An Artificial Intelligence-Based Model to Predict Pregnancy After Intrauterine Insemination: A Retrospective Analysis of 9501 Cycles
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approval
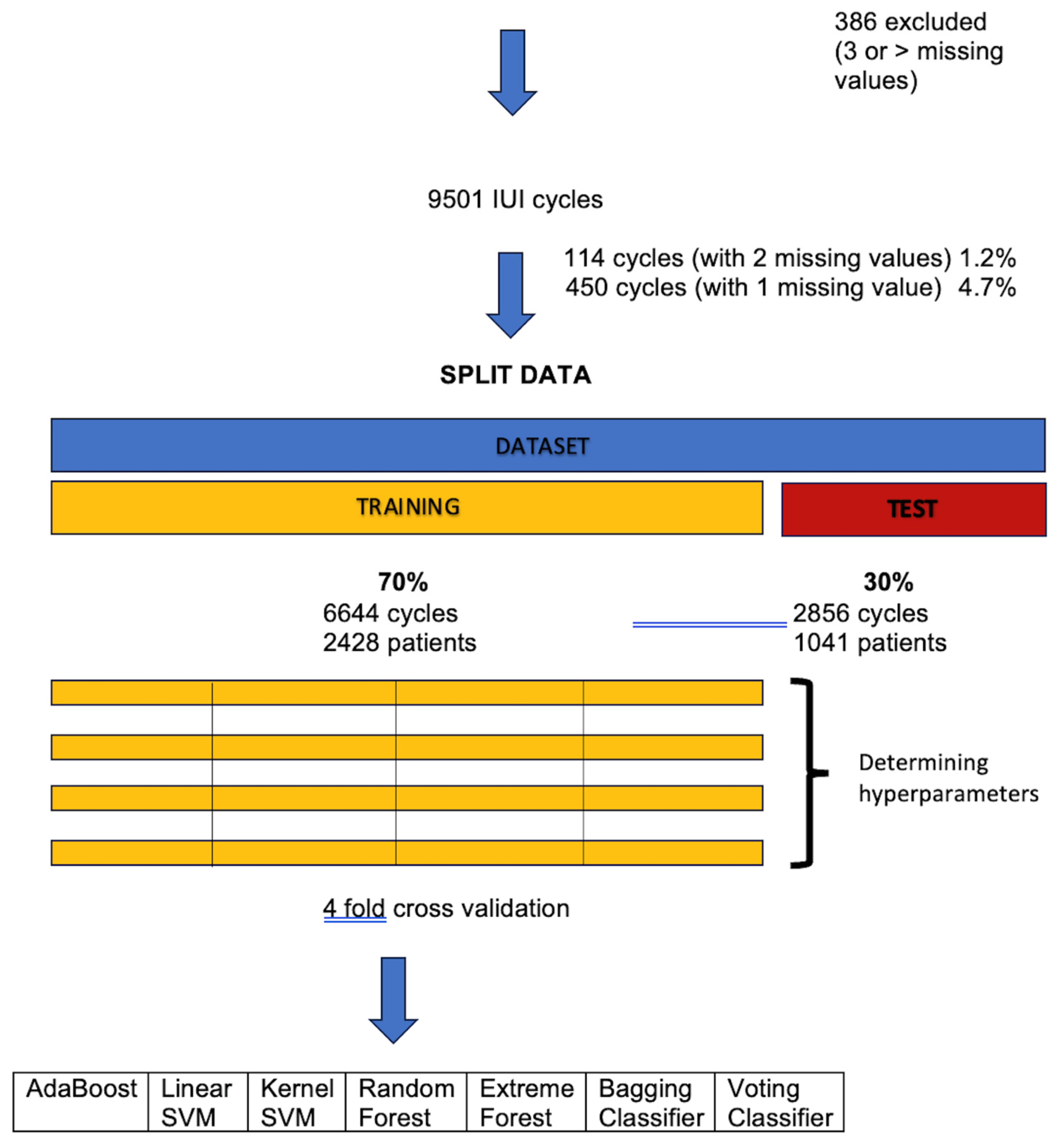
2.2. Study Design
2.3. Eligibility Criteria
2.4. Ovarian Stimulation Protocols
2.5. Sperm Analysis and Preparation
2.6. Intrauterine Insemination and Confirmation of Pregnancy
2.7. Data Pre-Processing and Feature Normalization
2.8. ML Model Training
2.9. ML Model Selection
2.10. Feature Ranking
2.11. Data Availability
3. Results
3.1. Baseline Characteristics
3.2. ML Model Selection
3.3. Feature Ranking
4. Discussion
Limitations and Future Work
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ayeleke, R.O.; Asseler, J.D.; Cohlen, B.J.; Veltman-Verhulst, S.M. Intra-uterine insemination for unexplained subfertility. Cochrane Database Syst Rev. 2020, 3, CD001838. [Google Scholar] [CrossRef] [PubMed]
- Jeong, M.; Kim, S.K.; Kim, H.; Lee, J.R.; Jee, B.C.; Kim, S.H. Predictive value of sperm motility before and after preparation for the pregnancy outcomes of intrauterine insemination. Clin. Exp. Reprod. Med. 2021, 48, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Grysole, C.; Phillips, S.; Minano, J.; Velez, M.P.; Antaki, R.; Sylvestre, C.; Bissonnette, F.; Kadoch, I.J. Series of 9386 IUI Cycles: The Impact of the Number of Motile Spermatozoa Inseminated Varies According to the Female Age. J. Vitr. Fertil. 2021, 3, 10–16. [Google Scholar]
- Yu, C.; Bai, L.; Mei-Zhou, J.; Yu-Wang, X.; Chen, L.; Zhang, J. Analysis of factors associated with IUI pregnancy outcomes in elderly and young patients. BMC Womens Health 2024, 24, 86. [Google Scholar] [CrossRef]
- Huang, C.; Shi, Q.; Xing, J.; Yan, Y.; Shen, X.; Shan, H.; Sun, H.; Mei, J. The relationship between duration of infertility and clinical outcomes of intrauterine insemination for younger women: A retrospective clinical study. BMC Pregnancy Childbirth 2024, 24, 199. [Google Scholar]
- He, W.; Chen, S.; Huang, J.; Zhang, X.; Hu, L.; Xue, Z.; Qiu, Y. Association Between Type of Infertility and Live Birth in Couples With a Single Intrauterine Insemination Resulting in Pregnancy: A Propensity Score Matching Cohort Study. Front. Endocrinol. 2022, 13, 926183. [Google Scholar] [CrossRef]
- Thijssen, A.; Creemers, A.; Van der Elst, W.; Creemers, E.; Vandormael, E.; Dhont, N.; Ombelet, W. Predictive value of different covariates influencing pregnancy rate following intrauterine insemination with homologous semen: A prospective cohort study. Reprod. Biomed. Online 2017, 34, 463–472. [Google Scholar] [CrossRef]
- Danhof, N.A.; van Eekelen, R.; Repping, S.; Mol, B.W.J.; van der Veen, F.; van Wely, M.; Mochtar, M.H.; SUPER Study group. Follicle stimulating hormone or clomiphene citrate in intrauterine insemination with ovarian stimulation for unexplained subfertility: A role for treatment selection markers? Reprod. Biomed. Online 2019, 38, 938–942. [Google Scholar] [CrossRef]
- Ejzenberg, D.; Callado, G.Y.; de Oliveira Gomes, T.J.; Cavalcanti, G.S.; Soares, J.M., Jr.; Baracat, E.C.; Monteleone, P.A. A new accurate model to assess intrauterine insemination success based on clinical parameters: Optimizing fertility treatment. Int. J. Gynaecol. Obstet. 2025. [Google Scholar] [CrossRef]
- Bagherian, E.; Jokari, S.; Borjian Boroujeni, P.; Haratian, K.; Sabbaghian, M.; Mohseni Meybodi, A. Role of genetic variations and protein expression of β-Microsemino protein in intrauterine insemination outcome of unexplained infertile men: A case-control study. Int. J. Reprod. Biomed. 2024, 22, 481–494, Erratum in Int. J. Reprod. Biomed. 2024, 22, 682. [Google Scholar] [CrossRef]
- Legro, R.S.; Hansen, K.R.; Diamond, M.P.; Steiner, A.Z.; Coutifaris, C.; Cedars, M.I.; Hoeger, K.M.; Usadi, R.; Johnstone, E.B.; Haisenleder, D.J.; et al. Reproductive Medicine Network. Effects of preconception lifestyle intervention in infertile women with obesity: The FIT-PLESE randomized controlled trial. PLoS Med. 2022, 19, e1003883. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Wu, S.; Yuan, J.; Zhou, H.; Zhong, Y.; Zhang, M.; Li, Q.; Xu, X.; Sun, X.; Zhu, D. Evaluation of Prognostic Factors for Clinical Pregnancy Rate Following Artificial Insemination by Husband in the Chinese Population. Front. Med. 2021, 8, 638560. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Zhong, Y.; Li, R.; Li, Y.; Zhong, Z.; Liu, T.; Wang, Q.; Lv, Z.; Huang, S.; Duan, Y.G.; et al. Association between exposure to ambient air pollution and semen quality: A systematic review and meta-analysis. Sci. Total Environ. 2023, 20, 161892. [Google Scholar] [CrossRef]
- Salih, M.; Austin, C.; Warty, R.R.; Tiktin, C.; Rolnik, D.L.; Momeni, M.; Rezatofighi, H.; Reddy, S.; Smith, V.; Vollenhoven, B.; et al. Embryo selection through artificial intelligence versus embryologists: A systematic review. Hum. Reprod. Open 2023, 2023, hoad031. [Google Scholar] [CrossRef]
- Correa, N.; Cerquides, J.; Arcos, J.L.; Vassena, R. Supporting first FSH dosage for ovarian stimulation with machine learning. Reprod. Biomed. Online 2022, 45, 1039–1045. [Google Scholar] [PubMed]
- Fjeldstad, J.; Qi, W.; Mercuri, N.; Siddique, N.; Meriano, J.; Krivoi, A.; Nayot, D. An artificial intelligence tool predicts blastocyst development from static images of fresh mature oocytes. Reprod. Biomed. Online 2024, 48, 103842. [Google Scholar] [CrossRef]
- Shi, X.; Prins, C.; Van Pottelbergh, G.; Mamouris, P.; Vaes, B.; De Moor, B. An automated data cleaning method for Electronic Health Records by incorporating clinical knowledge. BMC Med. Inform. Decis. Mak. 2021, 21, 267. [Google Scholar] [CrossRef]
- Pedregosa, P.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. JMLR 12 Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Kuhn, M.; Johnson, K. Applied Predictive Modeling; Spinger Nature: New York, NY, USA, 2013. [Google Scholar] [CrossRef]
- Bergstra, J.; Bengio, Y. Random Search for Hyper-Parameter Optimization. J. Mach. Learn. Res. 2012, 13, 281–305. [Google Scholar]
- Yoav, F.; Robert, E.S. A Decision-Theoretic Generalization of On-Line Learning and an Application to Boosting. J. Comput. Syst. Sci. 1997, 55, 119–139. [Google Scholar]
- Cristianini, N.; Shawe-Taylor, J. Support Vector Machines. In An Introduction to Support Vector Machines and Other Kernel-Based Learning Methods; Cambridge University Press: Cambridge, UK, 2000; pp. 93–124. [Google Scholar]
- Khyathi, G.; Indumathi, K.P.; Hasin, J.; Siluvai, S.; Krishnaprakash, G. Support Vector Machines: A Literature Review on Their Application in Analyzing Mass Data for Public Health. Cureus 2025, 17, e77169. [Google Scholar]
- Couronné, R.; Probst, P.; Boulesteix, A.L. Random forest versus logistic regression: A large-scale benchmark experiment. BMC Bioinform. 2018, 19, 270. [Google Scholar]
- Geurts, P.; Ernst, D.; Wehenkel, L. Extremely Randomized Trees. Mach. Learn. 2006, 63, 3–42. [Google Scholar]
- Zhang, C.; Ma, Y. Ensemble Machine Learning: Methods and Applications; Springer Nature: New York, NY, USA, 2012; p. 10. [Google Scholar]
- Raschka, S. Python Machine Learning; Packt Publishing: Birmingham, UK, 2015. [Google Scholar]
- Kursa, M.; Rudnicki, W. Feature Selection with Boruta Package. J. Stat. Softw. 2010, 36, 1–13. [Google Scholar] [CrossRef]
- Starosta, A.; Gordon, C.E.; Hornstein, M.D. Predictive factors for intrauterine insemination outcomes: A review. Fertil. Res. Pract. 2020, 6, 23. [Google Scholar]
- Huang, X.; Sun, Q.; Tang, X.; Li, M.; Zhou, C.; Cheng, X.; Yao, B.; Chen, L. Factors Influencing the Pregnancy Outcome of Intrauterine Insemination and Follow-up Treatment. J. Hum. Reprod. Sci. 2023, 16, 42–49. [Google Scholar]
- Ghaffari, F.; Sadatmahalleh, S.J.; Akhoond, M.R.; Eftekhari Yazdi, P.; Zolfaghari, Z. Evaluating The Effective Factors in Pregnancy after Intrauterine Insemination: A Retrospective Study. Int. J. Fertil. Steril. 2015, 9, 300–308. [Google Scholar]
- Bengio, Y.; Ian, G.; Aaron, C. Deep Learning. MIT Press: Cambridge, MA, USA, 2017. [Google Scholar]
- Moon, M.; Nakai, K. Stable feature selection based on the ensemble L1-norm support vector machine for biomarker discovery. BMC Genom. 2016, 17, 1026. [Google Scholar] [CrossRef]
- Farabet, C.; Pirtea, P.; Benammar, A.; De Ziegler, D.; Marchiori, C.; Vallée, A.; Ayoubi, J.M. The impact of paternal age on cumulative assisted reproductive technology outcomes. Front. Med. 2024, 10, 1294242. [Google Scholar] [CrossRef]
- Bakkensen, J.B.; Christou, G.; Dimitriadis, I.; James, K.; Souter, I. The effect of follicular phase length on cycle outcomes and endometrial development in gonadotrophin ovarian stimulation/intrauterine insemination cycles. Reprod. Biomed. Online 2020, 40, 362–368. [Google Scholar] [CrossRef]
- Moro, F.; Tropea, A.; Scarinci, E.; Leoncini, E.; Boccia, S.; Federico, A.; Alesiani, O.; Lanzone, A.; Apa, R. Anti-Müllerian hormone concentrations and antral follicle counts for the prediction of pregnancy outcomes after intrauterine insemination. Int. J. Gynaecol. Obstet. 2016, 133, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Diamond, M.P.; Legro, R.S.; Coutifaris, C.; Alvero, R.; Robinson, R.D.; Casson, P.; Christman, G.M.; Ager, J.; Huang, H.; Hansen, K.R.; et al. Letrozole, Gonadotropin, or Clomiphene for Unexplained Infertility. N. Engl. J. Med. 2015, 373, 1230–1240. [Google Scholar] [PubMed]
- Goldman, R.H.; Batsis, M.; Petrozza, J.C.; Souter, I. Patient-specific predictions of outcome after gonadotropin ovulation induction/intrauterine insemination. Fertil. Steril. 2014, 101, 1649–1655. [Google Scholar] [CrossRef] [PubMed]
- Erdem, M.; Erdem, A.; Mutlu, M.F.; Ozisik, S.; Yildiz, S.; Guler, I.; Karakaya, C. The impact of sperm morphology on the outcome of intrauterine insemination cycles with gonadotropins in unexplained and male subfertility. Eur. J. Obstet. Gynecol. Reprod. Biol. 2016, 197, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Lemmens, L.; Kos, S.; Beijer, C.; Brinkman, J.W.; van der Horst, F.A.; van den Hoven, L.; Kieslinger, D.C.; van Trooyen-van Vrouwerff, N.J.; Wolthuis, A.; Hendriks, J.C.; et al. Predictive value of sperm morphology and progressively motile sperm count for pregnancy outcomes in intrauterine insemination. Fertil. Steril. 2016, 105, 1462–1468. [Google Scholar] [CrossRef] [PubMed]
- Ranjbari, S.; Khatibi, T.; Vosough Dizaji, A.; Sajadi, H.; Totonchi, M.; Ghaffari, F. CNFE-SE: A novel approach combining complex network-based feature engineering and stacked ensemble to predict the success of intrauterine insemination and ranking the features. BMC Med. Inform. Decis. Mak. 2021, 21, 1. [Google Scholar] [CrossRef] [PubMed]
- Zippl, A.L.; Wachter, A.; Rockenschaub, P.; Toth, B.; Seeber, B. Predicting success of intrauterine insemination using a clinically based scoring system. Arch. Gynecol. Obstet. 2022, 306, 1777–1786. [Google Scholar] [CrossRef] [PubMed]
| IUI Attempt | Total Number of Cycles | Ovarian Stimulation Protocol | Positive Pregnancy Test | |||
|---|---|---|---|---|---|---|
| Oral Agents | Exogenous Gonadotropins | Combined Therapy | Natural Cycles | |||
| 1 | 3559 (37.45%) | 2954 (83.00%) | 82 (2.30%) | 469 (13.20%) | 54 (15.10%) | 512 (14.38%) |
| 2 | 2614 (27.51%) | 2236 (85.50%) | 46 (1.76%) | 296 (11.32%) | 36 (1.37%) | 325 (12.43%) |
| 3 | 1841 (19.37%) | 1576 (85.60%) | 32 (1.70%) | 217 (11.80%) | 16 (0.80%) | 189 (10.26%) |
| 4 | 774 (8.14%) | 420 (54.26%) | 28 (3.60%) | 317 (40.95%) | 9 (1.10%) | 107 (13.82%) |
| 5 | 713 (7.50%) | 376 (52.73%) | 28 (3.90%) | 305 (42.80%) | 4 (0.50%) | 97 (13.60%) |
| Statistically significant differences were found between IUI 1 and IUI 2 (p = 0.001), IUI 1 and IUI 3 (p = 0.001), IUI 2 and IUI 3 (p = 0.001), IUI 2 and IUI 4 (p = 0.046), IUI 3 and IUI 4 (p = 0.001), and IUI 3 and IUI 5 (p = 0.001). No significant differences were observed between IUI 1 and IUI 4 (p = 0.488), IUI 1 and IUI 5 (p = 0.235), IUI 2 and IUI 5 (p = 0.163), and IUI 4 and IUI 5 (p = 0.9). | ||||||
| Model | Parameters | Time (min) | AUC | |||||
|---|---|---|---|---|---|---|---|---|
| 21 Features | Top 4 | |||||||
| CV | Train | Test | ||||||
| 1 | AdaBoost | Learning rate | 1.0 | 14.7 | 0.585 ± 0.045 | 0.628 ± 0.02 | 0.57 ± 0.03 | 0.58 ± 0.05 |
| N_estimators | 500 | |||||||
| 2 | Linear SVM | C | 0.001 | 4.8 | 0.760 ± 0.045 | 0.790 ± 0.06 | 0.76 ± 0.04 | 0.78 ± 0.04 |
| Class weight | 1:10 | |||||||
| 3 | Kernel SVM | Kernel rbf Gamma 0.001 Class weight 1:10 C: 1000 | 52.1 | 0.584 ± 0.057 | 0.676 ± 0.042 | 0.56 ± 0.05 | 0.57 ± 0.05 | |
| 4 | Random Forest | N_estimators 50 Max_depth 15 Bootstrap true Min_samples_split 0.1 Min_samples_leaf 0.01 Max_features none Class_weight: balanced_subsample | 32.4 | 0.597 ± 0.044 | 0.689 ± 0.056 | 0.56 ± 0.03 | 0.58 ± 0.04 | |
| 5 | Extreme Forest | N_estimators 200 Max_depth none Bootstrap true Min_samples_split 2 Min_samples_leaf 10 Max_features auto Class_weight 1.25 Oob_score true | 49.9 | 0.590 ± 0.054 | 0.872 ± 0.045 | 0.55 ± 0.04 | 0.56 ± 0.06 | |
| 6 | Bagging Classifier | N_estimators 400 Max_samples 0.25 Max_features 0.1 Bootstrap true Oob_score true | 76.5 | 0.576 ± 0.040 | 0.883 ± 0.043 | 0.53 ± 0.05 | 0.54 ± 0.04 | |
| 7 | Voting Classifier | C 0.01 W 1:10 | 28.8 | 0.579 ± 0.047 | 0.853 ± 0.034 | 0.56 ± 0.05 | 0.57 ± 0.05 | |
| Rank | Feature | Accuracy |
|---|---|---|
| 1 | Pre-wash sperm concentration |  |
| 2 | Ovarian stimulation |  |
| 3 | Cycle length |  |
| 4 | Maternal age |  |
| 5 | Post-wash sperm A + B + C |  |
| 6 | IUI history |  |
| 7 | Post-wash sperm C |  |
| 8 | Post-wash sperm concentration |  |
| 9 | Pre-wash sperm B |  |
| 10 | Post-wash sperm C |  |
| 11 | Post-wash sperm A + B |  |
| 12 | Pre-wash sperm A |  |
| 13 | Pre-wash sperm D |  |
| 14 | Pre-wash sperm C |  |
| 15 | NMSI |  |
| 16 | Pre-wash sperm A + B + C |  |
| 17 | Pre-wash sperm A + B |  |
| 18 | Number of days between menses and pregnancy test |  |
| 19 | Post-wash sperm B |  |
| 20 | Post-wash sperm A |  |
| 21 | Paternal age |  |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Minano Masip, J.; Grysole, C.; Borduas, P.; Kadoch, I.-J.; Phillips, S.; Precup, D.; Dufort, D. An Artificial Intelligence-Based Model to Predict Pregnancy After Intrauterine Insemination: A Retrospective Analysis of 9501 Cycles. J. Pers. Med. 2025, 15, 308. https://doi.org/10.3390/jpm15070308
Minano Masip J, Grysole C, Borduas P, Kadoch I-J, Phillips S, Precup D, Dufort D. An Artificial Intelligence-Based Model to Predict Pregnancy After Intrauterine Insemination: A Retrospective Analysis of 9501 Cycles. Journal of Personalized Medicine. 2025; 15(7):308. https://doi.org/10.3390/jpm15070308
Chicago/Turabian StyleMinano Masip, Jaume, Camille Grysole, Penelope Borduas, Isaac-Jacques Kadoch, Simon Phillips, Doina Precup, and Daniel Dufort. 2025. "An Artificial Intelligence-Based Model to Predict Pregnancy After Intrauterine Insemination: A Retrospective Analysis of 9501 Cycles" Journal of Personalized Medicine 15, no. 7: 308. https://doi.org/10.3390/jpm15070308
APA StyleMinano Masip, J., Grysole, C., Borduas, P., Kadoch, I.-J., Phillips, S., Precup, D., & Dufort, D. (2025). An Artificial Intelligence-Based Model to Predict Pregnancy After Intrauterine Insemination: A Retrospective Analysis of 9501 Cycles. Journal of Personalized Medicine, 15(7), 308. https://doi.org/10.3390/jpm15070308






