Roles of Type 10 17β-Hydroxysteroid Dehydrogenase in Health and Disease
Abstract
1. Introduction
2. What Are ERAB and ABAD?
2.1. ERAB Is an Artificial Protein That Is Not Present in Any Tissues
2.2. 17β-HSD10 Is a Member of the Alcohol Dehydrogenase Family
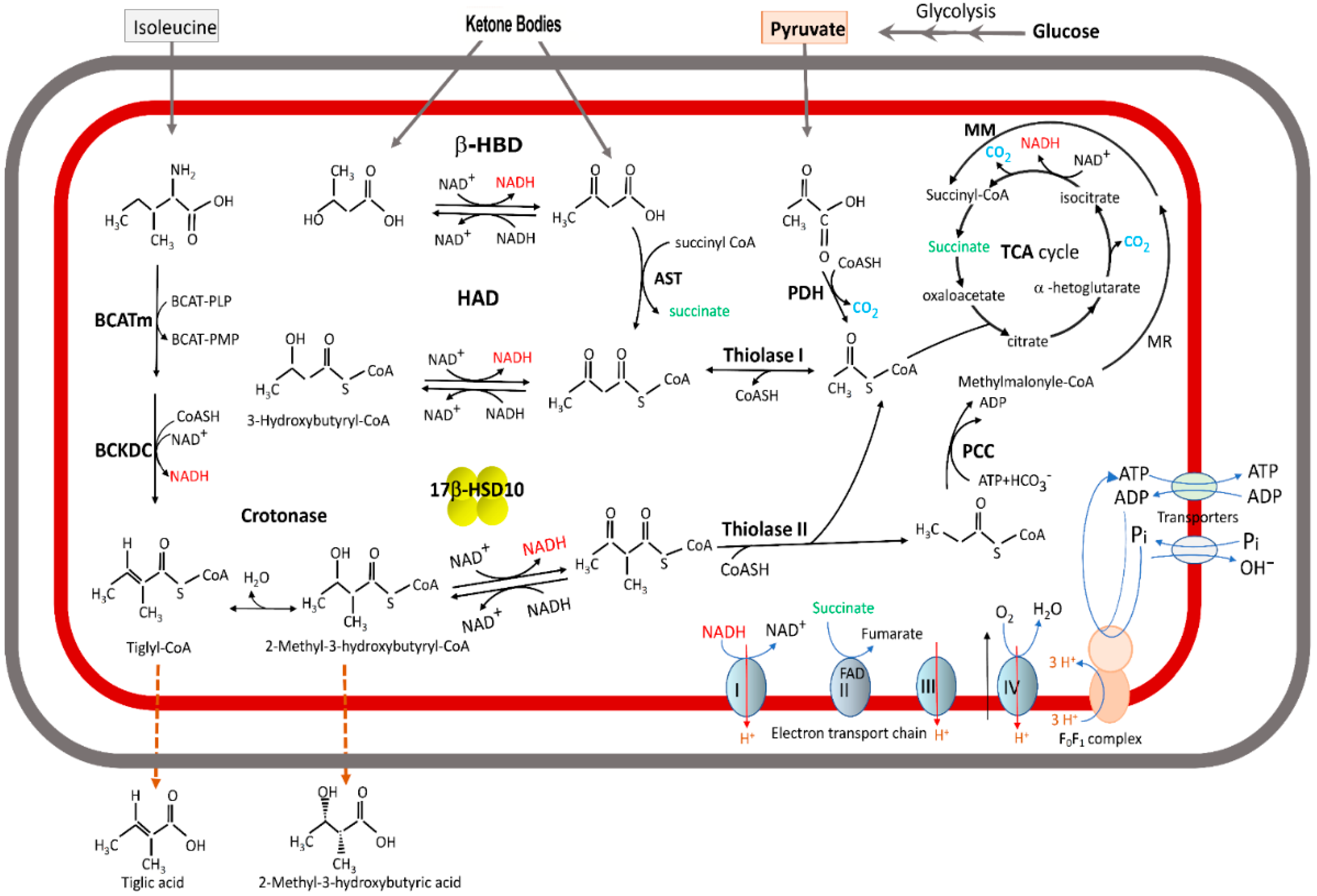
2.3. From 2-Methyl-3-Hydroxybutryl-CoA Dehydrogenase (MHBD) to 17β-HSD10
2.4. Short-Chain/Medium-Chain 3-Hydroxyacyl-CoA Dehydrogenase Deficiency Is Distinct from MHBD/HADHII/HSD10 Deficiency
2.5. Aβ Binds 17β-HSD10 to Inhibit Its Enzyme Activity
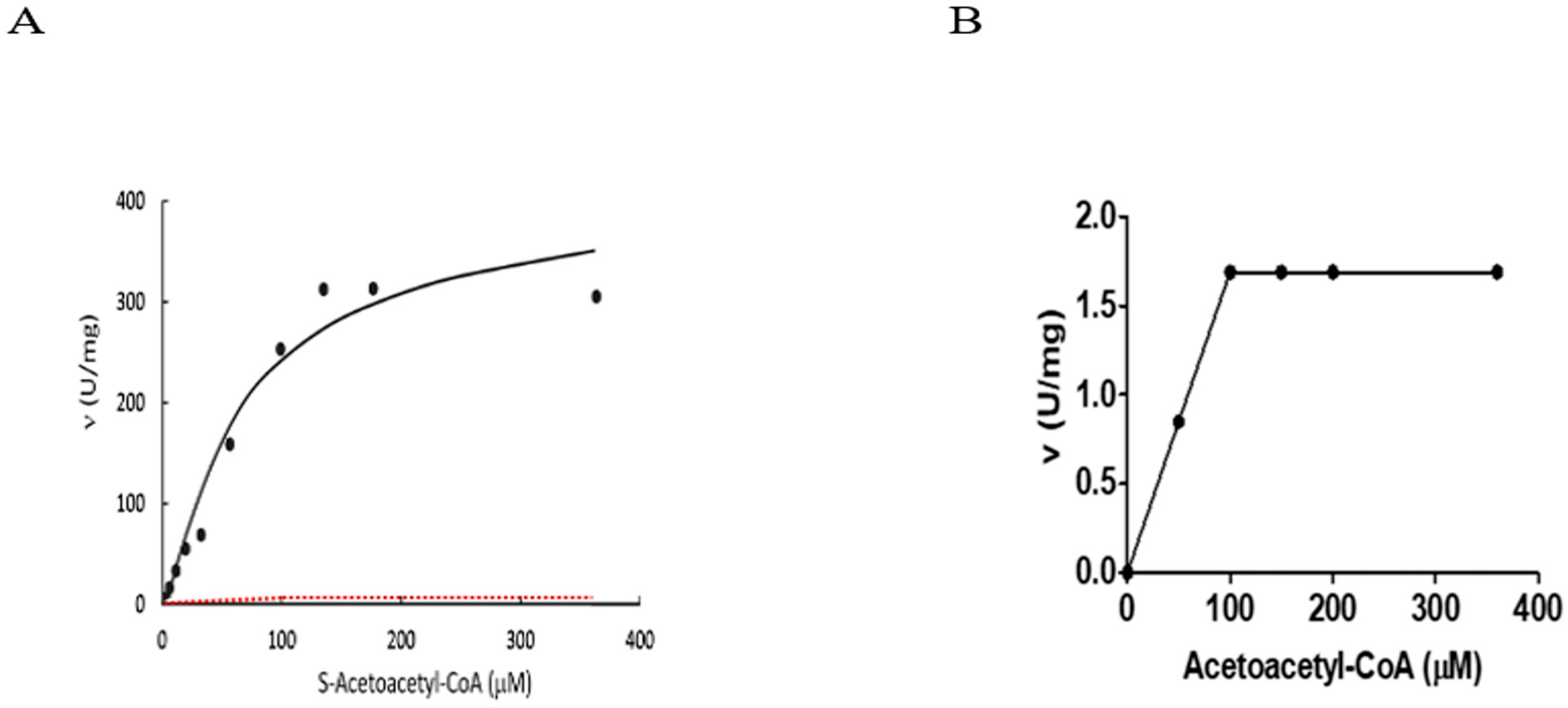
3. Kinetic Constants of ABAD/ERAB Not Derived from Experiments
‘The assay for reduction of S-acetoacetyl-CoA employed ERAB/HADH II (333 ng/mL), a range of S-acetoacetyl-CoA concentrations (0.0015–0.36 mM; Sigma, St. Louis MO, USA), and NADH (0.1 mM; Sigma) in 97 mM potassium phosphate (pH 7.3). The reaction was run for a total of 2 h at 25 °C under steady-state conditions (34)†, and the change in NADH absorbance at 340 nm was determined every 5 min.’
‘Alcohol dehydrogenase assays employed ERAB/HADH II (20 μg/mL), a range of alcohol substrates and concentrations (methanol, ethanol, n-propanol, isopropanol, n-butanol, isobutanol, n-pentanol, (±)-2-octanol, (+)-2-octanol, (−)-2-octanol, and n-decanol; Sigma), and NAD+ (7.5 mM) in 22 mM sodium pyrophosphate, 0.3 mM sodium phosphate (pH 8.8). The reaction was run for 2 h at 25 °C, and the absorbance at 340 nm was monitored every 5 min as described above.’
‘The alcohol dehydrogenase (ADH) activity of human short-chain L-3-hydroxyacyl-CoA dehydrogenase (SCHAD) has been characterized kinetically. The k(cat) of the purified enzyme was estimated to be 2.2 min−1, with apparent K(m) values of 280 mM and 22 mM for 2-propanol and NAD+, respectively. The kcat of the ADH activity was three orders of magnitude less than the L3-hydroxyacyl-CoA dehydrogenase activity but was comparable with that of the enzyme’s hydroxysteroid dehydrogenase (HSD) activity for oxidizing 17beta-oestradiol [He, Merz, Mehta, Schulz and Yang (1999) J. Biol. Chem. 274, 15014-15019]. However, the kcat values of intrinsic ADH and HSD activities of human SCHAD were found to be two orders of magnitude less than those reported for endoplasmic-reticulum-associated amyloid beta-peptide-binding protein (ERAB) [Yan, Shi, Zhu, Fu, Zhu, Zhu, et al. (1999) J. Biol. Chem. 274, 2145-2156].Since human SCHAD and ERAB apparently possess identical amino acid sequences, their catalytic properties should be identical. The recombinant SCHAD has been confirmed to be the right gene product and not a mutant variant. Steady-state kinetic measurements and quantitative analyses reveal that assay conditions such as pH and concentrations of coenzyme and substrate do not account for the kinetic differences reported for ERAB and SCHAD. Rather problematic experimental procedures appear to be responsible for the unrealistically high catalytic rate constants of ERAB. Eliminating the confusion surrounding the catalytic properties of this important multifunctional enzyme paves the way for exploring its role(s) in the pathogenesis of Alzheimer’s disease.’
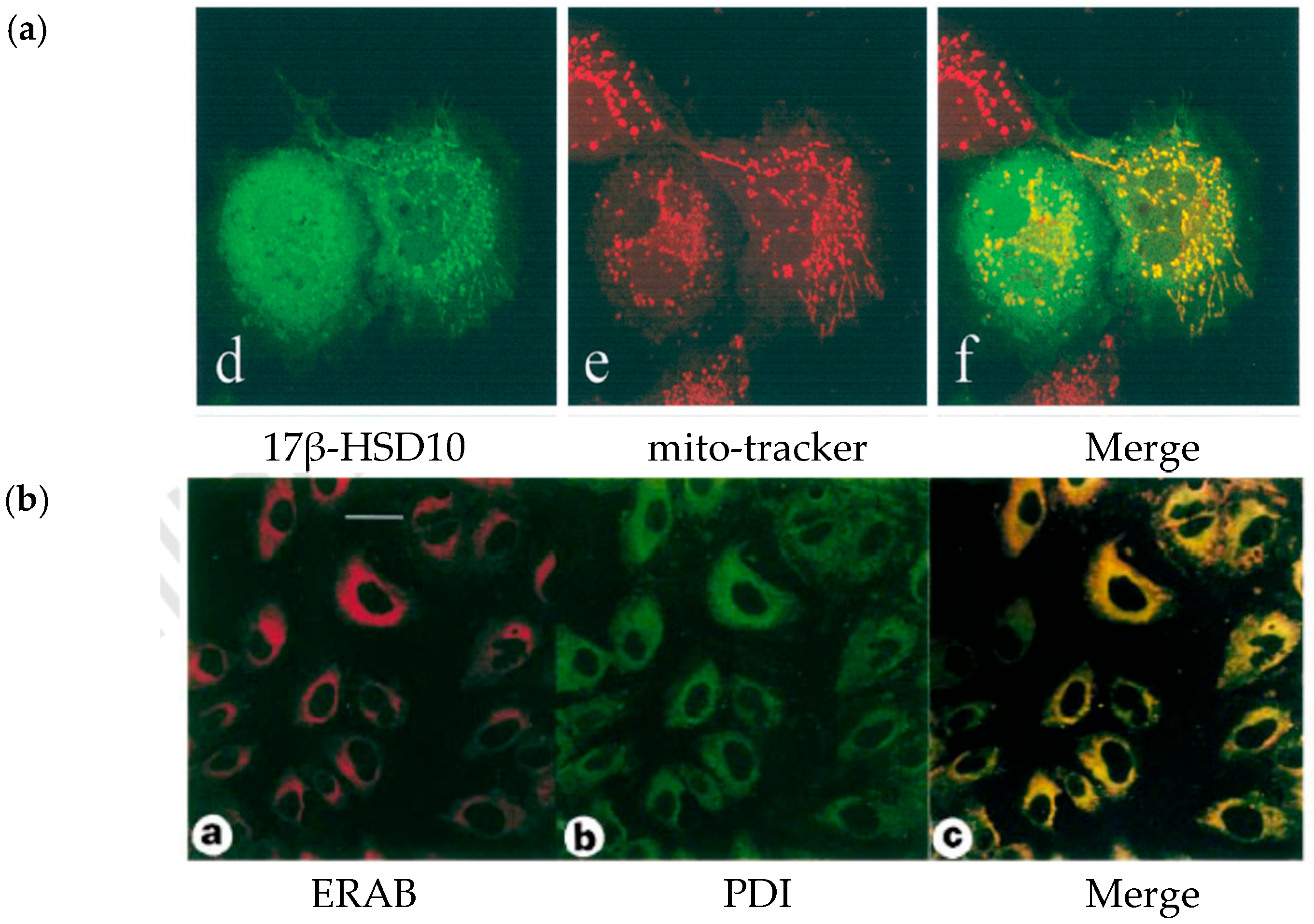
4. Re-Discovery of ABAD/ERAB in Mitochondria
5. Can Competitive Inhibition Be Defined by a Single Concentration of Substrate?
6. What Is the Scientific Basis to Designate a So-Called “Aβ-Binding Alcohol Dehydrogenase”?
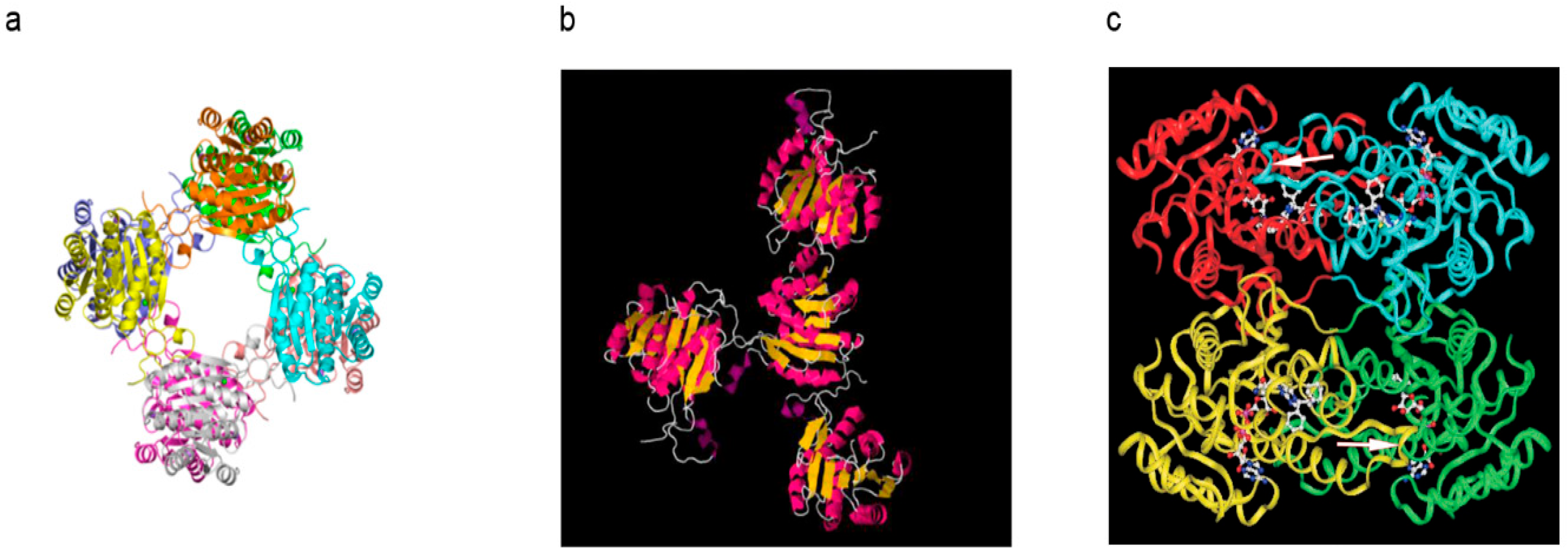
6.1. How Does the Aβ-Binding Alter the 3D Structure of 17β-HSD10?
6.2. Reported Alcohol Dehydrogenase Activity Data of ABAD Are Non-Reproducible
7. Roles of 17β-HSD10 in Neurosteroidogenesis
8. Mitochondrial 17β-HSD10 Associated with ER by a “Modified” Cell Fractionation
9. Neurodegeneration Results from Qualitative or Quantitative Alteration of 17β-HSD10
10. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Glossary
| ABAD | Aβ-binding protein alcohol dehydrogenase |
| AD | Alzheimer’s disease |
| ADH | Alcohol dehydrogenase |
| BCAT | Branched-chain amino-transferase |
| BCKDC | Branched-chain alpha keto acid dehydrogenase |
| COX | Cytochrome C oxidase |
| ERAB | Endoplasmic reticulum-associated Aβ-binding protein |
| HAD | l-3-hydroxyacyl dehydrogenase |
| HADH2 | 3-hydroxyacyl-CoA dehydrogenase type 2 |
| HBD | Hydroxybutyric acid dehydrogenase |
| 17β-HSD10 | 17β-hydroxysteroid dehydrogenase type 10 |
| MHBD | Methylhydroxybutyryl-CoA dehydrogenase |
| MRPP2 | Mitochondrial ribonuclease P protein 2 |
| OMIM | Online Mandelian Inheritance in Man (see NIH website) |
| PDI | Protein disulfide isomerase |
| SCHAD | Short-chain 3-hydroxyacyl-CoA dehydrogenase |
| SDR5C1 | Short-chain dehydrogenase/reductase 5C1 |
| PD | Parkinson’s disease |
| TCA | Tricarboxylic acid |
| VDAC | Voltage-dependent anion channel |
References
- He, X.Y.; Frackowiak, J.; Dobkin, C.; Brown, W.T.; Yang, S.Y. Involvement of type 10 17beta-hydroxysteroid dehydrogenase in the pathogenesis of infantile neurodegeneration and Alzheimer’s disease. Int. J. Mol. Sci. 2023, 24, 17604. [Google Scholar] [CrossRef]
- He, X.Y.; Schulz, H.; Yang, S.Y. A human brain L-3-hydroxyacyl coenzyme Adehydrogenase is identical with an amyloid (-peptide binding protein involved in Alzheimer’s disease. J. Biol. Chem. 1998, 273, 10741–10746. [Google Scholar] [CrossRef]
- He, X.Y.; Merz, G.; Mehta, P.; Schulz, H.; Yang, S.Y. Human brain short chain L-3-hydro-xyacyl coenzyme A dehydrogenase is a single-domain multifunctional enzyme. Characterization of a novel 17(-hydroxysteroid dehydrogenase. J. Biol. Chem. 1999, 274, 15014–15019. [Google Scholar] [CrossRef]
- Hiltunen, J.K.; Kastaniotis, A.J.; Autio, K.J.; Jiang, G.; Chen, Z.; Glumoff, T. 17β-Hydroxysteroid dehydrogenases as acyl thioester metabolizing enzymes. Mol. Cell Endocrinol. 2019, 489, 107–118. [Google Scholar] [CrossRef]
- Adamski, J.; Jacob, F.J. A guide to 17β-hydroxysteroid dehydrogenase. Mol. Cell Endocrinol. 2001, 171, 1–4. [Google Scholar] [CrossRef] [PubMed]
- He, X.Y.; Dobkin, C.; Brown, W.T.; Yang, S.Y. 3-Hydroxy-CoA dehydrogenase activities of mitochondrial type 10 17beta-hydroxysteroid dehydrogenase in neurodegeneration study. J. Alzheimer’s Dis. 2022, 88, 1487–1497. [Google Scholar] [CrossRef]
- Kissinger, C.R.; Rejto, P.A.; Pelletier, L.A.; Thomson, J.A.; Showalter, R.E.; Abreo, M.A.; Agree, C.S.; Margosiak, S.; Meng, J.J.; Aust, R.M.; et al. Crystal structure of human ABAD/HSD10 with a bound inhibitor: Implications for design of Alzheimer’s disease therapeutics. J. Mol. Biol. 2004, 342, 943–952. [Google Scholar] [CrossRef]
- Wohlfarter, Y.; Eidelpes, R.; Yu, R.D.; Sailer, S.; Koch, J.; Karall, D.; Scholl-Bürgi, S.; Amberger, A.; Hillen, H.S.; Zschocke, J.; et al. Lost in promiscuity? An evolutionary and biochemical evaluation of HSD10 function in cardiolipin metabolism. Cell Mol. Life Sci. 2022, 79, 562. [Google Scholar] [CrossRef]
- He, X.Y.; Dobkin, C.; Brown, W.T.; Yang, S.Y. Infantile neurodegeneration results from mutants of 17β-hydroxysteroid dehydrogenase type 10 rather than Aβ-binding alcohol dehydrogenase. Int. J. Mol. Sci. 2023, 24, 8487. [Google Scholar] [CrossRef]
- Yang, S.-Y.; He, X.-Y.; Olpin, S.E.; Sutton, V.R.; McMenamin, J.; Philipp, M.; Denman, R.B.; Malik, M. Mental retardation linked to mutations in the HSD17B10 gene interfering with neuro-steroid and isoleucine metabolism. Proc. Natl. Acad. Sci. USA 2009, 106, 14820–14824. [Google Scholar] [CrossRef]
- He, X.Y.; Yang, Y.Z.; Schulz, H.; Yang, S.Y. Intrinsic alcohol dehydrogenase and hydroxysteroid dehydrogenase activities of human mitochondrial short chain L-3-hydroxyacyl-CoA dehydrogenase. Biochem. J. 2000, 345, 139–143. [Google Scholar] [CrossRef]
- He, X.Y.; Merz, G.; Yang, Y.Z.; Schulz, H.; Yang, S.Y. Characterization localization of human type 10 17(-hydroxysteroid dehydrogenase. Eur. J. Biochem. 2001, 268, 4899–4907. [Google Scholar] [CrossRef]
- He, X.Y.; Yang, Y.Z.; Peehl, D.M.; Lauderdale, A.; Schulz, H.; Yang, S.Y. Oxidative 3α-hydroxysteroid dehydrogenase activity of human type 10 17β-hydroxysteroid dehydrogenase. J. Steroid Biochem. Mol. Biol. 2003, 87, 191–198. [Google Scholar] [CrossRef]
- BPT. Biochemistry: Mental retardation linked to mutations in the HSD17B10 gene. Proc. Nat. Acad. Sci. USA 2009, 106, 14735–14736. [Google Scholar]
- Melcangi, R.C.; Panzica, G.C. Allopregnanolone: State of the art. Prog. Neurobiol. 2014, 113, 1–5. [Google Scholar] [CrossRef]
- Melcangi, R.C. Roles of neuroactive steroids in health and disease. Biomolecules 2024, 14, 941. [Google Scholar] [CrossRef] [PubMed]
- Tsachaki, M.; Odermatt, A. Subcellular localization and membrane topology of 17β-hydroxysteroid dehydrogenases. Mol. Cell. Endocrinol. 2020, 489, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Amar, D.; Gay, N.R.; Jamenez-Morales, D.; Beltran, P.M.J.; Ramaker, M.; Zhao, B.; Sun, Y.; Marwaha, S.; Gaul, D.A.; Hershman, S.G.; et al. The mitochondrial multi-omic response to exercise training across rat tissues. Cell Metab. 2024, 36, 1411–1429. [Google Scholar] [CrossRef] [PubMed]
- MoTrPAC Stddy Group. Lead Analysts Temporal dynamics of the multi-omic response to endurance exercise training. Nature 2024, 629, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Roussel, E.; Drolet, M.C.; Lavigne, A.M.; Arsenault, M.; Couet, J. Multiple short-chain dehydrogenases/reductases are regulated in pathological cardiac hypertrophy. FEBS Open Bio 2018, 8, 1624–1635. [Google Scholar] [CrossRef]
- Yang, S.Y.; He, X.Y.; Schulz, H. Multiple functions of type 10 17beta-hydroxysteroid dehydrogenase. Trends Endocrinol. Metab. 2005, 16, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Boynton, T.O.; Shimkets, L.J. Myxococcus CsgA, Drosophila Sniffer, and human HSD10 are cardiolipin phospholipases. Genes. Dev. 2015, 29, 1903–1914. [Google Scholar] [CrossRef]
- Holzmann, J.; Frank, P.; Löffler, E.; Bennett, K.L.; Gerner, C.; Rossmanith, W. RNase P without RNA: Identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell 2008, 135, 462–474. [Google Scholar] [CrossRef]
- Bhatta, A.; Dienemann, C.; Cramer, P.; Hillen, H.S. Structural basis of RNA processing by human mitochondrial RNase P. Nat. Struct. Mol. Biol. 2021, 28, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Rossmanith, W.; Glege, P.; Hartmann, R.K. Discovery, mechanisms, and evolution of protein-only RNase P enzymes. J. Biol. Chem. 2024, 300, 105731. [Google Scholar] [CrossRef]
- Oerum, S.; Roovers, M.; Rambo, R.P.; Kopec, J.; Bailey, H.J.; Fitzpatrick, F.; Newman, J.A.; Newman, W.G.; Amberger, A.; Zschocke, J.; et al. Structural insight into the human mitochondrial tRNA purine N1-methyltransferase and ribonuclease P complexs. J. Biol. Chem. 2018, 293, 12862–12876. [Google Scholar] [CrossRef] [PubMed]
- Reinhard, L.; Sridhara, S.; Hällberg, B.M. The MRPP1/MRPP2 complex is a tRNA-maturation platform in human mitochondria. Nucleic Acids Res. 2017, 45, 12469–12480. [Google Scholar] [CrossRef]
- Oerum, S.; Roovers, M.; Leichsenring, M.; Acquaviva-Bourdain, C.; Beermann, F.; Gemperle-Britschgi, C.; Fouilhoux, A.; Korwitz-Reichelt, A.; Bailey, H.J.; Droogmans, L.; et al. Novel patient missense mutations in the HSD17B10 gene affect dehydrogenase and mitochondrial tRNA modification functions of the encoded protein. Biochim. Biophys. Acta 2017, 1863, 3294–3302. [Google Scholar] [CrossRef] [PubMed]
- Falk, M.J.; Gai, X.; Shigematsu, M.; Vilardo, E.; Takase, R.; McCormick, E.; Christian, T.; Place, E.; Pierce, E.A.; Consugar, M.; et al. A novel HSD17B10 mutation impairing the activities of the mitochondrial RNase P complex causes X-linked intractable epilepsy and neurodevelopmental regression. RNA Biol. 2016, 13, 477–485. [Google Scholar] [CrossRef]
- Vilardo, E.; Rossmanith, W. Molecular insights into HSD10 disease: Impact of SDR5C1 mutations on the human mitochondrial RNase P complex. Nucleic Acids Res. 2015, 43, 5112–5119. [Google Scholar] [CrossRef]
- Ofman, R.; Ruiter, J.P.N.; Feenstra, M.; Dura, N.; Poll-The, B.T.; Zschocke, J.; Ensenauer, R.; Lehnert, W.; Sass, J.O.; Sperl, W.; et al. 2-Methyl-3-hydroxybutyryl-CoA dehydrogenase deficiency is caused by mutations in the HADH2 gene. Am. J. Hum. Genet. 2003, 72, 1300–1307. [Google Scholar] [CrossRef]
- Ensenauer, R.; Niederhoff, H.; Ruiter, J.P.N.; Wanders, R.J.A.; Schwab, K.O.; Brandis, M.; Lehnert, W. Clinical variability in 3-hydroxy-2-methylbutyryl-CoA dehydrogenase deficiency. Ann. Neurol. 2002, 51, 477–482. [Google Scholar] [CrossRef]
- Rauschenberger, K.; Schöler, K.; Sass, J.O.; Sauer, S.; Djuric, Z.; Rumig, C.; Wolf, N.I.; Okun, J.G.; Kölker, S.; Schwarz, H.; et al. A non-enzymatic function of 17beta-hydroxysteroid dehydrogenase type 10 is required for mitochondrial integrity and cell survival. EMBO Mol. Med. 2010, 2, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Shafik, A.M.; Zhou, H.; Lim, J.; Dickinson, B.; Jin, P. Dysregulated mitochondrial and cytosolic tRNA m1A methylation in Alzheimer’s disease. Hum. Mol. Genet. 2022, 31, 1673–1680. [Google Scholar] [CrossRef]
- Kanavin, O.J.; Woldseth, B.; Jellum, E.; Tvedt, B.; Andersen, B.S.; Stromme, P. 2-methylbutyryl-CoA dehydrogenase deficiency associated with autism and mental retardation: A case report. J. Med. Case Rep. 2007, 1, 98. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Ouyang, W.; Yang, H.; Li, S. A novel c.59 C > T variant of the HSD17B10 gene as a possible cause of the neonatal form of HSD10 mitochondrial disease with hepatic dysfunction: A case report and review of the literature. Orphanet J. Rare Dis. 2025, 7, 108. [Google Scholar] [CrossRef] [PubMed]
- Akagawa, S.; Fukao, T.; Akagawa, Y.; Sasai, H.; Kohdera, U.; Kino, M.; Shigematsu, Y.; Aoyama, Y.; Kaneko, K. Japanese male siblings with 2-methyl-3-hydroxybutyryl-CoA dehydrogenase deficiency (HSD10 disease) without neurological regression. JIMD Rep. 2017, 32, 81–85. [Google Scholar]
- Upadia, J.; Walano, N.; Noh, G.S.; Liu, J.; Li, Y.; Deputy, S.; Elliott, L.T.; Wong, J.; Lee, J.A.; Caylor, R.C.; et al. HSD10 disease in a female: A case report and review of literature. JIMD Rep. 2021, 62, 35–43. [Google Scholar] [CrossRef]
- Su, L.; Li, X.; Lin, R.; Sheng, H.; Feng, Z.; Liu, L. Clinical and molecular analysis of 6 Chinese patients with isoleucine metabolism defects: Identification of 3 novel mutations in the HSD17B10 and ACAT1 gene. Metab. Brain Dis. 2017, 32, 2063–2071. [Google Scholar] [CrossRef]
- Richardson, A.; Berry, G.T.; Garganta, C.; Abbott, M.A. Hydroxysteroid 17-Beta Dehydrogenase Type 10 Disease in Siblings. JIMD Rep. 2017, 32, 25–32. [Google Scholar]
- García-Villoria, J.; Gort, L.; Madrigal, I.; Fons, C.; Fernández, C.; Navarro-Sastre, A.; Milà, M.; Briones, P.; García-Cazorla, A.; Campistol, J.; et al. X-inactivation of HSD17B10 revealed by cDNA analysis in two female patients with 17beta-hydroxysteroid dehydrogenase 10 deficiency. Eur. J. Hum. Genet. 2010, 18, 1353–1355. [Google Scholar] [CrossRef][Green Version]
- Hochberg, I.; Demain, L.A.; Richer, J.; Thompson, K.; Urquhart, J.E.; Rea, A.; Pagarkar, W.; Rodríguez-Palmero, A.; Schlüter, A.; Verdura, E.; et al. Bi-allelic variants in the mitochondrial RNase P subunit PRORP cause mitochondrial tRNA processing defects and pleiotropic multisystem presentations. Am. J. Hum. Genet. 2021, 108, 2195–2204. [Google Scholar] [CrossRef]
- Waters, P.J.; Lace, B.; Buhas, D.; Gravel, S.; Cyr, D.; Boucher, R.M.; Bernard, G.; Lévesque, S.; Maranda, B. HSD10 mitochondrial disease: P.Leu122Val variant, mild clinical phenotype, and founder effect in French-Canadian patients from Quebec. Mol. Genet. Genom. Med. 2019, 7, e1000. [Google Scholar] [CrossRef] [PubMed]
- Zschocke, J. Clinical consequences of mutations in the HSD17B10 gene. J. Inherit. Metab. Dis. 2012, 35, 81–89. [Google Scholar] [CrossRef]
- Seaver, L.H.; He, X.-Y.; Abe, K.; Cowan, T.; Enns, G.M.; Sweetman, L.; Philipp, M.; Lee, S.; Malik, M.; Yang, S.-Y.; et al. A novel mutation in the HSD17B10 gene of a 10-year-old boy with refractory epilepsy, choreoathetosis and learning disability. PLoS ONE 2011, 6, e27348. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sass, J.O.; Forstner, R.; Sperl, W. 2-Methyl-3-hydroxybutyryl-CoA dehydrogenase deficiency: Impaired catabolism of isoleucine presenting as neurodegenerative disease. Brain Dev. 2004, 26, 12–14. [Google Scholar] [CrossRef] [PubMed]
- Perez-Cerda, C.; García-Villoria, J.; Ofman, R.; Sala, P.R.; Merinero, B.; Ramos, J.; García-Silva, M.T.; Beseler, B.; Dalmau, J.; Wanders, R.J.; et al. 2-Methyl-3-hydroxybutyryl-CoA dehydrogenase (MHBD) deficiency: An X-linked inborn error of isoleucine metabolism that may mimic a mitochondrial disease. Pediatr. Res. 2005, 58, 488–491. [Google Scholar] [CrossRef]
- García-Villoria, J.; Navarro-Sastre, A.; Fons, C.; Pérez-Cerdá, C.; Baldellou, A.; Fuentes-Castelló, M.Á.; González, I.; Hernández-Gonzalez, A.; Fernández, C.; Campistol, J.; et al. A study of patients and carriers with 2-methyl-3-hydroxybutyryl-CoA dehydrogenase (MHBD) deficiency: Difficulties in the diagnosis. Clin. Biochem. 2009, 42, 27–33. [Google Scholar] [CrossRef]
- Ciki, K.; Alavanda, C.; Kara, M. Novel Mutation in the HSD17B10 Gene Accompanied by Dysmorphic Findings in Female Patients. Mol. Syndromol. 2024, 15, 211–216. [Google Scholar] [CrossRef]
- He, X.Y.; Isaacs, C.E.; Yang, S.Y. Roles of 17β-hydroxysteroid dehydrogenase type 10 in Alzheimer’s disease. J. Alz. Dis. 2018, 62, 665–673. [Google Scholar] [CrossRef]
- Yan, S.D.; Fu, J.; Soto, C.; Chen, X.; Zhu, H.; Al-Mohanna, F.; Collison, K.; Zhu, A.; Stern, E.; Saido, T.; et al. An intracellular protein that binds amyloid-beta peptide and mediates neurotoxicity in Alzheimer’s disease. Nature 1997, 389, 689–695. [Google Scholar] [CrossRef]
- Deyreuther, K.; Marsters, C.L. The ins and outs of amyloid-β. Nature 1997, 389, 677–678. [Google Scholar] [CrossRef] [PubMed]
- Du Yan, S.; Shi, Y.; Zhu, A.; Fu, J.; Zhu, H.; Zhu, Y.; Gibson, L.; Stern, E.; Collison, K.; Al-Mohanna, F.; et al. Role of ERAB/L-3-hydroxyacyl-coenzyme A dehydrogenase type II activity in Abeta-induced cytotoxicity. J. Biol. Chem. 1999, 274, 2145–2156. [Google Scholar] [CrossRef] [PubMed]
- Du Yan, S.; Zhu, Y.; Stern, E.D.; Hwang, Y.C.; Hori, O.; Ogawa, S.; Frosch, M.P.; Connolly, E.S.; McTaggert, R.; Pinsky, D.J.; et al. Amyloid β-peptide-binding alcohol dehydrogenase is a component of the cellular response to nutritional stress. J. Biol. Chem. 2000, 275, 27100–27109. [Google Scholar] [CrossRef]
- Sambamurti, K.; Lahiri, D.K. ERAB contains a putative non-cleavable signal peptide. Biochim. Biophys. Res. Commun. 1998, 249, 546–549. [Google Scholar] [CrossRef]
- Morsy, A.; Trippier, P.C. Amyloid-Binding Alcohol Dehydrogenase (ABAD) Inhibitors for the Treatment of Alzheimer’s Disease. J. Med. Chem. 2019, 62, 4252–4264. [Google Scholar] [CrossRef]
- Ye, Z.; Liu, Y.; Jin, X.; Wu, Y.; Zhao, H.; Gao, T.; Deng, Q.; Cheng, J.; Lin, J.; Tong, Z. Aβ—binding with alcohol dehydrogenase drives Alzheimer’s pathogenesis: A review. Int. J. Biol. Macromol. 2024, 264, 130580. [Google Scholar] [CrossRef]
- Lustbader, J.W.; Cirilli, M.; Lin, C.; Xu, H.W.; Takuma, K.; Wang, N.; Caspersen, C.; Chen, X.; Pollak, S.; Chaney, M.; et al. ABAD directly links Abeta to mitochondrial toxicity in Alzheimer’s disease. Science 2004, 304, 448–452. [Google Scholar] [CrossRef]
- Xiao, X.; Chen, Q.; Zhu, X.; Wang, Y. ABAD/17beta-HSD10 reduction contributes to the protective mechanism of huperzine a on the cerebral mitochondrial function in APP/PS1 mice. Neurobiol. Aging 2019, 81, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Reyniers, E.; Van Bogaert, P.; Peeters, N.; Vits, L.; Pauly, F.; Fransen, E.; Van Regemorter, N.; Kooy, R.F. A new neurological syndrome with mental retardation, choreoathetosis, and abnormal behavior maps to chromosome Xp11. Am. J. Hum. Genet. 1999, 65, 1406–1412. [Google Scholar] [CrossRef]
- Lenski, C.; Kooy, R.F.; Reyniers, E.; Loessner, D.; Wanders, R.J.; Winnepenninckx, B.; Hellebrand, H.; Engert, S.; Schwartz, C.E.; Meindl, A.; et al. The reduced expression of the HADH2 protein causes X-linked mental retardation, choreoathetosis, and abnormal behavior. Am. J. Hum. Genet. 2007, 80, 372–377. [Google Scholar] [CrossRef] [PubMed]
- Tieu, K.; Perier, C.; Vila, M.; Caspersen, C.; Zhang, H.P.; Teismann, P.; Jackson-Lewis, V.; Stern, D.M.; Yan, S.D.; Przedborski, S. L-3-hydroxyacyl-CoA dehydrogenase II protects a model of Parkinson’s disease. Ann. Neurol. 2004, 56, 51–60. [Google Scholar] [CrossRef]
- Bertolin, G.; Jacoupy, M.; Traver, S.; Ferrando-Miguel, R.; Georges, T.S.; Grenier, K.; Ardila-Osorio, H.; Muriel, M.-P.; Takahashi, H.; Lees, A.J.; et al. Parkin maintains mitochondrial levels of the protective Parkinson’s disease-related enzyme 17-β hydroxysteroid dehydrogenase type 10. Cell Death Differ. 2015, 22, 1563–1576. [Google Scholar] [CrossRef]
- Qi, T.; Chang, X.; Wang, W. Multi-omics pan-cancer profiling of HSD17B10 unveils its prognostic potential, metabolic regulation, and immune microenviroment interactions. Biology 2025, 14, 567. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhou, M.; Gao, X.; Xie, Y.; Xiao, J.; Liu, T.; Zeng, X. Estrogen-related genes for thyroid cancer prognosis, immune infiltration, staging, and drug sensitivity. BMC Cancer 2023, 23, 1048. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Xu, Y.; Xiong, H.H. Mechanism of salidroside promoting testosteronr secretion induced by H2O2 in TM3 leydig cells based on metabolomics and network pharmacology. Front. Chem. 2025, 13, 1544876. [Google Scholar] [CrossRef]
- Zuo, Q.; Yoo, J.Y.; Nelson, E.R.; Sikora, M.J.; Riggins, R.B.; Madak-Erdogan, Z. Co-targeting of metabolism using dietary and pharmacologic approaches reduces breast cancer metastatic burden. NPJ Breast Cancer 2025, 11, 3. [Google Scholar] [CrossRef]
- Zhu, L.; Liu, F.; Shi, C.; Su, X.; Tan, M.; Xie, S.; Yu, M.; Zou, S.; Tan, Y.; Xie, S.; et al. Hepatic micropeptide modulates 722 mitochondrial RNA processing machinery in hepatocellular carcinoma. Mol. Cell 2025, 85, 2303–2319. [Google Scholar] [CrossRef]
- He, X.Y.; Wen, G.Y.; Merz, G.; Lin, D.; Yang, Y.Z. Abundant short chain L-3-hydroxyacyl-CoA dehydrogenase/type 10 17beta-hydroxysteroid dehydrogenase in the hippocampus of mouse Alzheimer’s disease model. Mol. Brain Res. 2002, 99, 46–53. [Google Scholar] [CrossRef]
- Bian, Y.; Shi, J.; Chen, Z.; Fang, J.; Chen, W.; Zou, Y.; Yao, H.; Tu, J.; Liao, Y.; Xie, X.; et al. A diagnostic signature developed based on the necroptosis-related genes and its association with immune infiltration in osteosarcoma. Heliyon 2024, 10, e35719. [Google Scholar] [CrossRef]
- Wang, M.; Li, Q.; Wang, S.; Zuo, L.; Hai, Y.; Yuan, S.; Li, X.; Huang, X.; Yang, C.; Yao, L.; et al. Astragaloside IV protects renal tubular epithelial cells against oxidative stress-induced injury by upregulating CPT1A-mediated HSD17B10 lysine succinylation in diabetic kidney disease. Phytother. Res. 2024, 38, 4519–4540. [Google Scholar] [CrossRef]
- Oppermann, U.C.; Salim, S.; Tjernberg, L.O.; Terenius, L.; Jornvall, H. Binding of amyloid β-peptide to mitochondrial hydroxyacyl-CoA (ERAB): Regulation of an SDR enzyme activity with implication for apoptosis in Alzheimer’s disease. FEBS Lett. 1999, 451, 238–242. [Google Scholar] [CrossRef]
- He, X.Y.; Wegiel, J.; Yang, S.Y. Intracellular oxidation of allopregnanolone by human brain type 10 17beta-hydroxysteroid dehydrogenase. Brain Res. 2005, 1040, 29–35. [Google Scholar] [CrossRef]
- He, X.Y.; Yang, S.Y. Why so far the best microbiol converting glucose to long chain fatty acids is the, E. coli strain RB03? Nature 2011, 476. [Google Scholar] [CrossRef]
- Carlson, E.A.; Marquez, R.T.; Du, F.; Wang, Y.F.; Xu, L.; Yan, S.D. Overexpression of 17β-hydroxysteroid dehydrogenase type 10 increases pheochromocytoma cell growth and resistance to cell death. BMC Cancer 2015, 15, 166. [Google Scholar] [CrossRef][Green Version]
- Luo, M.J.; Mao, L.F.; Schulz, H. Short chain L-3-hydroxy-2-methylacyl-CoA dehydrogenase from rat liver: Purification and characterization of a novel enzyme of isoleucine metabolism. Arch. Biochem. Biophys. 1995, 321, 214–220. [Google Scholar] [CrossRef]
- Schulz, H. Beta oxidation of fatty acids. Biochim. Biophys. Acta 1991, 1081, 109–120. [Google Scholar] [CrossRef]
- Binstock, J.F.; Schulz, H. Fatty acid oxidation complex from escherichia coli. Methods Enzymol. 1981, 71, 403–411. [Google Scholar] [PubMed]
- Powell, A.J.; Read, J.A.; Banfield, M.J.; Gunn-Moore, F.; Yan, S.D.; Lustbader, J.; Stern, A.R.; Stern, D.M.; Brady, R.L. Recognition of structurally diverse substrates by type II 3-hydroxyacyl-CoA dehydrogenase (HADH II)/amyloid-beta binding alcohol dehydrogenase (ABAD). J. Mol. Biol. 2000, 303, 311–327. [Google Scholar] [CrossRef] [PubMed]
- Jörnvall, H.; Persson, M.; Jeffery, J. Alcohol polyol dehydrogenases are both divided into two protein types structural properties cross-relate the different enzyme activities within each type. Proc. Natl. Acad. Sci. USA 1981, 78, 4226–4230. [Google Scholar] [CrossRef] [PubMed]
- Beck, K.R.; Kaserer, T.; Schuster, D.; Odermatt, A. Virtual screening applications in short-chain dehydrogenase/reductase research. J. Steroid Biochem. Mol. Biol. 2017, 171, 157–177. [Google Scholar] [CrossRef]
- He, X.Y.; Yang, Y.X.; Yang, S.Y. Changes of the HSD17B10 gene expression levels in ulcerative colitis. Inflamm. Bowel Dis. 2013, 19, E23–E24. [Google Scholar] [CrossRef]
- Poppe, J.; Boesmans, L.; Vieira-Silva, S.; Deroover, L.; Tito, R.; Vandeputte, D.; Vandermeulen, G.; De Preter, V.; Raes, J.; Vermeire, S.; et al. Differential contributions of the gut microbiota and metabolome to pathomechanisms in ulcerative colitis: An in vitro analysis. Gut Microbes 2024, 16, 2424913. [Google Scholar] [CrossRef]
- Zhang, W.; Sang, Y.M. Genetic pathogenesis, diagnosis, and treatment of short-chain 3-hydroxyacyl-coenzyme A dehydrogenase hyperinsulinism. Orphanet J. Rare Dis. 2021, 16, 467. [Google Scholar] [CrossRef] [PubMed]
- Segel, I.H. Enzyme Kinetics, Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme System; Wiley-Interscience: New York, NY, USA, 1975. [Google Scholar]
- Cornish-Bowden, A. Analysis of Enzyme Kinetic Data; Oxford University Press: New York, NY, USA, 1995. [Google Scholar]
- Vangavaragu, J.R.; Valasani, K.R.; Fang, D.; Williams, T.D.; Yan, S.S. Determination of small molecule ABAD inhibitors crossing blood brain barrier and pharmacokinetics. J. Alzheimer’s Dis. 2014, 42, 333–344. [Google Scholar] [CrossRef]
- Wegiel, J.; Flory, M.; Kuchna, I.; Nowicki, K.; Wegiel, J.; Ma, S.Y.; Zhong, N.; Bobrowicz, T.W.; de Leon, M.; Lai, F.; et al. Developmental deficits and staging of dynamics of age associated Alzheimer’s disease neurodegeneration and neuronal loss in subjects with Down syndrome. Acta Neuropathol. Commun. 2022, 10, 2. [Google Scholar] [CrossRef]
- Iqbal, K. Tau and Alzheimer’s disease: Past, present and future. Cytoskeleton 2024, 81, 116–121. [Google Scholar] [CrossRef]
- Ayan, D.; Maltais, R.; Poirier, D. Identification of a 17β-hydroxysteroid dehydrogenase type 10 steroidal inhibitor: A tool to investigate the role of type 10 in Alzheimer’s disease and prostate cancer. ChemMedChem 2012, 7, 1181–1184. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.Y.; Dobkin, C.; He, X.Y.; Brown, W.T. Transcription start sites and epigenetic analysis of the HSD17B10 proximal promoter. BMC Biochem. 2013, 14, 17. [Google Scholar] [CrossRef] [PubMed]
- Kristofikova, Z.; Ricny, J.; Vyhnalek, M.; Hort, J.; Laczo, J.; Sirova, J.; Klaschka, J.; Ripova, D. Levels of 17β-Hydroxysteroid Dehydrogenase Type 10 in Cerebrospinal Fluid of People with Mild Cognitive Impairment and Various Types of Dementias. J. Alzheimer’s Dis. 2015, 48, 105–114. [Google Scholar] [CrossRef]
- Vinklarova, L.; Schmidt, M.; Benek, O.; Kuca, K.; Gunn-Moore, F.; Musilek, K. Friend or enemy? Review of 17beta-HSD10 and its role in human health or disease. J. Neurochem. 2020, 155, 231–249. [Google Scholar] [CrossRef] [PubMed]
- Labrie, F. All sex steroids are made intracellularly in peripheral tissues by mechanisms of intracrinology after menopause. J. Steroid Biochem. Mol. Biol. 2015, 145, 133–138. [Google Scholar] [CrossRef]
- Valaasani, K.R.; Sun, Q.; Hu, G.; Li, J.; Du, F.; Guo, Y.; Carlson, E.A.; Gan, X.; Yan, S.S.; Valasani, K.R. Identification of human ABAD inhibitors for rescuing Aβ-mediated mitochondrial dysfunction. Curr. Alzheimer Res. 2014, 11, 128–136. [Google Scholar] [CrossRef]
- Hroch, L.; Guest, P.; Benek, O.; Soukup, O.; Janockova, J.; Dolezal, R.; Kuca, K.; Aitken, L.; Smith, T.K.; Gunn-Moore, F.; et al. Synthesis and evaluation of frentizole-based indolyl thiourea analogues as MAO/ABAD inhibitors for AlzhePimer’s disease treatment. Bioorg. Med. Chem. 2017, 25, 1143–1152. [Google Scholar] [CrossRef]
- Metodieva, V.; Smith, T.; Gum-Moore, F. The mitochondrial enzyme 17β-HSD10 mediates ischemic and amyloid-βinduced stress in primary mouse astrocytes. eNeuro 2022, 9, 0040–22.2022. [Google Scholar] [CrossRef]
- Liu, L.; Chen, S.; Yu, M.; Ge, C.; Ren, M.; Liu, B.; Yang, X.; Christian, T.W.; Hou, Y.-M.; Zou, J.; et al. Deacetylation of HSD17B10 by SIRT3 regulates cell growth and cell resistance under oxidative and starvation stresses. Cell Death Dis. 2020, 11, 563. [Google Scholar] [CrossRef]
- Schmidt, M.; Benek, O.; Vinklarova, L.; Hrabinova, M.; Zemanova, L.; Chribek, M.; Kralova, V.; Hroch, L.; Dolezal, R.; Lycka, A.; et al. Benzothiazolyl ureas are low micromolar and uncompetitive inhibitors of 17β-HSD10 with implications to Alzheimer’s disease treatment. Int. J. Mol. Sci. 2020, 21, 2059. [Google Scholar] [CrossRef]
- Kuwabara, K.; Matsumoto, M.; Ikeda, J.; Hori, O.; Ogawa, S.; Maeda, Y.; Kitagawa, K.; Imuta, N.; Kinoshita, T.; Stern, D.M.; et al. Purification and characterization of a novel stress protein, the 150-kDa oxygen-regulated protein (ORP150), from cultured rat astrocytes and its expression in ischemic mouse brain. J. Biol. Chem. 1996, 271, 5025–5032. [Google Scholar] [CrossRef] [PubMed]
- Ortez, C.; Villar, C.; Fons, C.; Duarte, S.T.; Pérez, A.; García-Villoria, J.; Ribes, A.; Ormazábal, A.; Casado, M.; Campistol, J.; et al. Undetectable levels of CSF amyloid-beta peptide in a patient with 17β-hydroxysteroid dehydrogenase deficiency. J. Alzheimer’s Dis. 2011, 27, 253–257. [Google Scholar]

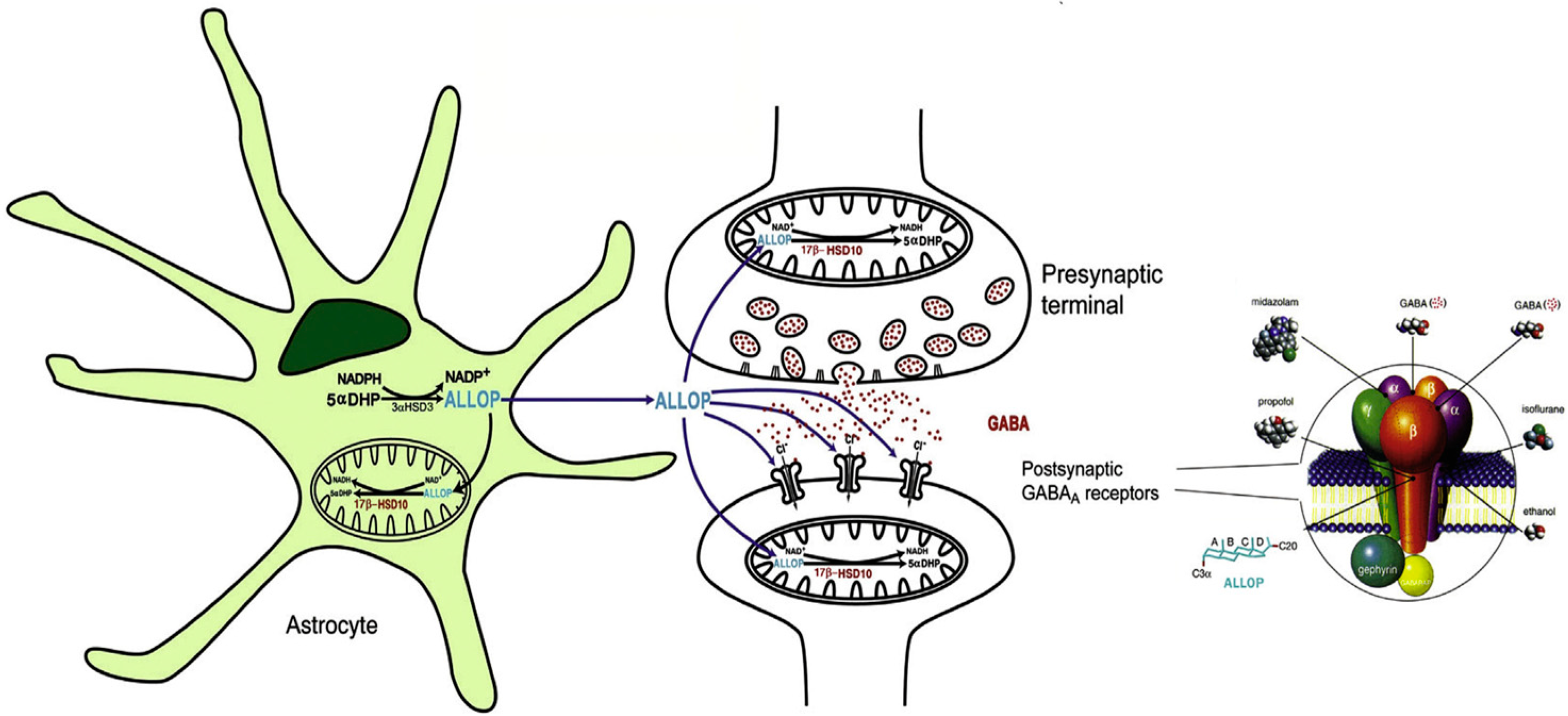
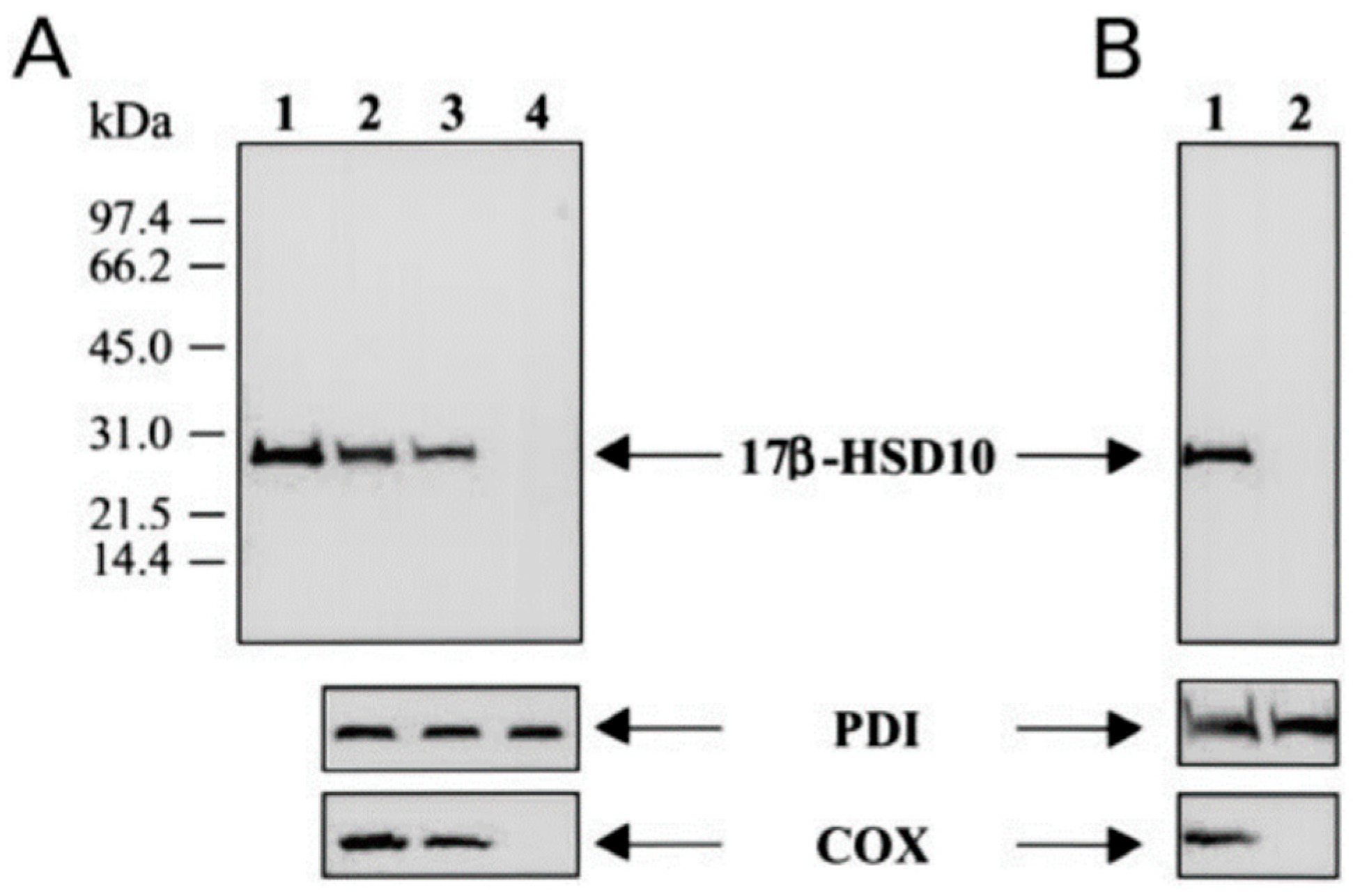
| Year | Accession Number | Name | Acronym | Comments | Ref. | |
|---|---|---|---|---|---|---|
| cDNA | Gene | |||||
| 1997–1998 | AF035555 AF037438 [11/21/97] [12/9/97] Deposited into the Genbank | Short-chain 3-hydroxyacyl-CoA dehydrogenase | SCHAD | MW = 108 kDa, composed of 1044 residues. Homotetrameric enzyme exhibited HAD and 17β-HSD activity proposed to reside in mitochondria | [2] | |
| U96132 n/a | Endoplasmic reticulum-associated Aβ-binding protein | ERAB | MW = 27 kDa, composed of 262 amino acid residues and associated with the endoplasmic reticulum | [51,52,53] | ||
| 1999 | Novel 17β-Hydroxysteroid dehydrogenase | Novel 17β-HSD | Mitochondrial, multifunctional protein inactivates 17β-estradiol to estrone | [3,12] | ||
| Amyloid β-peptide binding alcohol dehydrogenase | ABAD | Substitution of ABAD to ERAB but maintains association with ER and further conveys incongruous data of generalized alcohol dehydrogenase (C2-C10) activities | [54] | |||
| 2000 | AF035555 AF037438 | 2-methyl-3-hydroxyacyl-CoA dehydrogenase | MHBD | This term is appropriate especially for isoleucine metabolism | [31,32,46,47,48,76] | |
| 2001 | OMIM300256: 17β -Hydroxysteroid dehydrogenase X | Type 10 17β-Hydroxysteroid dehydrogenase | 17β-HSD10 | Involved in neurosteroid, such as allopregnanolone, metabolism, and identification of its N-terminal mitochondrial-targeting signal | [1,2,3,4,10,11,45] | |
| 2004 | Amyloid β-peptide binding alcohol dehydrogenase | ABAD | Renames ER-associated ABAD to be a mitochondrial ABAD but still ignores reported activities of 17β-HSD10 | [58] | ||
| 2007 | NM_004493, Gene symbol: HSD17B10 * | 3-Hydroxyacyl-CoA dehydro-genase type 2 | HADH2 | A silent mutation was found in MRXS10 ** patients | [61] | |
| 2008 | Mitochondrial RNase P protein 2 | MRPP2 | A component of the RNA-free RNase P complex | [23,24,25,26] | ||
| 2015 | Short-chain de- hydrogenase/re-ductase 5C1 | SDR5C1 | Shown in a short-chain dehydrogenase/reductase (SDR) evolution tree | [30] | ||
| Substrate or Alcohol | Km | Vmax | kcat a | Catalytic Efficiency kcat/Km |
|---|---|---|---|---|
| mM | Units/mg | S−1 | M−1 S−1 | |
| Reduction of S-acetoacetyl-CoA b | 0.068 ± 0.020 | 430 ± 45 | 190 | 2.8 × 106 |
| Oxidation of alcohol substrates b | ||||
| 17beta-Estradiol | 0.014 ± 0.006 | 23 ± 3 | 10 | 7.4 × 105 |
| Methanol | No activity | No activity | No activity | No activity |
| Ethanol | 1210 ± 260 | 2.2 ± 0.4 | 1.0 | 0.82 |
| Isopropanol | 150 ± 17 | 36 ± 2 | 16 | 110 |
| n-Propanol | 272 ± 62 | 4.2 ± 0.5 | 1.9 | 6.9 |
| n-Butanol | 53 ± 6 | 9.0 ± 0.3 | 4.0 | 76 |
| Isobutanol | 56 ± 16 | 8.0 ± 0.7 | 3.6 | 64 |
| n-Pentanol | 18 ± 5 | 6.9 ± 0.4 | 3.1 | 170 |
| (±)-2-Octanol | 85 ± 17 | 245 ± 20 | 110 | 1300 |
| (+)-2-Octanol | 84 ± 16 | 102 ± 8 | 46 | 540 |
| (−)-2-Octanol | 43 ± 9.0 | 133 ± 23 | 60 | 1400 |
| n-Decanol | 14 ± 6.3 | 2.8 ± 0.5 | 1.3 | 90 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, X.-Y.; Frackowiak, J.; Yang, S.-Y. Roles of Type 10 17β-Hydroxysteroid Dehydrogenase in Health and Disease. J. Pers. Med. 2025, 15, 346. https://doi.org/10.3390/jpm15080346
He X-Y, Frackowiak J, Yang S-Y. Roles of Type 10 17β-Hydroxysteroid Dehydrogenase in Health and Disease. Journal of Personalized Medicine. 2025; 15(8):346. https://doi.org/10.3390/jpm15080346
Chicago/Turabian StyleHe, Xue-Ying, Janusz Frackowiak, and Song-Yu Yang. 2025. "Roles of Type 10 17β-Hydroxysteroid Dehydrogenase in Health and Disease" Journal of Personalized Medicine 15, no. 8: 346. https://doi.org/10.3390/jpm15080346
APA StyleHe, X.-Y., Frackowiak, J., & Yang, S.-Y. (2025). Roles of Type 10 17β-Hydroxysteroid Dehydrogenase in Health and Disease. Journal of Personalized Medicine, 15(8), 346. https://doi.org/10.3390/jpm15080346






