0247 |
| 57 | Glucuronic Acid | 3.37± 3.71 | 2.59 ± 2.88 | 7.06 ± 11.11 | 6.43 ± 5.96 | 1.19 ± 2.67 | 0.5053 |
| 58 | Threonic Acid Deriv. | 66.70 ± 123.42 | 15.65 ± 15.43 | 33.12 ± 22.20 | 67.12 ± 81.69 | 31.79 ± 37.23 | 0.1031 |
| 59 | Threonic Acid | 48.34 ± 34.19 | 193.47 ± 193.11 | 95.82 ± 65.08 | 100.25 ± 114.15 | 974.05 ± 1182.63 | 0.8799 |
| 60 | 2-Ketoglutaric Acid | 2.04 ± 2.02 | 2.24 ± 0.75 | 2.08 ± 2.61 | 6.50 ± 7.55 | 5.70 ± 5.44 | 0.5845 |
| 61 | Arabino- Hexaric Acid | 5.59 ± 5.12 | 7.28 ± 10.57 | 5.44 ± 2.67 | 3.30 ± 3.88 | 4.52 ± 3.49 | 0.5592 |
| 62 | Unk Sugar Acid 333 | 75.24 ± 92.05 | 26.66 ± 38.38 | 132.95 ± 202.05 | 165.58 ± 185.82 | 34.11 ± 42.22 | 0.2685 |
| 63 | Glycerol Glycoside | 170.89 ± 251.21 | 256.32 ± 375.03 | 180.57 ± 177.95 | 33.23 ± 55.60 | 34.83 ± 27.99 | 0.3235 |
| Selection/Species | Eggs Laid | Adults Counted | Total Adults Emerged | % Adults Emerged from Egg (95% CI) | |||
|---|---|---|---|---|---|---|---|
| Day 10 1 | Day 20 2 | Day 24 | Day 27 | Day 29 | |||
| Mandarin 2 | 578 | 12 | 139 | 66 | 19 | 236 | 40.83% (36.9–44.9%) b |
| Mandarin 19 | 87 | 0 | 3 | 1 | 0 | 4 | 4.60% (1.5–10.7%) c |
| Mandarin 26 | 76 | 0 | 0 | 3 | 0 | 3 | 3.95% (1.1–10.3%) c |
| Mandarin 31 | 29 | 2 | 3 | 6 | 6 | 27 | 93.10% (79.3–100%) a |
| Pummelo | 457 | 42 | 241 | 78 | 11 | 371 | 81.18% (77.4–85.6%) a |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shugart, H.; Ebert, T.; Gmitter, F.; Rogers, M. The Power of Electropenetrography in Enhancing Our Understanding of Host Plant-Vector Interactions. Insects 2019, 10, 407. https://doi.org/10.3390/insects10110407
Shugart H, Ebert T, Gmitter F, Rogers M. The Power of Electropenetrography in Enhancing Our Understanding of Host Plant-Vector Interactions. Insects. 2019; 10(11):407. https://doi.org/10.3390/insects10110407
Chicago/Turabian StyleShugart, Holly, Timothy Ebert, Frederick Gmitter, and Michael Rogers. 2019. "The Power of Electropenetrography in Enhancing Our Understanding of Host Plant-Vector Interactions" Insects 10, no. 11: 407. https://doi.org/10.3390/insects10110407
APA StyleShugart, H., Ebert, T., Gmitter, F., & Rogers, M. (2019). The Power of Electropenetrography in Enhancing Our Understanding of Host Plant-Vector Interactions. Insects, 10(11), 407. https://doi.org/10.3390/insects10110407





