Methodologies for the Emulation of Biomarker-Guided Trials Using Observational Data: A Systematic Review
Abstract
1. Introduction
2. Materials and Methods
2.1. Eligibility Criteria
2.2. Search Strategy
2.3. Data Collection
2.4. Data Extraction
- Basic information about the study;
- Study design, including specifics of the trial design emulated;
- Statistical analysis, including details of statistical tests applied;
- Methods to control for confounding/biases, including specific details and rationale of methods to adjust for baseline, time-varying, unmeasured and residual confounding, alongside immortal time bias
- Outcomes measured;
- Use of biomarkers, including details of any biomarkers measured, and whether the study would be identified as a biomarker-guided trial, as defined above.
2.5. Data Analysis
3. Results
3.1. Observations from the Included Studies
3.1.1. Specification of the Target Trial Protocol
3.1.2. Specification of Causal Contrasts of Interest
3.1.3. Specification of Follow-Up
3.2. Methods to Control for Confounding
3.2.1. Baseline Confounding
3.2.2. Time-Varying Confounding
3.2.3. Residual and Unmeasured Confounding
3.2.4. Use of Biomarker-Guided Trial Designs
4. Discussion
4.1. Overview
4.2. Applicability of Methods in a Biomarker-Guided Target Trial Setting
5. Recommendations and Future Directions
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| RCT | Randomised controlled trial |
| ITT | Intention-to-treat |
| PP | Per-protocol |
| IPW | Inverse probability weighting |
| IPTW | Inverse probability of treatment weighting |
| MSM | Marginal structural model |
| IPW-MSM | Inverse probability weighting as part of a marginal structural model |
| PSM | Propensity score matching |
| ATE | Average treatment effect |
| ATT | Average treatment effect on the treated |
| IPCW | Inverse probability of censoring weighting |
| SMD | Standardised mean difference |
| MI | Multiple imputation |
| MCMC | Markov chain Monte Carlo |
| BART | Bayesian additive regression trees |
| MAR | Missing at Random |
| MNAR | Missing Not at Random |
| EHR | Electronic health record |
| eGFR | Estimated glomerular filtration rate |
| DAG | Directed acyclic graph |
| TARGET | TrAnsparent ReportinG of observational studies Emulating a Target trial reporting guideline |
| AI | Artificial intelligence |
References
- Hernán, M.A.; Robins, J.M. Using Big Data to Emulate a Target Trial When a Randomized Trial Is Not Available. Am. J. Epidemiol. 2016, 183, 758–764. [Google Scholar] [CrossRef]
- Hernán, M.A.; Sauer, B.C.; Hernández-Díaz, S.; Platt, R.; Shrier, I. Specifying a target trial prevents immortal time bias and other self-inflicted injuries in observational analyses. J. Clin. Epidemiol. 2016, 79, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Mansournia, M.A.; Etminan, M.; Danaei, G.; Kaufman, J.S.; Collins, G. Handling time varying confounding in observational research. BMJ 2017, 359, j4587. [Google Scholar] [CrossRef]
- Brown, L.C.; Jorgensen, A.L.; Antoniou, M.; Wason, J. Biomarker-Guided Trials. In Principles and Practice of Clinical Trials; Piantadosi, S., Meinert, C.L., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 1145–1170. [Google Scholar]
- Le Tourneau, C.; Delord, J.-P.; Gonçalves, A.; Gavoille, C.; Dubot, C.; Isambert, N.; Campone, C.; Trédan, O.; Massiani, M.-A.; Mauborgne, C.; et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): A multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol. 2015, 16, 1324–1334. [Google Scholar] [CrossRef] [PubMed]
- Duijkers, R.; Prins, H.J.; Kross, M.; Snijders, D.; van den Berg, J.W.K.; Werkman, G.M.; van der Veen, N.; Schoorl, M.; Bonten, M.J.M.; van Werkhoven, C.H.; et al. Biomarker guided antibiotic stewardship in community acquired pneumonia: A randomized controlled trial. PLoS ONE 2024, 19, e0307193. [Google Scholar] [CrossRef] [PubMed]
- O’Dwyer, P.J.; Gray, R.J.; Flaherty, K.T.; Chen, A.P.; Li, S.; Wang, V.; McShane, L.M.; Patton, D.R.; Tricoli, J.V.; Williams, P.M.; et al. The NCI-MATCH trial: Lessons for precision oncology. Nat. Med. 2023, 29, 1349–1357. [Google Scholar] [CrossRef]
- Antoniou, M.; Kolamunnage-Dona, R.; Wason, J.; Bathia, R.; Billingham, C.; Bliss, J.M.; Brown, L.C.; Gillman, A.; Paul, J.; Jorgensen, A.L.; et al. Biomarker-guided trials: Challenges in practice. Contemp. Clin. Trials Commun. 2019, 16, 100493. [Google Scholar] [CrossRef]
- Papp, M.; Kiss, N.; Baka, M.; Trásy, D.; Zubek, L.; Fehérvári, P.; Harnos, A.; Turan, C.; Heygi, P.; Molnár, Z.; et al. Procalcitonin-guided antibiotic therapy may shorten length of treatment and may improve survival-a systematic review and meta-analysis. Crit. Care 2023, 27, 394. [Google Scholar] [CrossRef]
- Jorgensen, A.L.; Khalaf, R.K.; Kolamunnage-Dona, R.; Baldwin, F.D. Methodologies for the Emulation of Biomarker-Guided Trials Using Observational Data: A Systematic Review Protocol [Internet]; Open Science Framework: Charlottesville, VA, USA, 2025. [Google Scholar] [CrossRef]
- Ouzzani, M.; Hammady, H.; Fedorowicz, Z.; Elmagarmid, A. Rayyan—A web and mobile app for systematic reviews. Syst. Rev. 2016, 5, 210. [Google Scholar] [CrossRef]
- Ahn, N.; Nolde, M.; Günter, A.; Güntner, F.; Gerlach, R.; Tauscher, M.; Amann, U.; Linseisen, J.; Meisinger, C.; Rückert-Eheberg, I.M.; et al. Emulating a target trial of proton pump inhibitors and dementia risk using claims data. Eur. J. Neurol. 2022, 29, 1335–1343. [Google Scholar] [CrossRef]
- Althunian, T.A.; de Boer, A.; Groenwold, R.H.H.; Rengerink, K.O.; Souverein, P.C.; Klungel, O.H. Rivaroxaban was found to be noninferior to warfarin in routine clinical care: A retrospective noninferiority cohort replication study. Pharmacoepidemiol. Drug Saf. 2020, 29, 1263–1272. [Google Scholar] [CrossRef]
- Aubert, C.E.; Sussman, J.B.; Hofer, T.P.; Cushman, W.C.; Ha, J.K.; Min, L. Adding a New Medication Versus Maximizing Dose to Intensify Hypertension Treatment in Older Adults: A Retrospective Observational Study. Ann. Intern. Med. 2021, 174, 1666–1673. [Google Scholar] [CrossRef] [PubMed]
- Barbulescu, A.; Askling, J.; Saevarsdottir, S.; Kim, S.C.; Frisell, T. Combined Conventional Synthetic Disease Modifying Therapy vs. Infliximab for Rheumatoid Arthritis: Emulating a Randomized Trial in Observational Data. Clin. Pharmacol. Ther. 2022, 112, 836–845. [Google Scholar] [CrossRef] [PubMed]
- Becker, W.C.; Li, Y.; Caniglia, E.C.; Vickers-Smith, R.; Feinberg, T.; Marshall, B.D.L.; Edelman, E.J. Cannabis use, pain interference, and prescription opioid receipt among persons with HIV: A target trial emulation study. AIDS Care 2022, 34, 469–477. [Google Scholar] [CrossRef]
- Börnhorst, C.; Reinders, T.; Rathmann, W.; Bongaerts, B.; Haug, U.; Didelez, V.; Kollhorst, B. Avoiding Time-Related Biases: A Feasibility Study on Antidiabetic Drugs and Pancreatic Cancer Applying the Parametric g-Formula to a Large German Healthcare Database. Clin. Epidemiol. 2021, 13, 1027–1038. [Google Scholar] [CrossRef]
- Bosch, N.A.; Law, A.C.; Vail, E.A.; Gillmeyer, K.R.; Gershengorn, H.B.; Wunsch, H.; Walkey, A. Inhaled Nitric Oxide vs Epoprostenol During Acute Respiratory Failure: An Observational Target Trial Emulation. Chest 2022, 162, 1287–1296. [Google Scholar] [CrossRef] [PubMed]
- Boyne, D.J.; Brenner, D.R.; Gupta, A.; Mackay, E.; Arora, P.; Wasiak, R.; Cheung, W.Y.; Hernán, M.A. Head-to-head comparison of FOLFIRINOX versus gemcitabine plus nab-paclitaxel in advanced pancreatic cancer: A target trial emulation using real-world data. Ann. Epidemiol. 2023, 78, 28–34. [Google Scholar] [CrossRef]
- Cain, L.E.; Saag, M.S.; Petersen, M.; May, M.T.; Ingle, S.M.; Logan, R.; Robins, J.M.; Abgrall, S.; Shepherd, B.E.; Deeks, S.G.; et al. Using observational data to emulate a randomized trial of dynamic treatment-switching strategies: An application to antiretroviral therapy. Int. J. Epidemiol. 2016, 45, 2038–2049. [Google Scholar] [CrossRef]
- Caniglia, E.C.; Robins, J.M.; Cain, L.E.; Sabin, C.; Logan, R.; Abgrall, S.; Mugavero, M.J.; Hernández-Díaz, S.; Meyer, L.; Seng, R.; et al. Emulating a trial of joint dynamic strategies: An application to monitoring and treatment of HIV-positive individuals. Stat. Med. 2019, 38, 2428–2446. [Google Scholar] [CrossRef]
- Caniglia, E.C.; Rojas-Saunero, L.P.; Hilal, S.; Licher, S.; Logan, R.; Stricker, B.; Ikram, M.A.; Swanson, S.A. Emulating a target trial of statin use and risk of dementia using cohort data. Neurology 2020, 95, e1322–e1332. [Google Scholar] [CrossRef]
- Caniglia, E.C.; Zash, R.; Jacobson, D.L.; Diseko, M.; Mayondi, G.; Lockman, S.; Chen, J.Y.; Mmalane, M.; Makhema, J.; Hernán, M.A.; et al. Emulating a target trial of antiretroviral therapy regimens started before conception and risk of adverse birth outcomes. Aids 2018, 32, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Cheng-Lai, A.; Prlesi, L.; Murthy, S.; Bellin, E.Y.; Sinnett, M.J.; Goriacko, P. Evaluating Pharmacist-Led Heart Failure Transitions of Care Clinic: Impact of Analytic Approach on Readmission Rate Endpoints. Curr. Probl. Cardiol. 2023, 48, 101507. [Google Scholar] [CrossRef]
- Danaei, G.; García Rodríguez, L.A.; Cantero, O.F.; Logan, R.W.; Hernán, M.A. Electronic medical records can be used to emulate target trials of sustained treatment strategies. J. Clin. Epidemiol. 2018, 96, 12–22. [Google Scholar] [CrossRef]
- Dickerman, B.A.; García-Albéniz, X.; Logan, R.W.; Denaxas, S.; Hernán, M.A. Avoidable flaws in observational analyses: An application to statins and cancer. Nat. Med. 2019, 25, 1601–1606. [Google Scholar] [CrossRef]
- Dickerman, B.A.; García-Albéniz, X.; Logan, R.W.; Denaxas, S.; Hernán, M.A. Emulating a target trial in case-control designs: An application to statins and colorectal cancer. Int. J. Epidemiol. 2020, 49, 1637–1646. [Google Scholar] [CrossRef] [PubMed]
- Dickerman, B.A.; Gerlovin, H.; Madenci, A.L.; Kurgansky, K.E.; Ferolito, B.R.; Figueroa Muñiz, M.J.; Gagnon, D.R.; Gaziano, J.M.; Cho, K.; Casas, J.P.; et al. Comparative Effectiveness of BNT162b2 and mRNA-1273 Vaccines in U.S. Veterans. N. Engl. J. Med. 2022, 386, 105–115. [Google Scholar] [CrossRef]
- Franklin, J.M.; Patorno, E.; Desai, R.J.; Glynn, R.J.; Martin, D.; Quinto, K.; Pawar, A.; Bessette, L.G.; Lee, H.; Garry, E.M.; et al. Emulating Randomized Clinical Trials with Nonrandomized Real-World Evidence Studies. Circulation 2021, 143, 1002–1013. [Google Scholar] [CrossRef] [PubMed]
- Fu, E.L.; Evans, M.; Clase, C.M.; Tomlinson, L.A.; van Diepen, M.; Dekker, F.W.; Carrero, J.J. Stopping Renin-Angiotensin System Inhibitors in Patients with Advanced CKD and Risk of Adverse Outcomes: A Nationwide Study. J. Am. Soc. Nephrol. 2021, 32, 424–435. [Google Scholar] [CrossRef]
- Hernán, M.A.; Alonso, A.; Logan, R.; Grodstein, F.; Michels, K.B.; Willett, W.C.; Manson, J.E.; Robins, J.M. Observational studies analyzed like randomized experiments: An application to postmenopausal hormone therapy and coronary heart disease. Epidemiology 2008, 19, 766–779. [Google Scholar] [CrossRef]
- Ioannou, G.N.; Bohnert, A.S.B.; O’Hare, A.M.; Boyko, E.J.; Maciejewski, M.L.; Smith, V.A.; Bowling, C.A.; Viglianti, E.; Iwashyna, T.J.; Hynes, D.M.; et al. Effectiveness of mRNA COVID-19 Vaccine Boosters Against Infection, Hospitalization, and Death: A Target Trial Emulation in the Omicron (B.1.1.529) Variant Era. Ann. Intern. Med. 2022, 175, 1693–1706. [Google Scholar] [CrossRef]
- Ioannou, G.N.; Locke, E.R.; Green, P.K.; Berry, K. Comparison of Moderna versus Pfizer-BioNTech COVID-19 vaccine outcomes: A target trial emulation study in the U.S. Veterans Affairs healthcare system. EClinicalMedicine 2022, 45, 101326. [Google Scholar] [CrossRef] [PubMed]
- Kirchgesner, J.; Desai, R.J.; Beaugerie, L.; Kim, S.C.; Schneeweiss, S. Calibrating Real-World Evidence Studies Against Randomized Trials: Treatment Effectiveness of Infliximab in Crohn’s Disease. Clin. Pharmacol. Ther. 2022, 111, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Kirchgesner, J.; Desai, R.J.; Schneeweiss, M.C.; Beaugerie, L.; Kim, S.C.; Schneeweiss, S. Emulation of a randomized controlled trial in ulcerative colitis with US and French claims data: Infliximab with thiopurines compared to infliximab monotherapy. Pharmacoepidemiol. Drug Saf. 2022, 31, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Kirchgesner, J.; Desai, R.J.; Schneeweiss, M.C.; Beaugerie, L.; Schneeweiss, S.; Kim, S.C. Decreased risk of treatment failure with vedolizumab and thiopurines combined compared with vedolizumab monotherapy in Crohn’s disease. Gut 2022, 71, 1781–1789. [Google Scholar] [CrossRef]
- Kraglund, F.; Christensen, D.H.; Eiset, A.H.; Villadsen, G.E.; West, J.; Jepsen, P. Effects of statins and aspirin on HCC risk in alcohol-related cirrhosis: Nationwide emulated trials. Hepatol. Commun. 2023, 7, e0013. [Google Scholar] [CrossRef]
- Kuehne, F.; Arvandi, M.; Hess, L.M.; Faries, D.E.; Matteucci Gothe, R.; Gothe, H.; Beyrer, J.; Zeimet, A.G.; Stojkov, I.; Mühlberger, N.; et al. Causal analyses with target trial emulation for real-world evidence removed large self-inflicted biases: Systematic bias assessment of ovarian cancer treatment effectiveness. J. Clin. Epidemiol. 2022, 152, 269–280. [Google Scholar] [CrossRef]
- Kwee, S.A.; Wong, L.L.; Ludema, C.; Deng, C.K.; Taira, D.; Seto, T.; Landsittel, D. Target Trial Emulation: A Design Tool for Cancer Clinical Trials. JCO Clin. Cancer Inform. 2023, 7, e2200140. [Google Scholar] [CrossRef]
- Lyu, B.; Chan, M.R.; Yevzlin, A.S.; Gardezi, A.; Astor, B.C. Arteriovenous Access Type and Risk of Mortality, Hospitalization, and Sepsis Among Elderly Hemodialysis Patients: A Target Trial Emulation Approach. Am. J. Kidney Dis. 2022, 79, 69–78. [Google Scholar] [CrossRef]
- Massol, J.; Simon-Tillaux, N.; Tohme, J.; Hariri, G.; Dureau, P.; Duceau, B.; Belin, L.; Hajage, D.; De Rycke, Y.; Charfeddine, A.; et al. Levosimendan in patients undergoing extracorporeal membrane oxygenation after cardiac surgery: An emulated target trial using observational data. Crit. Care 2023, 27, 51. [Google Scholar] [CrossRef]
- Mazzotta, V.; Cozzi-Lepri, A.; Colavita, F.; Lanini, S.; Rosati, S.; Lalle, E.; Mastrorosa, I.; Cimaglia, C.; Vergori, A.; Bevilacqua, N.; et al. Emulation of a Target Trial From Observational Data to Compare Effectiveness of Casirivimab/Imdevimab and Bamlanivimab/Etesevimab for Early Treatment of Non-Hospitalized Patients with COVID-19. Front. Immunol. 2022, 13, 868020. [Google Scholar] [CrossRef]
- McGrath, L.J.; Nielson, C.; Saul, B.; Breskin, A.; Yu, Y.; Nicolaisen, S.K.; Kilpatrick, K.; Ghanima, W.; Christiansen, C.F.; Bahmanyar, S.; et al. Lessons Learned Using Real-World Data to Emulate Randomized Trials: A Case Study of Treatment Effectiveness for Newly Diagnosed Immune Thrombocytopenia. Clin. Pharmacol. Ther. 2021, 110, 1570–1578. [Google Scholar] [CrossRef] [PubMed]
- Nolde, M.; Ahn, N.; Dreischulte, T.; Rückert-Eheberg, I.M.; Güntner, F.; Günter, A.; Gerlach, R.; Tauscher, M.; Amann, U.; Linseisen, J.; et al. The long-term risk for myocardial infarction or stroke after proton pump inhibitor therapy (2008–2018). Aliment. Pharmacol. Ther. 2021, 54, 1033–1040. [Google Scholar] [CrossRef]
- Puéchal, X.; Iudici, M.; Perrodeau, E.; Bonnotte, B.; Lifermann, F.; Le Gallou, T.; Karras, A.; Blanchard-Delaunay, C.; Quéméneur, T.; Aouba, A.; et al. Rituximab vs Cyclophosphamide Induction Therapy for Patients with Granulomatosis with Polyangiitis. JAMA Netw. Open 2022, 5, e2243799. [Google Scholar] [CrossRef]
- Schroeder, E.B.; Neugebauer, R.; Reynolds, K.; Schmittdiel, J.A.; Loes, L.; Dyer, W.; Pimental, N.; Desai, J.R.; Vazquez-Benitez, G.; Ho, P.M.; et al. Association of Cardiovascular Outcomes and Mortality with Sustained Long-Acting Insulin Only vs Long-Acting Plus Short-Acting Insulin Treatment. JAMA Netw. Open 2021, 4, e2126605. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.H.; García-Albéniz, X.; Chan, J.M.; Zhao, S.; Cowan, J.E.; Broering, J.M.; Cooperberg, M.R.; Carroll, P.R.; Hernán, M.A. Emulation of a target trial with sustained treatment strategies: An application to prostate cancer using both inverse probability weighting and the g-formula. Eur. J. Epidemiol. 2022, 37, 1205–1213. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, Y.; Kumamaru, H.; Hagiwara, Y.; Matsui, H.; Yasunaga, H.; Miyata, H.; Matsuyama, Y. Sodium-glucose cotransporter-2 inhibitors and the risk of urinary tract infection among diabetic patients in Japan: Target trial emulation using a nationwide administrative claims database. Diabetes Obes. Metab. 2021, 23, 1379–1388. [Google Scholar] [CrossRef]
- Talmor-Barkan, Y.; Yacovzada, N.S.; Rossman, H.; Witberg, G.; Kalka, I.; Kornowski, R.; Segal, E. Head-to-head efficacy and safety of rivaroxaban, apixaban, and dabigatran in an observational nationwide targeted trial. Eur. Heart J. Cardiovasc. Pharmacother. 2022, 9, 26–37. [Google Scholar] [CrossRef]
- Trevisan, M.; Fu, E.L.; Xu, Y.; Savarese, G.; Dekker, F.W.; Lund, L.H.; Clase, C.M.; Sjölander, A.; Carrero, J.J. Stopping mineralocorticoid receptor antagonists after hyperkalaemia: Trial emulation in data from routine care. Eur. J. Heart Fail. 2021, 23, 1698–1707. [Google Scholar] [CrossRef] [PubMed]
- van Santen, D.K.; Boyd, A.; Matser, A.; Maher, L.; Hickman, M.; Lodi, S.; Prins, M. The effect of needle and syringe program and opioid agonist therapy on the risk of, H.I.V.; hepatitis B and C virus infection for people who inject drugs in Amsterdam, the Netherlands: Findings from an emulated target trial. Addiction 2021, 116, 3115–3126. [Google Scholar] [CrossRef]
- van Santen, D.K.; Lodi, S.; Dietze, P.; van den Boom, W.; Hayashi, K.; Dong, H.; Cui, Z.; Maher, L.; Hickman, M.; Boyd, A.; et al. Comprehensive needle and syringe program and opioid agonist therapy reduce HIV and hepatitis c virus acquisition among people who inject drugs in different settings: A pooled analysis of emulated trials. Addiction 2023, 118, 1116–1126. [Google Scholar] [CrossRef]
- Xie, Y.; Bowe, B.; Gibson, A.K.; McGill, J.B.; Maddukuri, G.; Yan, Y.; Al-Aly, Z. Comparative Effectiveness of SGLT2 Inhibitors, GLP-1 Receptor Agonists, DPP-4 Inhibitors, and Sulfonylureas on Risk of Kidney Outcomes: Emulation of a Target Trial Using Health Care Databases. Diabetes Care 2020, 43, 2859–2869. [Google Scholar] [CrossRef]
- Xu, Y.; Fu, E.L.; Trevisan, M.; Jernberg, T.; Sjölander, A.; Clase, C.M.; Carrero, J.J. Stopping renin-angiotensin system inhibitors after hyperkalemia and risk of adverse outcomes. Am. Heart J. 2022, 243, 177–186. [Google Scholar] [CrossRef]
- Yarnell, C.J.; Angriman, F.; Ferreyro, B.L.; Liu, K.; De Grooth, H.J.; Burry, L.; Munshi, L.; Mehta, S.; Celi, L.; Elbers, P.; et al. Oxygenation thresholds for invasive ventilation in hypoxemic respiratory failure: A target trial emulation in two cohorts. Crit. Care 2023, 27, 67. [Google Scholar] [CrossRef] [PubMed]
- Yiu, Z.Z.N.; Mason, K.J.; Hampton, P.J.; Reynolds, N.J.; Smith, C.H.; Lunt, M.; Griffiths, C.E.M.; Warren, R.B.; BADBIR Study Group. Randomized Trial Replication Using Observational Data for Comparative Effectiveness of Secukinumab and Ustekinumab in Psoriasis: A Study From the British Association of Dermatologists Biologics and Immunomodulators Register. JAMA Dermatol. 2021, 157, 66–73. [Google Scholar] [CrossRef]
- Young, J.; Wong, S.; Janjua, N.Z.; Klein, M.B. Comparing direct acting antivirals for hepatitis C using observational data—Why and how? Pharmacol. Res. Perspect 2020, 8, e00650. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Young, J.G.; Thamer, M.; Hernán, M.A. Comparing the Effectiveness of Dynamic Treatment Strategies Using Electronic Health Records: An Application of the Parametric g-Formula to Anemia Management Strategies. Health Serv. Res. 2018, 53, 1900–1918. [Google Scholar] [CrossRef] [PubMed]
- Admon, A.J.; Donnelly, J.P.; Casey, J.D.; Janz, D.R.; Russell, D.W.; Joffe, A.M.; Vonderhaar, D.J.; Dischert, K.M.; Stempek, S.B.; Dargin, J.M.; et al. Emulating a Novel Clinical Trial Using Existing Observational Data. Predicting Results of the PreVent Study. Ann. Am. Thorac. Soc. 2019, 16, 998–1007. [Google Scholar] [CrossRef]
- Bakker, L.; Goossens, L.; O’Kane, M.; Groot, C.; Redekop, W. Analysing Electronic Health Records: The Benefits of Target Trial Emulation. Health Policy Technol. 2021, 10, 100545. [Google Scholar] [CrossRef]
- Cain, L.E.; Robins, J.M.; Lanoy, E.; Logan, R.; Costagliola, D.; Hernán, M.A. When to start treatment? A systematic approach to the comparison of dynamic regimes using observational data. Int. J. Biostat. 2010, 6, 18. [Google Scholar] [CrossRef]
- Hernán, M.A. How to estimate the effect of treatment duration on survival outcomes using observational data. BMJ 2018, 360, k182. [Google Scholar] [CrossRef]
- Hernández-Díaz, S.; Huybrechts, K.F.; Chiu, Y.H.; Yland, J.J.; Bateman, B.T.; Hernán, M.A. Emulating a Target Trial of Interventions Initiated During Pregnancy with Healthcare Databases: The Example of COVID-19 Vaccination. Epidemiology 2023, 34, 238–246. [Google Scholar] [CrossRef] [PubMed]
- Kalia, S.; Saarela, O.; Escobar, M.; Moineddin, R.; Greiver, M. Estimation of marginal structural models under irregular visits and unmeasured confounder: Calibrated inverse probability weights. BMC Med. Res. Methodol. 2023, 23, 4. [Google Scholar] [CrossRef] [PubMed]
- Kuehne, F.; Jahn, B.; Conrads-Frank, A.; Bundo, M.; Arvandi, M.; Endel, F.; Popper, N.; Endel, G.; Urach, C.; Gyimesi, M.; et al. Guidance for a causal comparative effectiveness analysis emulating a target trial based on big real world evidence: When to start statin treatment. J. Comp. Eff. Res. 2019, 8, 1013–1025. [Google Scholar] [CrossRef]
- Lodi, S.; Phillips, A.; Lundgren, J.; Logan, R.; Sharma, S.; Cole, S.R.; Babiker, A.; Law, M.; Chu, H.; Byrne, D.; et al. Effect Estimates in Randomized Trials and Observational Studies: Comparing Apples with Apples. Am. J. Epidemiol. 2019, 188, 1569–1577. [Google Scholar] [CrossRef] [PubMed]
- Maringe, C.; Benitez Majano, S.; Exarchakou, A.; Smith, M.; Rachet, B.; Belot, A.; Leyrat, C. Reflection on modern methods: Trial emulation in the presence of immortal-time bias. Assessing the benefit of major surgery for elderly lung cancer patients using observational data. Int. J. Epidemiol. 2020, 49, 1719–1729. [Google Scholar] [CrossRef]
- Schnitzer, M.E.; Guerra, S.F.; Longo, C.; Blais, L.; Platt, R.W. A potential outcomes approach to defining and estimating gestational age-specific exposure effects during pregnancy. Stat. Methods Med. Res. 2022, 31, 300–314. [Google Scholar] [CrossRef]
- Wendling, T.; Jung, K.; Callahan, A.; Schuler, A.; Shah, N.H.; Gallego, B. Comparing methods for estimation of heterogeneous treatment effects using observational data from health care databases. Stat. Med. 2018, 37, 3309–3324. [Google Scholar] [CrossRef]
- Wintzell, V.; Svanström, H.; Pasternak, B. Selection of Comparator Group in Observational Drug Safety Studies: Alternatives to the Active Comparator New User Design. Epidemiology 2022, 33, 707–714. [Google Scholar] [CrossRef]
- Sterne, J.A.C.; Hernán, M.A.; McAleenan, A.; Reeves, B.C.; Higgins, J.P.T. Chapter 25: Assessing risk of bias in a non-randomized study [last updated October 2019]. In Cochrane Handbook for Systematic Reviews of Interventions Version 6.5. Cochrane; Higgins, J.P.T., Thomas, J., Chandler, J., Cumpston, M., Li, T., Page, M.J., Welch, V.A., Eds.; Cochrane: London, UK, 2024; Available online: www.training.cochrane.org/handbook (accessed on 10 March 2025).
- Fu, E.L. Target Trial Emulation to Improve Causal Inference from Observational Data: What, Why, and How? J. Am. Soc. Nephrol. 2023, 34, 1305–1314. [Google Scholar] [CrossRef]
- Chesnaye, N.C.; Stel, V.S.; Tripepi, G.; Dekker, F.W.; Fu, E.L.; Zoccali, C.; Jager, K.J. An introduction to inverse probability of treatment weighting in observational research. Clin. Kidney J. 2022, 15, 14–20. [Google Scholar] [CrossRef]
- Austin, P.C. The performance of different propensity score methods for estimating marginal hazard ratios. Stat. Med. 2013, 32, 2837–2849. [Google Scholar] [CrossRef] [PubMed]
- Hernan, M.A.; Robins, J.M. Causal Inference: What If; CRC Press: Boca Raton, FL, USA, 2024. [Google Scholar]
- Austin, P.C. An Introduction to Propensity Score Methods for Reducing the Effects of Confounding in Observational Studies. Multivar. Behav. Res. 2011, 46, 399–424. [Google Scholar] [CrossRef] [PubMed]
- Lu, B. Propensity Score Matching with Time-Dependent Covariates. Biometrics 2005, 61, 721–728. [Google Scholar] [CrossRef]
- Bykov, K.; Patorno, E.; D’Andrea, E.; He, M.; Lee, H.; Graff, J.S.; Franklin, J.M. Prevalence of Avoidable and Bias-Inflicting Methodological Pitfalls in Real-World Studies of Medication Safety and Effectiveness. Clin. Pharmacol. Ther. 2022, 111, 209–217. [Google Scholar] [CrossRef]
- Keil, A.P.; Edwards, J.K.; Richardson, D.B.; Naimi, A.I.; Cole, S.R. The parametric g-formula for time-to-event data: Intuition and a worked example. Epidemiology 2014, 25, 889–897. [Google Scholar] [CrossRef]
- McGrath, S.; Lin, V.; Zhang, Z.; Petito, L.C.; Logan, R.W.; Hernán, M.A.; Young, J.G. gfoRmula: An R Package for Estimating the Effects of Sustained Treatment Strategies via the Parametric g-formula. Patterns 2020, 1, 100008. [Google Scholar] [CrossRef]
- Williamson, T.; Ravani, P. Marginal structural models in clinical research: When and how to use them? Nephrol. Dial. Transplant. 2017, 32 (Suppl. 2), ii84–ii90. [Google Scholar] [CrossRef] [PubMed]
- Sterne, J.A.C.; White, I.R.; Carlin, J.B.; Spratt, M.; Royston, P.; Kenward, M.G.; Wood, A.M.; Carpenter, J.R. Multiple imputation for missing data in epidemiological and clinical research: Potential and pitfalls. BMJ 2009, 338, b2393. [Google Scholar] [CrossRef]
- Huque, M.H.; Carlin, J.B.; Simpson, J.A.; Lee, K.J. A comparison of multiple imputation methods for missing data in longitudinal studies. BMC Med. Res. Methodol. 2018, 18, 168. [Google Scholar] [CrossRef]
- Daniel, R.M.; Cousens, S.N.; De Stavola, B.L.; Kenward, M.G.; Sterne, J.A.C. Methods for dealing with time-dependent confounding. Stat. Med. 2013, 32, 1584–1618. [Google Scholar] [CrossRef]
- Greifer, N.; Stuart, E.A. Matching Methods for Confounder Adjustment: An Addition to the Epidemiologist’s Toolbox. Epidemiol. Rev. 2022, 43, 118–129. [Google Scholar] [CrossRef] [PubMed]
- Hansford, H.J.; Cashin, A.G.; Jones, M.D.; Swanson, S.A.; Islam, N.; Dahabreh, I.J.; Dickerman, B.A.; Egger, M.; Garcia-Albeniz, X.; Golub, R.M.; et al. Development of the TrAnsparent ReportinG of observational studies Emulating a Target trial (TARGET) guideline. BMJ Open 2023, 13, e074626. [Google Scholar] [CrossRef] [PubMed]
- Lipsitch, M.; Tchetgen, E.T.; Cohen, T. Negative controls: A tool for detecting confounding and bias in observational studies. Epidemiology 2010, 21, 383–388. [Google Scholar] [CrossRef]
- VanderWeele, T.J.; Ding, P. Sensitivity Analysis in Observational Research: Introducing the E-Value. Ann. Intern. Med. 2017, 167, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Keogh, R.H.; Gran, J.M.; Seaman, S.R.; Davies, G.; Vansteelandt, S. Causal inference in survival analysis using longitudinal observational data: Sequential trials and marginal structural models. Stat. Med. 2023, 42, 2191–2225. [Google Scholar] [CrossRef]
- Su, L.; Rezvani, R.; Seaman, S.R.; Starr, C.; Gravestock, I. TrialEmulation: An R Package to Emulate Target Trials for Causal Analysis of Observational Time-to-Event Data. arXiv 2024, arXiv:2402.12083. [Google Scholar]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Desai, D.; Kantliwala, S.V.; Vybhavi, J.; Ravi, R.; Patel, H.; Patel, J. Review of AlphaFold 3: Transformative Advances in Drug Design and Therapeutics. Cureus 2024, 16, e63646. [Google Scholar] [CrossRef]
- Johnson, K.B.; Wei, W.Q.; Weeraratne, D.; Frisse, M.E.; Misulis, K.; Rhee, K.; Zhao, J.; Snowdon, J.L. Precision Medicine, AI, and the Future of Personalized Health Care. Clin. Transl. Sci. 2021, 14, 86–93. [Google Scholar] [CrossRef]

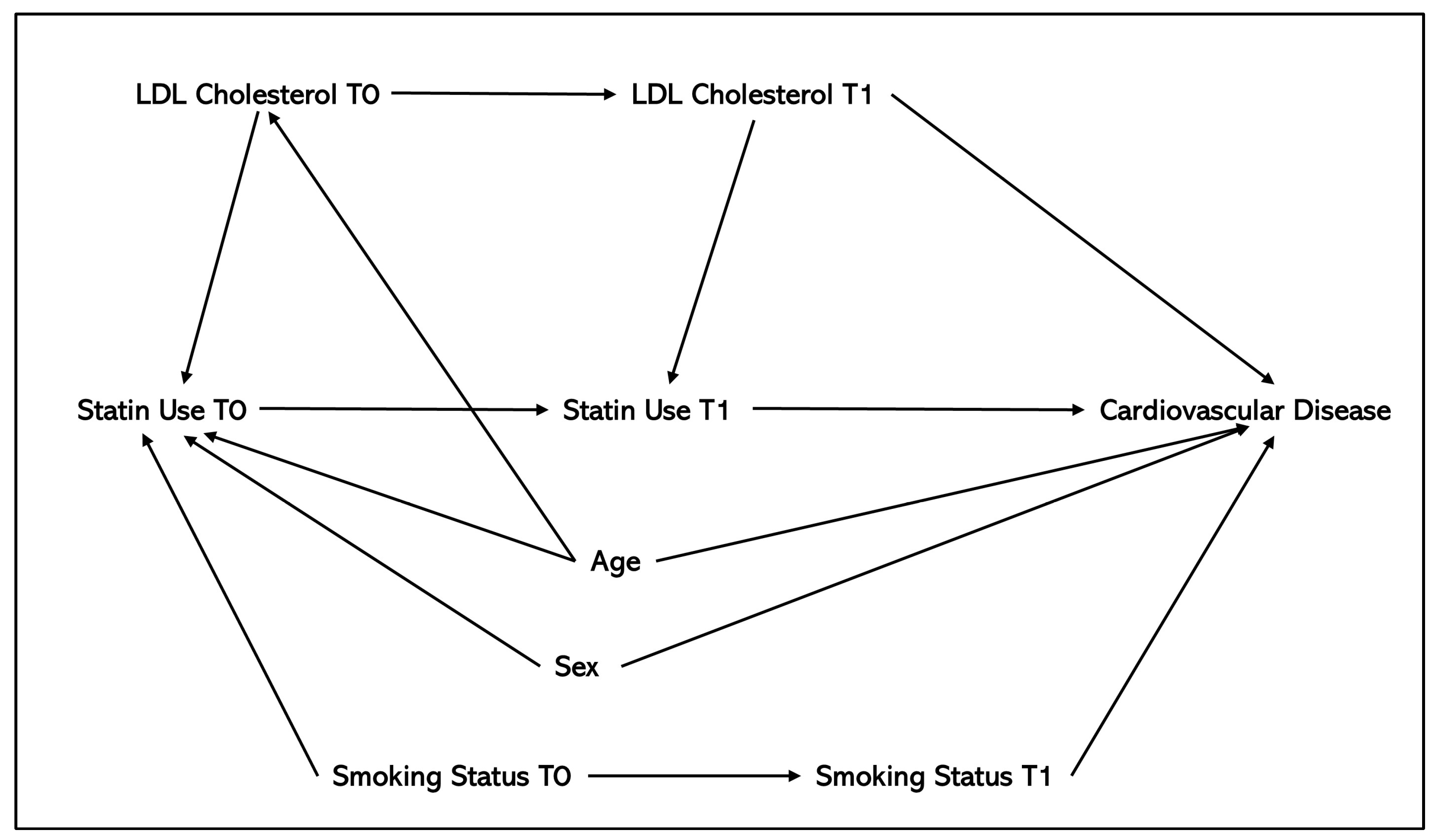
| Method | Overview of Method | Types of Confounding Adjusted for | Number of Application Studies (N = 47) | References of Application Papers | Number of Methodological Studies (N = 12) | References of Methods Papers | Advantages | Disadvantages |
|---|---|---|---|---|---|---|---|---|
| Standard regression adjustment (adjusting for covariates in a regression model) | Inclusion of confounders as covariates in a regression model, e.g., logistic, linear, and Cox regression. | Baseline confounding only. | 18 | [13,16,17,18,19,20,23,25,26,27,31,32,33,37,38,41,42,45] | 2 | [60,67] |
|
|
| Inverse probability of treatment weighting (IPTW) | Weighting individuals by the inverse probability of being assigned the treatment they were actually assigned to conditionally on the individual’s baseline covariates [73]. The weighted observations are then adjusted in a regression model (e.g., weighted logistic regression, weighted Cox model) for estimation of the average treatment effect (ATE). | Baseline and time-varying confounding | 7 | [15,40,44,45,48,53,56] | 0 | Not Applicable * |
|
|
| Propensity score matching (PSM) | Participants receiving the intervention are matched with those receiving the comparator based on similar propensity scores, which represent the probability of treatment assignment given observed covariates [76]. This approach estimates the Average Treatment Effect on the Treated (ATT). Covariate balance is assessed using the standardised mean difference (SMD). | Primarily baseline confounding; however, time-dependent propensity scores have been developed [77]. | 10 | [29,32,33,34,35,36,39,49,56,57] | 2 | [59,70] ** |
|
|
| Clone-censor-weight | Each individual is assigned to all treatment strategies compatible with their data at time zero, creating clones for each strategy. Clones deviating from their assigned strategy are artificially censored, and inverse probability of censoring weighting adjusts for the resulting selection bias [62]. | Baseline, time-varying confounding and immortal-time bias. | 9 | [19,20,21,30,38,41,47,50,54] | 4 | [61,62,65,67] |
|
|
| Inverse probability of censoring weighting (IPCW) | To account for selection bias from artificial censoring, inverse probability of censoring weights are calculated for each individual at all time points, based on the probability of being uncensored, given prior exposure and censoring-related characteristics [62]. These weights are then used in the outcome model (e.g., weighted linear, logistic, or Cox regression). | Time-varying confounding only, but can be combined with IPTW in a marginal structural model or included as part of clone-censor-weighting to adjust for baseline and time-varying confounding. | 10 | [12,22,23,25,26,27,31,43,48,56] | 3 | [55,58,63] |
|
|
| Parametric G-Formula | The g-formula adjusts for time-varying confounders affected by prior exposures using a three-step algorithm. First, it models conditional probabilities from the observed data. Next, it uses these probabilities and baseline covariates to simulate time-varying covariates and outcomes via Monte Carlo sampling for each treatment group. Finally, the datasets are combined, and treatment effects are estimated by comparing hazard ratios across groups using a Cox model [79]. | Baseline and time-varying confounding. | 4 | [17,21,47,58] | 3 | [64,66,68] |
|
|
| Inverse probability weighting as part of a marginal structural model (IPW-MSM) | Rather than using a single treatment weight for baseline confounding, separate treatment weights are calculated at each time point, based on prior treatment, time-varying confounders, and baseline covariates, using a pooled logistic regression model [81]. A separate pooled logistic regression is used to calculate inverse probability of censoring weights for informative censoring. The final weights are obtained by multiplying treatment and censoring weights, which are then adjusted for in the outcome model (e.g., weighted logistic or Cox regression) [81]. | Baseline, time-varying confounding and immortal time bias. | 19 | [14,20,21,22,25,26,27,30,31,36,38,42,46,47,48,51,52,53,58] | 4 | [61,64,65,68] |
|
|
| Multiple imputation (MI) | Multiple imputation (MI) handles missing data by generating multiple datasets, where missing values are replaced with imputed values drawn from their predicted distribution based on available data [82]. | Can be used to adjust for time-varying confounding by imputing missing confounder values, but is primarily used as a missing data technique. | 1 | [55] | 1 | [60] |
|
|
| Nonparametric Bayesian G-computation | Like the parametric G-formula, nonparametric Bayesian G-computation models conditional probabilities from observed data. Using Markov chain Monte Carlo sampling (MCMC), it simulates time-varying covariates and outcomes, with the flexibility to employ models such as Bayesian additive regression trees (BART) and Hilbert S pace Gaussian Processes to capture complex, high-dimensional relationships between covariates, outcome and treatment [69,84]. | Baseline and time-varying confounding | 1 | [55] | 1 | [69] |
|
|
| Exact matching | Matching treated individuals to controls with identical values for all covariates of interest [85]. | Baseline confounding only | 2 | [32,33] | 1 | [63] |
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baldwin, F.D.; Khalaf, R.K.S.; Kolamunnage-Dona, R.; Jorgensen, A.L. Methodologies for the Emulation of Biomarker-Guided Trials Using Observational Data: A Systematic Review. J. Pers. Med. 2025, 15, 195. https://doi.org/10.3390/jpm15050195
Baldwin FD, Khalaf RKS, Kolamunnage-Dona R, Jorgensen AL. Methodologies for the Emulation of Biomarker-Guided Trials Using Observational Data: A Systematic Review. Journal of Personalized Medicine. 2025; 15(5):195. https://doi.org/10.3390/jpm15050195
Chicago/Turabian StyleBaldwin, Faye D., Rukun K. S. Khalaf, Ruwanthi Kolamunnage-Dona, and Andrea L. Jorgensen. 2025. "Methodologies for the Emulation of Biomarker-Guided Trials Using Observational Data: A Systematic Review" Journal of Personalized Medicine 15, no. 5: 195. https://doi.org/10.3390/jpm15050195
APA StyleBaldwin, F. D., Khalaf, R. K. S., Kolamunnage-Dona, R., & Jorgensen, A. L. (2025). Methodologies for the Emulation of Biomarker-Guided Trials Using Observational Data: A Systematic Review. Journal of Personalized Medicine, 15(5), 195. https://doi.org/10.3390/jpm15050195






