Updated Insights into the Molecular Pathophysiology of Olfactory Neuroblastoma Using Multi-Omics Analysis
Abstract
1. Introduction
2. Pathogenesis
2.1. Neural Crest Origin
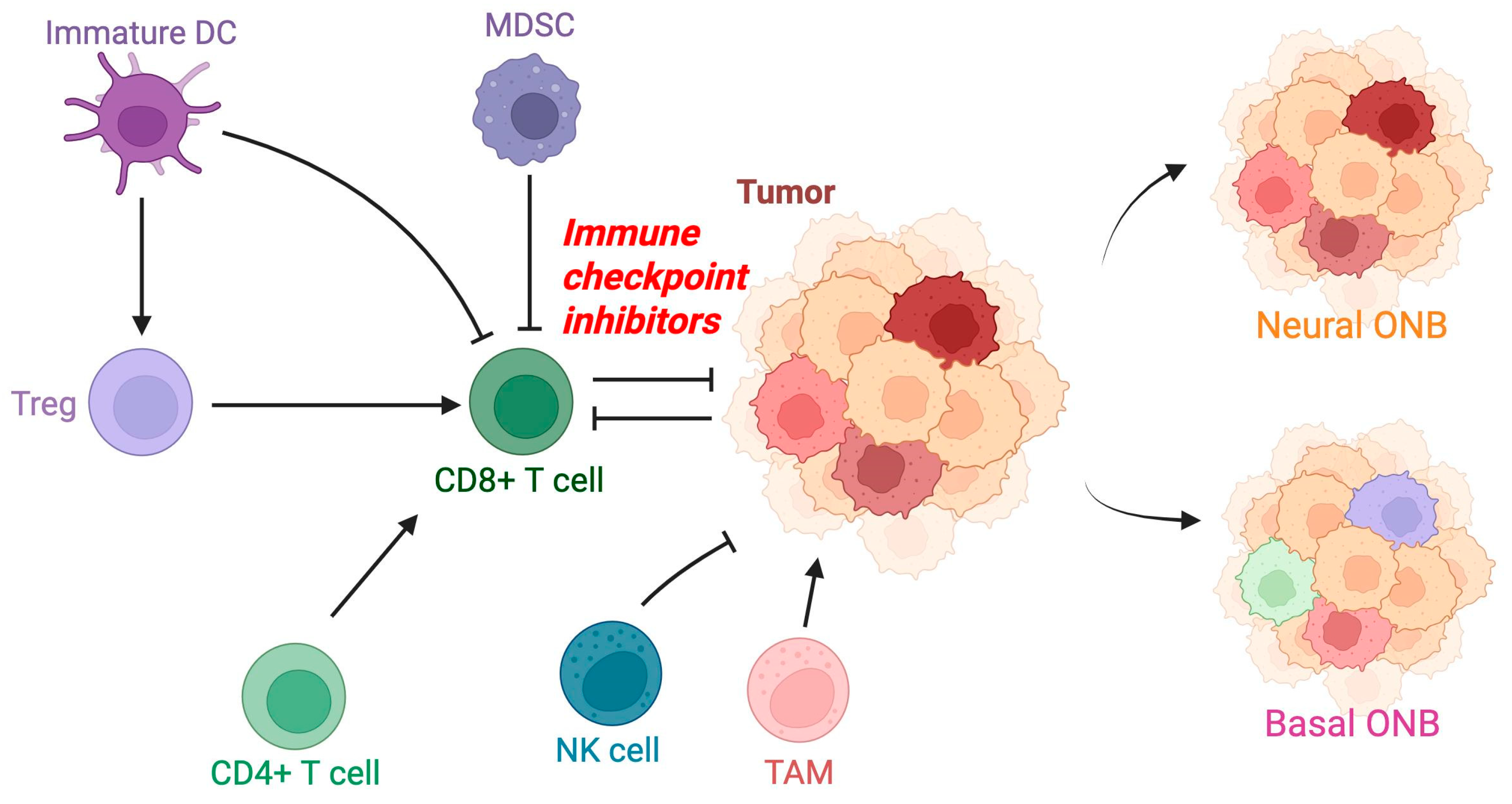
2.2. Tumor Microenvironment
2.3. Immune Cell Infiltration
2.4. Myeloid-Derived Suppressor Cells and Immune Suppression
2.5. Natural Killer Cells and Immune Evasion
2.6. Major Histocompatibility Complex Downregulation and Chemokine Signaling
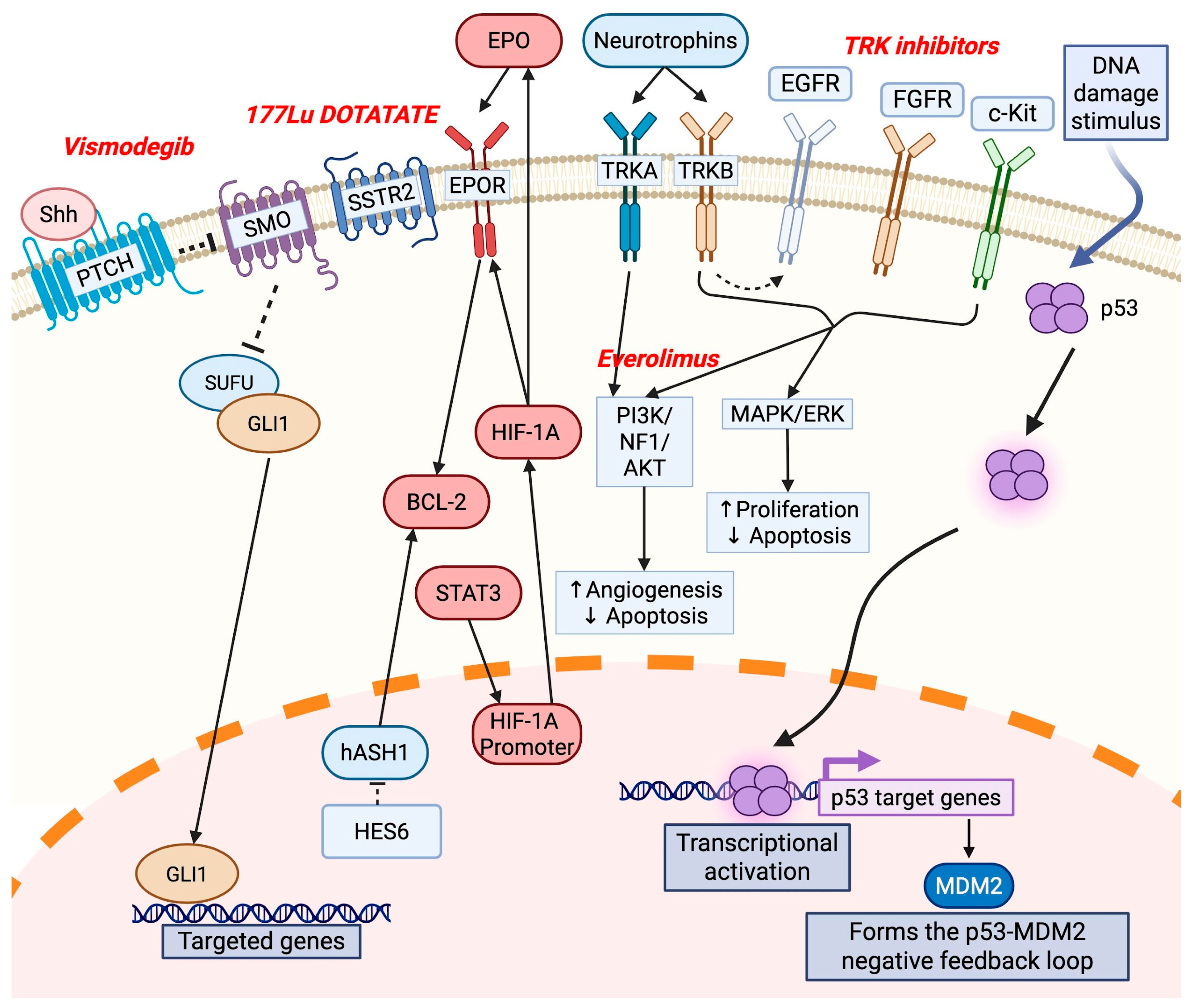
3. Molecular Factors and Structures
4. Genomic Profiling of ONB
5. Genomic Subclassification of ONB
6. Current Management, Knowledge Gaps, and Implications for Future Management
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MDSC | Myeloid-derived suppressor cells |
| NK | Natural killer |
| ONB | Olfactory neuroblastoma |
| TAM | Tumor-associated macrophage |
| TME | Tumor microenvironment |
References
- Berger LLuc, R. L’esthesioneuroepitheliome olfactif. Bull. Assoc. Fr. Etude Cancer 1924, 13, 410–421. [Google Scholar]
- Faragalla, H.; Weinreb, I. Olfactory neuroblastoma: A review and update. Adv. Anat. Pathol. 2009, 16, 322–331. [Google Scholar] [CrossRef]
- de la Vega, L.L.; McHugh, J.B.; Cani, A.K.; Kunder, K.; Walocko, F.M.; Liu, C.-J.; Hovelson, D.H.; Robinson, D.; Chinnaiyan, A.M.; Tomlins, S.A.; et al. Comprehensive Molecular Profiling of Olfactory Neuroblastoma Identifies Potentially Targetable FGFR3 Amplifications. Mol. Cancer Res. 2017, 15, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Sideris, G.; Solomos, P.; Gogoulos, P.; Margaris, I.; Panagoulis, E.; Vlastarakos, P.; Karamagkiolas, S.; Tzagkaroulakis, M.; Nikolopoulos, T.; Delides, A. Neuroendocrine and undifferentiated sinonasal and skull base tumors: An up-to-date narrative review. Oral Maxillofac. Surg. 2024, 28, 1009–1017. [Google Scholar] [CrossRef] [PubMed]
- Finlay, J.B.; Ireland, A.S.; Hawgood, S.B.; Reyes, T.; Ko, T.; Olsen, R.R.; Hachem, R.A.; Jang, D.W.; Bell, D.; Chan, J.M.; et al. Olfactory neuroblastoma mimics molecular heterogeneity and lineage trajectories of small-cell lung cancer. Cancer Cell 2024, 42, 1086–1105.e13. [Google Scholar] [CrossRef]
- Matayoshi, R.; Otaki, J.M. Immunohistochemical detection of olfactory-specific sensory transduction proteins in olfactory neuroblastoma. Neurosci. Res. 2011, 69, 258–262. [Google Scholar] [CrossRef]
- Zunitch, M.J.; Fisch, A.S.; Lin, B.; Barrios-Camacho, C.M.; Faquin, W.C.; Tachie-Baffour, Y.; Louie, J.D.; Jang, W.; Curry, W.T.; Gray, S.T.; et al. Molecular Evidence for Olfactory Neuroblastoma as a Tumor of Malignant Globose Basal Cells. Mod. Pathol. 2023, 36, 100122. [Google Scholar] [CrossRef]
- Ghanem, A.; Finlay, J.B.; Jang, D.W.; Goldstein, B.J.; Abi Hachem, R. Recent developments in olfactory neuroblastoma research. Curr. Opin. Otolaryngol. Head Neck Surg. 2025, 33, 50–55. [Google Scholar] [CrossRef]
- Patel, A.; Im, E.; Kresak, J.; Olgaard, E.; Blatt, J.E.; Lobo, B.C.; Chapurin, N. Olfactory Neuroblastoma With Divergent Differentiation: Contemporary Management of Unusual Pathology and Literature Review. Ear Nose Throat J. 2024, 01455613241299684. [Google Scholar] [CrossRef]
- Lerner, D.K.; Palmer, J.N. Personalized Approach to Olfactory Neuroblastoma Care. J. Pers. Med. 2024, 14, 423. [Google Scholar] [CrossRef]
- Rao, K.R.; Upadhya, I.B. A Review on Esthesioneuroblastoma. Indian J. Otolaryngol. Head Neck Surg. 2022, 74 (Suppl. S2), 1584–1590. [Google Scholar] [CrossRef] [PubMed]
- Bell, D.; Brandea, A.I.; Hanna, E.Y. Olfactory Neuroblastoma: Morphological Reappraisal and Molecular Insights with Quantum Leap in Clinical Perspectives. Curr. Oncol. Rep. 2023, 25, 11–18. [Google Scholar] [CrossRef]
- Korra, H.; Gandi, J.B.; Nanuvala, P.; Ardha, A. Experiences and Outcomes in Olfactory Neuroblastoma Over A Decade at a Tertiary Cancer Center. South Asian J. Cancer 2022, 11, 336–339. [Google Scholar] [CrossRef]
- Kaur, R.P.; Izumchenko, E.; Blakaj, D.M.; Mladkova, N.; Lechner, M.; Beaumont, T.L.; Floudas, C.S.; Gallia, G.L.; London, N.R. The genomics and epigenetics of olfactory neuroblastoma: A systematic review. Laryngoscope Investig. Otolaryngol. 2021, 6, 721–728. [Google Scholar] [CrossRef] [PubMed]
- Tosoni, A.; Di Nunno, V.; Gatto, L.; Corradi, G.; Bartolini, S.; Ranieri, L.; Franceschi, E. Olfactory neuroblastoma: Diagnosis, management, and current treatment options. Front. Oncol. 2023, 13, 1242453. [Google Scholar] [CrossRef]
- Lopez, F.; Agaimy, A.; Franchi, A.; Suárez, C.; Poorten, V.V.; Mäkitie, A.A.; Homma, A.; Eisbruch, A.; Olsen, K.D.; Saba, N.F.; et al. Update on olfactory neuroblastoma. Virchows Arch. 2024, 484, 567–585. [Google Scholar] [CrossRef]
- Carney, M.E.; O’REilly, R.C.; Sholevar, B.; Buiakova, O.I.; Lowry, L.D.; Keane, W.M.; Margolis, F.L.; Rothstein, J.L. Expression of the humanAchaete-Scute 1 gene in olfactory neuroblastoma (esthesioneuroblastoma). J. Neuro-Oncol. 1995, 26, 35–43. [Google Scholar] [CrossRef]
- Larkin, R.M.; Lopez, D.C.; Robbins, Y.L.; Lassoued, W.; Canubas, K.; Warner, A.; Karim, B.; Vulikh, K.; Hodge, J.W.; Floudas, C.S.; et al. Augmentation of tumor expression of HLA-DR, CXCL9, and CXCL10 may improve olfactory neuroblastoma immunotherapeutic responses. J. Transl. Med. 2024, 22, 524. [Google Scholar] [CrossRef] [PubMed]
- Joyce, J.A.; Fearon, D.T. T cell exclusion, immune privilege, and the tumor microenvironment. Science 2015, 348, 74–80. [Google Scholar] [CrossRef]
- Sun, L.; Clavijo, P.E.; Robbins, Y.; Patel, P.; Friedman, J.; Greene, S.; Das, R.; Silvin, C.; Van Waes, C.; Horn, L.A.; et al. Inhibiting myeloid-derived suppressor cell trafficking enhances T cell immunotherapy. J. Clin. Investig. 2019, 4, e126853. [Google Scholar] [CrossRef]
- Wu, S.-Y.; Fu, T.; Jiang, Y.-Z.; Shao, Z.-M. Natural killer cells in cancer biology and therapy. Mol. Cancer 2020, 19, 120. [Google Scholar] [CrossRef] [PubMed]
- Reits, E.A.; Hodge, J.W.; Herberts, C.A.; Groothuis, T.A.; Chakraborty, M.; Wansley, E.K.; Camphausen, K.; Luiten, R.M.; De Ru, A.H.; Neijssen, J.; et al. Radiation modulates the peptide repertoire, enhances MHC class I expression, and induces successful antitumor immunotherapy. J. Exp. Med. 2006, 203, 1259–1271. [Google Scholar] [CrossRef]
- van Gerven, M.R.; Bozsaky, E.; Matser, Y.A.H.; Vosseberg, J.; Taschner-Mandl, S.; Koster, J.; Tytgat, G.A.M.; Molenaar, J.J.; van den Boogaard, M. Mutational spectrum of ATRX aberrations in neuroblastoma and associated patient and tumor characteristics. Cancer Sci. 2022, 113, 2167–2178. [Google Scholar] [CrossRef]
- Kim, J.; Kong, G.; Lee, C.H.; Kim, D.Y.; Rhee, C.; Min, Y.; Kim, C.W.; Chung, J. Expression of Bcl-2 in Olfactory Neuroblastoma and its Association with Chemotherapy and Survival. Otolaryngol.–Head Neck Surg. 2008, 139, 708–712. [Google Scholar] [CrossRef]
- Yang, J.; Song, X.; Zhang, H.; Liu, Q.; Wei, R.; Guo, L.; Yuan, C.; Chen, F.; Xue, K.; Lai, Y.; et al. Single-cell transcriptomic landscape deciphers olfactory neuroblastoma subtypes and intra-tumoral heterogeneity. Nat. Cancer 2024, 5, 1919–1939. [Google Scholar] [CrossRef] [PubMed]
- Topcagic, J.; Feldman, R.; Ghazalpour, A.; Swensen, J.; Gatalica, Z.; Vranic, S.; Kumar-Sinha, C. Comprehensive molecular profiling of advanced/metastatic olfactory neuroblastomas. PLoS ONE 2018, 13, e0191244. [Google Scholar] [CrossRef] [PubMed]
- Classe, M.; Yao, H.; Mouawad, R.; Creighton, C.J.; Burgess, A.; Allanic, F.; Wassef, M.; Leroy, X.; Verillaud, B.; Mortuaire, G.; et al. Integrated Multi-omic Analysis of Esthesioneuroblastomas Identifies Two Subgroups Linked to Cell Ontogeny. Cell Rep. 2018, 25, 811–821.e5. [Google Scholar] [CrossRef]
- Wang, L.; Ding, Y.; Wei, L.; Zhao, D.; Wang, R.; Zhang, Y.; Gu, X.; Wang, Z. Recurrent Olfactory Neuroblastoma Treated with Cetuximab and Sunitinib: A Case Report. Medicine 2016, 95, e3536. [Google Scholar] [CrossRef]
- Diensthuber, M.; Potinius, M.; Rodt, T.; Stan, A.C.; Welkoborsky, H.-J.; Samii, M.; Schreyögg, J.; Lenarz, T.; Stöver, T. Expression of bcl-2 is associated with microvessel density in olfactory neuroblastoma. J. Neuro-Oncol. 2008, 89, 131–139. [Google Scholar] [CrossRef]
- Peng, X.; Liu, Y.; Peng, X.; Wang, Z.; Zhang, Z.; Qiu, Y.; Jin, M.; Wang, R.; Kong, D. Clinical features and the molecular biomarkers of olfactory neuroblastoma. Pathol.-Res. Pract. 2018, 214, 1123–1129. [Google Scholar] [CrossRef] [PubMed]
- Finlay, J.B.; Hachem, R.A.; Jang, D.W.; Osazuwa-Peters, N.; Goldstein, B.J. Deconstructing Olfactory Epithelium Developmental Pathways in Olfactory Neuroblastoma. Cancer Res. Commun. 2023, 3, 980–990. [Google Scholar] [CrossRef] [PubMed]
- Weinreb, I.; Goldstein, D.; Irish, J.; Perez-Ordonez, B. Expression patterns of Trk-A, Trk-B, GRP78, and p75NRT in olfactory neuroblastoma. Hum. Pathol. 2009, 40, 1330–1335. [Google Scholar] [CrossRef] [PubMed]
- Gallia, G.L.; Zhang, M.; Ning, Y.; Haffner, M.C.; Batista, D.; Binder, Z.A.; Bishop, J.A.; Hann, C.L.; Hruban, R.H.; Ishii, M.; et al. Genomic analysis identifies frequent deletions of Dystrophin in olfactory neuroblastoma. Nat. Commun. 2018, 9, 5410. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, T.; Tatehara, S.; Teshima, M.; Shinomiya, H.; Inokuchi, G.; Komatsu, M.; Hara, S.; Zen, Y.; Nibu, K.-I. Expressions of NeuroD and GAP43 as diagnostic markers for olfactory neuroblastoma. Auris Nasus Larynx 2023, 50, 358–364. [Google Scholar] [CrossRef]
- Pirrone, C.; Chiaravalli, A.M.; Marando, A.; Conti, A.; Rainero, A.; Pistochini, A.; Curto, F.L.; Pasquali, F.; Castelnuovo, P.; Capella, C.; et al. OTX1 and OTX2 as possible molecular markers of sinonasal carcinomas and olfactory neuroblastomas. Eur. J. Histochem. 2017, 61, 2730. [Google Scholar] [CrossRef][Green Version]
- Yang, Y.; Wan, Z.; Zhang, E.; Piao, Y. Genomic profiling and immune landscape of olfactory neuroblastoma in China. Front. Oncol. 2023, 13, 1226494. [Google Scholar] [CrossRef] [PubMed]
- Bishop, J.A.; Antonescu, C.R.; Westra, W.H. SMARCB1 (INI-1)-deficient carcinomas of the sinonasal tract. Am. J. Surg. Pathol. 2014, 38, 1282–1289. [Google Scholar] [CrossRef]
- Agaimy, A.; Hartmann, A.; Antonescu, C.R.; Chiosea, S.I.; El-Mofty, S.K.; Geddert, H.; Iro, H.; Lewis, J.S.J.; Märkl, B.; Mills, S.E.; et al. SMARCB1 (INI-1)-deficient Sinonasal Carcinoma: A Series of 39 Cases Expanding the Morphologic and Clinicopathologic Spectrum of a Recently Described Entity. Am. J. Surg. Pathol. 2017, 41, 458–471. [Google Scholar] [CrossRef]
- Chitguppi, C.; Rabinowitz, M.R.; Johnson, J.; Bar-Ad, V.; Fastenberg, J.H.; Molligan, J.; Berman, E.; Nyquist, G.G.; Rosen, M.R.; Evans, J.E.; et al. Loss of SMARCB1 Expression Confers Poor Prognosis to Sinonasal Undifferentiated Carcinoma. J. Neurol. Surg. Part B Skull Base 2020, 81, 610–619. [Google Scholar] [CrossRef]
- Agaimy, A.; Koch, M.; Lell, M.; Semrau, S.; Dudek, W.; Wachter, D.L.; Knöll, A.; Iro, H.; Haller, F.; Hartmann, A. SMARCB1(INI1)-deficient sinonasal basaloid carcinoma: A novel member of the expanding family of SMARCB1-deficient neoplasms. Am. J. Surg. Pathol. 2014, 38, 1274–1281. [Google Scholar] [CrossRef]
- Agaimy, A.; Jain, D.; Uddin, N.; Rooper, L.M.; Bishop, J.A. SMARCA4-deficient Sinonasal Carcinoma: A Series of 10 Cases Expanding the Genetic Spectrum of SWI/SNF-driven Sinonasal Malignancies. Am. J. Surg. Pathol. 2020, 44, 703–710. [Google Scholar] [CrossRef]
- Agaimy, A.; Weichert, W. SMARCA4-deficient Sinonasal Carcinoma. Head Neck Pathol. 2017, 11, 541–545. [Google Scholar] [CrossRef]
- Papadaki, H.; Kounelis, S.; Kapadia, S.B.; Bakker, A.; Swalsky, P.A.; Finkelstein, S.D. Relationship of p53 gene alterations with tumor progression and recurrence in olfactory neuroblastoma. Am. J. Surg. Pathol. 1996, 20, 715–721. [Google Scholar] [CrossRef] [PubMed]
- Sriuranpong, V.; Borges, M.W.; Strock, C.L.; Nakakura, E.K.; Watkins, D.N.; Blaumueller, C.M.; Nelkin, B.D.; Ball, D.W. Notch Signaling Induces Rapid Degradation of Achaete-Scute Homolog 1. Mol. Cell. Biol. 2002, 22, 3129–3139. [Google Scholar] [CrossRef]
- Osada, H.; Tatematsu, Y.; Yatabe, Y.; Horio, Y.; Takahashi, T. ASH1 gene is a specific therapeutic target for lung cancers with neuroendocrine features. Cancer Res. 2005, 65, 10680–10685. [Google Scholar] [CrossRef] [PubMed]
- Krüttgen, A.; Schneider, I.; Weis, J. The dark side of the NGF family: Neurotrophins in neoplasias. Brain Pathol. 2006, 16, 304–310. [Google Scholar] [CrossRef] [PubMed]
- Lawn, S.; Krishna, N.; Pisklakova, A.; Qu, X.; Fenstermacher, D.A.; Fournier, M.; Vrionis, F.D.; Tran, N.; Chan, J.A.; Kenchappa, R.S.; et al. Neurotrophin signaling via TrkB and TrkC receptors promotes the growth of brain tumor-initiating cells. J. Biol. Chem. 2015, 290, 3814–3824. [Google Scholar] [CrossRef]
- Eggert, A.; Grotzer, M.A.; Ikegaki, N.; Liu, X.-G.; Evans, A.E.; Brodeur, G.M. Expression of neurotrophin receptor TrkA inhibits angiogenesis in neuroblastoma. Med. Pediatr. Oncol. 2000, 35, 569–572. [Google Scholar] [CrossRef]
- Mao, L.; Xia, Y.-P.; Zhou, Y.-N.; Dai, R.-L.; Yang, X.; Wang, Y.-J.; Duan, S.-J.; Qiao, X.; Mei, Y.-W.; Hu, B. Activation of sonic hedgehog signaling pathway in olfactory neuroblastoma. Oncology 2009, 77, 231–243. [Google Scholar] [CrossRef]
- Nibu, K.-I.; Li, G.; Kaga, K.; Rothstein, J.L. bFGF induces differentiation and death of olfactory neuroblastoma cells. Biochem. Biophys. Res. Commun. 2000, 279, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-Y.; Dakir, E.H.; Naizhen, X.; Jensen-Taubman, S.M.; DeMayo, F.J.; Linnoila, R.I. Achaete-scute homolog-1 linked to remodeling and preneoplasia of pulmonary epithelium. Mod. Pathol. 2007, 87, 527–539. [Google Scholar] [CrossRef] [PubMed]
- Koschny, R.; Holland, H.; Sykora, J.; Erdal, H.; Krupp, W.; Bauer, M.; Bockmuehl, U.; Ahnert, P.; Meixensberger, J.; Stremmel, W.; et al. Bortezomib sensitizes primary human esthesioneuroblastoma cells to TRAIL-induced apoptosis. J. Neuro-Oncol. 2010, 97, 171–185. [Google Scholar] [CrossRef] [PubMed]
- Weiss, G.J.; Liang, W.S.; Izatt, T.; Arora, S.; Cherni, I.; Raju, R.N.; Hostetter, G.; Kurdoglu, A.; Christoforides, A.; Sinari, S.; et al. Paired Tumor and Normal Whole Genome Sequencing of Metastatic Olfactory Neuroblastoma. PLoS ONE 2012, 7, e37029. [Google Scholar] [CrossRef] [PubMed]
- Cha, S.; Lee, J.; Shin, J.-Y.; Kim, J.-Y.; Sim, S.H.; Keam, B.; Kim, T.M.; Kim, D.-W.; Heo, D.S.; Lee, S.-H.; et al. Clinical application of genomic profiling to find druggable targets for adolescent and young adult (AYA) cancer patients with metastasis. BMC Cancer 2016, 16, 170. [Google Scholar] [CrossRef]
- Eischen, C.M.; Lozano, G. The Mdm network and its regulation of p53 activities: A rheostat of cancer risk. Hum. Mutat. 2014, 35, 728–737. [Google Scholar] [CrossRef]
- Subhasree, N.; Jiangjiang, Q.; Kalkunte, S.; Minghai, W.; Ruiwen, Z. The MDM2-p53 pathway revisited. J. Biomed. Res. 2013, 27, 254–271. [Google Scholar] [CrossRef]
- Qin, J.-J.; Li, X.; Hunt, C.; Wang, W.; Wang, H.; Zhang, R. Natural products targeting the p53-MDM2 pathway and mutant p53: Recent advances and implications in cancer medicine. Genes Dis. 2018, 5, 204–219. [Google Scholar] [CrossRef]
- Harris, S.L.; Levine, A.J. The p53 pathway: Positive and negative feedback loops. Oncogene 2005, 24, 2899–2908. [Google Scholar] [CrossRef]
- Gay, L.M.; Kim, S.; Fedorchak, K.; Kundranda, M.; Odia, Y.; Nangia, C.; Battiste, J.; Colon-Otero, G.; Powell, S.; Russell, J.; et al. Comprehensive Genomic Profiling of Esthesioneuroblastoma Reveals Additional Treatment Options. Oncologist 2017, 22, 834–842. [Google Scholar] [CrossRef] [PubMed]
- Cracolici, V.; Wang, E.W.; Gardner, P.A.; Snyderman, C.; Gargano, S.M.; Chiosea, S.; Singhi, A.D.; Seethala, R.R. SSTR2 Expression in Olfactory Neuroblastoma: Clinical and Therapeutic Implications. Head Neck Pathol. 2021, 15, 1185–1191. [Google Scholar] [CrossRef]
- Guled, M.; Myllykangas, S.; Frierson, H.F.; Mills, S.E.; Knuutila, S.; Stelow, E.B. Array comparative genomic hybridization analysis of olfactory neuroblastoma. Mod. Pathol. 2008, 21, 770–778. [Google Scholar] [CrossRef]
- Czapiewski, P.; Kunc, M.; Haybaeck, J. Genetic and molecular alterations in olfactory neuroblastoma: Implications for pathogenesis, prognosis and treatment. Oncotarget 2016, 7, 52584–52596. [Google Scholar] [CrossRef]
- Valli, R.; De Bernardi, F.; Frattini, A.; Volpi, L.; Bignami, M.; Facchetti, F.; Pasquali, F.; Castelnuovo, P.; Maserati, E. Comparative genomic hybridization on microarray (a-CGH) in olfactory neuroblastoma: Analysis of ten cases and review of the literature. Genes Chromosom. Cancer 2015, 54, 771–775. [Google Scholar] [CrossRef]
- Capper, D.; Engel, N.W.; Stichel, D.; Lechner, M.; Glöss, S.; Schmid, S.; Koelsche, C.; Schrimpf, D.; Niesen, J.; Wefers, A.K.; et al. DNA methylation-based reclassification of olfactory neuroblastoma. Acta Neuropathol. 2018, 136, 255–271. [Google Scholar] [CrossRef]
- Batchu, S.; Gill, A.S.; Karsy, M. Characterizing Immune Infiltration in Esthesioneuroblastoma Subtypes Through Gene Expression Deconvolution. World Neurosurg. 2024, 183, e928–e935. [Google Scholar] [CrossRef]
- Fiani, B.; Quadri, S.A.; Cathel, A.; Farooqui, M.; Ramachandran, A.; Siddiqi, I.; Ghanchi, H.; Zafar, A.; Berman, B.W.; Siddiqi, J. Esthesioneuroblastoma: A Comprehensive Review of Diagnosis, Management, and Current Treatment Options. World Neurosurg. 2019, 126, 194–211. [Google Scholar] [CrossRef]
- Preusser, M.; Hutterer, M.; Sohm, M.; Koperek, O.; Elandt, K.; Dieckmann, K.; Prayer, D.; Marosi, C. Disease stabilization of progressive olfactory neuroblastoma (esthesioneuroblastoma) under treatment with sunitinib mesylate. J. Neuro-Oncol. 2010, 97, 305–308. [Google Scholar] [CrossRef] [PubMed]
- Atkins, M.; Jones, C.A.; Kirkpatrick, P. Sunitinib maleate. Nat. Rev. Drug Discov. 2006, 5, 279–280. [Google Scholar] [CrossRef]
- Kim, S.; Atta, J.R.; Bergmann, L.; Ottmann, O.G. Imatinib mesylate as second-line treatment in a c-kit-positive Esthesioneuroblastoma. J. Clin. Oncol. 2011, 29, e12513. [Google Scholar] [CrossRef]
| Reference | Gene Alteration | Study Type | Function in ONB |
|---|---|---|---|
| [23] | ATRX | Human and cell lines | Chromatin remodeling protein; maintains telomere stability; ATRX loss leads to genomic instability, alternative lengthening of telomeres, and aggressive tumor behavior |
| [24] | BCL-2 | Human | Antiapoptotic protein; elevated expression in ONB; promotes angiogenesis in malignant tumors |
| [25] | BMP7 | Human | Member of the bone morphogenetic protein family involved in mesenchymal differentiation and extracellular matrix remodeling, characteristic of the mesenchymal ONB subtype |
| [26,27] | CDKN2A | Human | Encodes tumor suppressors p16INK4a and p14ARF; loss of CDKN2A leads to uncontrolled cellular proliferation |
| [26] | CTNNB1 | Human | Gene encodes beta-catenin; mutations result in constitutive activation of Wnt signaling and lead to enhanced cellular proliferation, reduced apoptosis, and increased oncogenic potential |
| [28] | EGFR | Human | May be activated via TRKB; promotes cell proliferation and inhibits cell apoptosis |
| [29] | EPO, EPOR | Human | Molecular marker present in ONB cells; binding with EPOR promotes angiogenesis |
| [30] | ERK | Human | Promotes transcription of factors that drive cell proliferation and prevent apoptosis; activated by TRKB |
| [7,31] | EZH2 | Integrated human–mouse single-cell atlas—human | Stemness marker; potential therapeutic target; silences tumor suppressor genes through trimethylation of histone H3 at lysine 27 (H3K27me3) as a histone methyltransferase |
| [28] | FGFR2 | Human | Receptor tyrosine kinase; drives tumor growth through activation of downstream signaling pathways such as MAPK and PI3K/AKT; alterations suggest potential for targeted therapy in ONB |
| [32] | GRP78 | Human | Chaperone protein; regulates the unfolded protein response, protecting cells from stress-induced apoptosis; overexpression indicates a role in tumor adaptation to hypoxic and metabolic stress conditions and promotes survival and therapeutic resistance |
| [17] | hASH-1 | Human | Encoded by ASCL1 gene; overexpression may act as a trigger for cancer formation in olfactory epithelial cells; involved in lineage specification, neuronal commitment, and differentiation; downregulated via Notch pathway; activates BCL-2 |
| [7] | HES6 | Integrated human–mouse single-cell atlas | Transcription factor; regulates neuronal differentiation; represses Notch signaling, promoting cell cycle exit and differentiation |
| [29] | HIF-1a/EPO/EPOR/Bcl-2 | Human | Leads to autocrine signaling, which promotes angiogenesis through Bcl-2 |
| [29] | HIF1A | Human | Transcription induced by phosphorylated STAT3; induces EPO and EPOR expression in ONB cells |
| [27,31] | IDH2 mutations | Human-Human | Identified in a subset of ONB cases with atypical epithelial differentiation, often associated with more aggressive behavior; leads to the production of the oncometabolite 2-hydroxyglutarate (2-HG), which inhibits α-KG-dependent dioxygenases, including histone demethylases and ten-eleven translocation (TET) family DNA demethylases; results in DNA hypermethylation, epigenetic reprogramming, and cellular differentiation blockade |
| [33] | LAMA2 | Human | Regulates extracellular matrix integrity and cell adhesion; potential role in tumor invasion |
| [30] | MAPK/ERK | Human | Enhances the maintenance of brain tumor-initiating cells (BTICs); prevents apoptosis and increases cell proliferation; promotes lung adenocarcinoma metastasis formation through expression of TRKB |
| [34] | NEUROD1 | Human | Transcription factor; enhances neuronal maturation by activating genes involved in synapse formation; plays a role in neural lineage commitment and synaptic development; helps in distinguishing ONB from SNUC |
| [30] | PI3K/AKT | Human | Activated via overexpression of TRKB; prevents apoptosis, causes an increase in cell growth, and promotes angiogenesis |
| [35] | OTX2 | Human | Homeobox gene; plays a critical role in embryonic brain and neural crest development; overexpression promotes tumor cell proliferation by activating neurodevelopmental pathways and leads to enhanced oncogenesis |
| [28] | RET | Human | Receptor tyrosine kinase involved in cell survival and proliferation; RET mutations have been implicated in ONB oncogenesis, and targeted therapies against RET are currently under investigation |
| [5] | Rb1 protein | Animal | Tumor suppressor protein that controls the G1/S transition of the cell cycle; binds to and inhibits E2F transcription factors, preventing excessive cell cycle progression when it is functional; loss of function leads to dysregulated cell division and is associated with neuroendocrine differentiation |
| [36] | SETD2 | Human | Histone methyltransferase; regulates gene expression by modifying chromatin structure; catalyzes trimethylation of histone H3 at lysine 36 (H3K36me3); mutations impair transcriptional regulation, DNA repair, and chromatin integrity |
| [30] | STAT3 | Human | Activated by phosphorylation in ONB; triggers increased transcription of HIF1A |
| [37,38,39,40] | SMARCB1 | Human | Critical component of the SWI/SNF chromatin remodeling complex; regulates gene expression by altering nucleosome positioning; mutations or deletions lead to epigenetic dysregulation, loss of differentiation, and increased tumorigenicity |
| [41,42] | SMARCA4 | Human | SWI/SNF component; encodes the ATPase Brg1, which is essential for chromatin modeling; loss-of-function mutations lead to transcriptional deregulation and are associated with highly aggressive ONB subtypes |
| [43] | TP53 mutations | Human | Implicated particularly in high-grade cases of ONB and correlates with poor prognosis; results in uncontrolled cell proliferation, increased genomic instability, and resistance to apoptosis; may confer susceptibility to WEE kinase inhibitors, which regulate the DNA damage response, providing a promising therapeutic approach |
| [32] | TrkA | Human | Strongly expressed in ONB; high-affinity neurotrophin receptor; not expressed in brain tumor-initiating cells; participates through the PI3/AKT pathway; promotes proapoptotic and antiangiogenic effects |
| [32] | TrkB | Human | Strongly expressed in ONB; high-affinity neurotrophin receptor; binds BDNF; participates in the MAPK/ERK and PI3/Akt pathways; induces tumorigenesis; enhances maintenance of brain tumor-initiating cells; promotes lung adenocarcinoma metastasis formation |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Demir, E.; Montgomery, D.; Naravetla, V.; Karsy, M. Updated Insights into the Molecular Pathophysiology of Olfactory Neuroblastoma Using Multi-Omics Analysis. J. Pers. Med. 2025, 15, 309. https://doi.org/10.3390/jpm15070309
Demir E, Montgomery D, Naravetla V, Karsy M. Updated Insights into the Molecular Pathophysiology of Olfactory Neuroblastoma Using Multi-Omics Analysis. Journal of Personalized Medicine. 2025; 15(7):309. https://doi.org/10.3390/jpm15070309
Chicago/Turabian StyleDemir, Enes, Deondra Montgomery, Varun Naravetla, and Michael Karsy. 2025. "Updated Insights into the Molecular Pathophysiology of Olfactory Neuroblastoma Using Multi-Omics Analysis" Journal of Personalized Medicine 15, no. 7: 309. https://doi.org/10.3390/jpm15070309
APA StyleDemir, E., Montgomery, D., Naravetla, V., & Karsy, M. (2025). Updated Insights into the Molecular Pathophysiology of Olfactory Neuroblastoma Using Multi-Omics Analysis. Journal of Personalized Medicine, 15(7), 309. https://doi.org/10.3390/jpm15070309






