Rapid and Label-Free Histopathology of Oral Lesions Using Deep Learning Applied to Optical and Infrared Spectroscopic Imaging Data
Abstract
1. Introduction
2. Materials and Methods
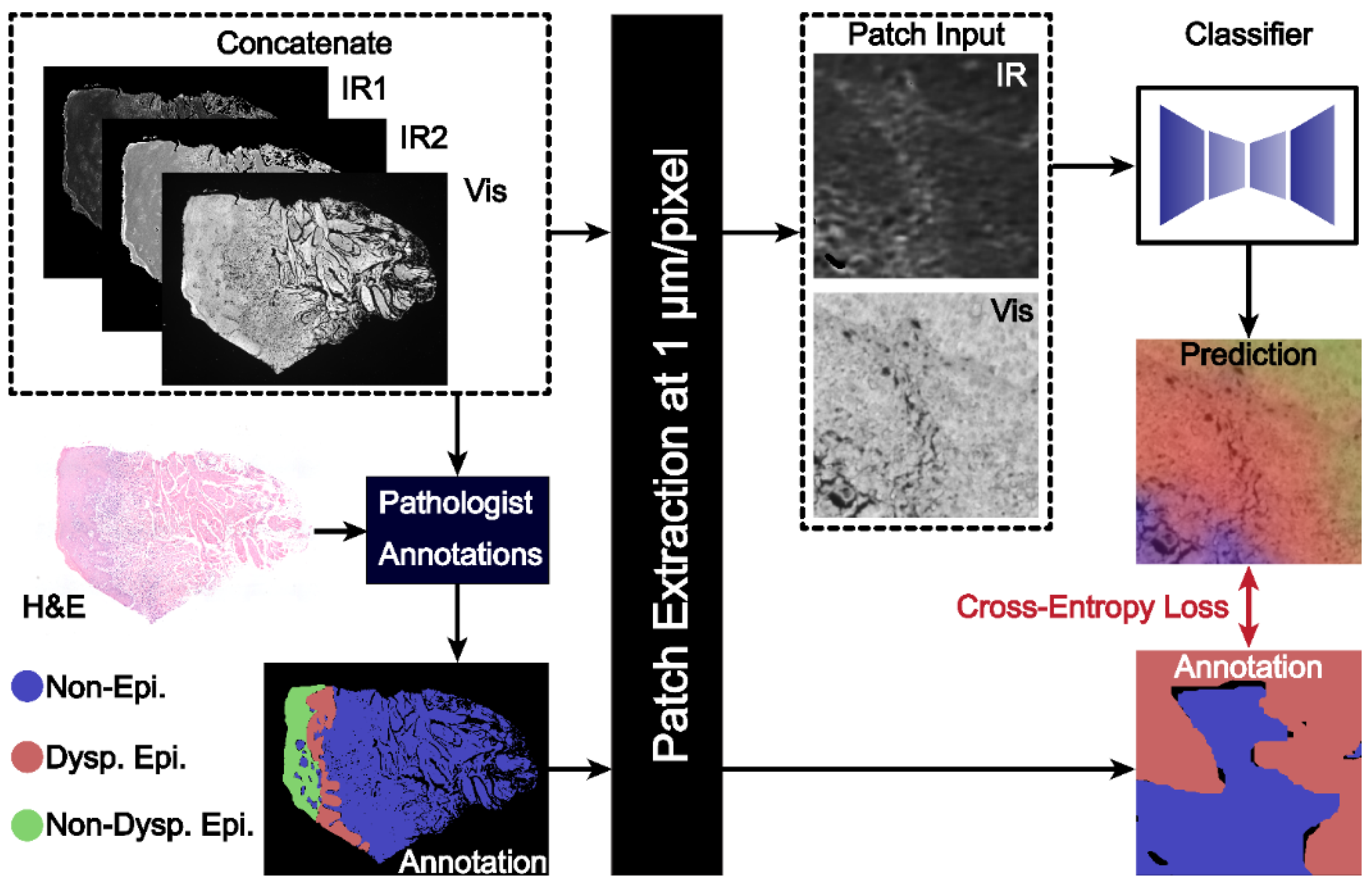
2.1. Sample Preparation and Data Acquisition
2.2. Dataset
2.3. Model Design
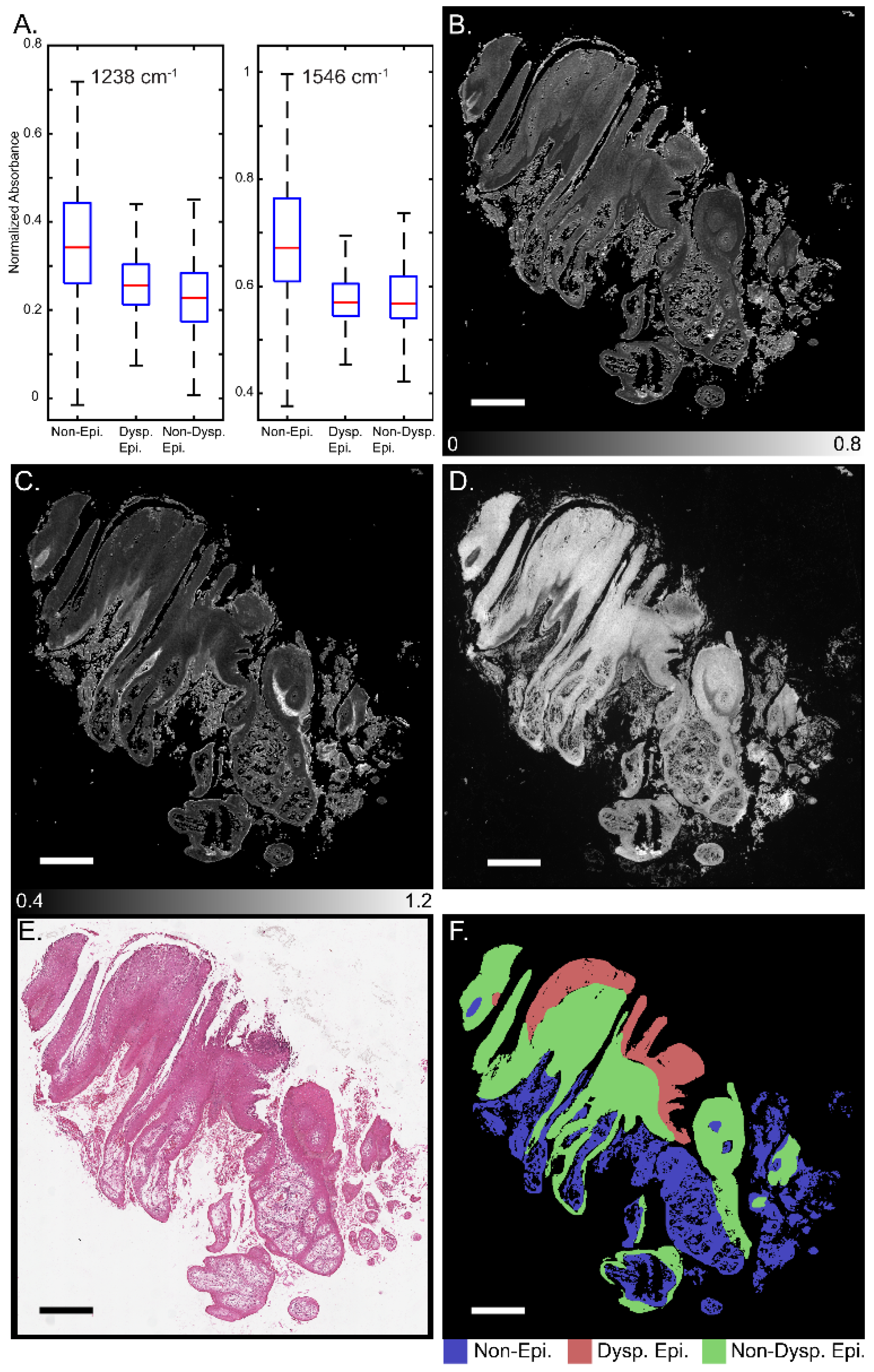
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Warnakulasuriya, S.; Johnson, N.W.; van der Waal, I. Nomenclature and classification of potentially malignant disorders of the oral mucosa. J. Oral. Pathol. Med. 2007, 36, 575–580. [Google Scholar] [CrossRef]
- Warnakulasuriya, S.; Ariyawardana, A. Malignant transformation of oral leukoplakia: A systematic review of observational studies. J. Oral. Pathol. Med. 2016, 45, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Condurache, H.; Oana, M.; Botez, A.E.; Olinici, D.T.; Onofrei, P.; Stoica, L.; Grecu, V.B.; Toader, P.M.; Gheucă Solovăstru, L.; Cotrutz, E.C. Molecular markers associated with potentially malignant oral lesions (Review). Exp. Ther. Med. 2021, 22, 447. [Google Scholar] [CrossRef] [PubMed]
- Slootweg, P.J.; Eveson, J.W. Tumours of the Oral Cavity and Oropharynx. In Pathology and Genetics of Head and Neck Tumours; Barnes, L., Evenson, J.W., Reichart, P., Sidransky, D., Eds.; IARC Press: Lyon, France, 2005. [Google Scholar]
- Takata, T.; Slootweg, P.J. Tumours of the oral cavity and mobile tongue: Epithelial precursor lesions. In WHO Classification of Head and Neck Tumours, 4th ed.; El-Naggar, A.K., Chan, J.K.C., Grandis, J.R., Takata, T., Slootweg, P.J., Eds.; IARC Press: Lyon, France, 2017; pp. 112–114. [Google Scholar]
- Kujan, O.; Oliver, R.J.; Khattab, A.; Roberts, S.A.; Thakker, N.; Sloan, P. Evaluation of a new binary system of grading oral epithelial dysplasia for prediction of malignant transformation. Oral. Oncol. 2006, 42, 987–993. [Google Scholar] [CrossRef] [PubMed]
- Muller, S.; Tilakaratne, W.M. Update from the 5th Edition of the World Health Organization Classification of Head and Neck Tumors: Tumours of the Oral Cavity and Mobile Tongue. Head. Neck Pathol. 2022, 16, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Speight, P.M.; Abram, T.J.; Floriano, P.N.; James, R.; Vick, J.; Thornhill, M.H.; Murdoch, C.; Freeman, C.; Hegarty, A.M.; D’Apice, K.; et al. Inter-observer agreement in dysplasia grading: Towards an enhanced gold standard for clinical pathology trials. Oral Surg. Oral. Med. Oral. Pathol. Oral. Radiol. 2015, 120, 474–482. [Google Scholar] [CrossRef] [PubMed]
- Birur, P.N.; Patrick, S.; Warnakulasuriya, S.; Gurushanth, K.; Raghavan, S.A.; Rath, G.K.; Chaturvedi, P.; Chandru, V.; Mathew, B.; Prabhash, K.; et al. Consensus guidelines on management of oral potentially malignant disorders. Indian J. Cancer 2022, 59, 442–453. [Google Scholar]
- Kujan, O.; Khattab, A.; Oliver, R.J.; Roberts, S.A.; Thakker, N.; Sloan, P. Why oral histopathology suffers inter-observer variability on grading oral epithelial dysplasia: An attempt to understand the sources of variation. Oral. Oncol. 2007, 43, 224–231. [Google Scholar] [CrossRef]
- Mello, F.W.; Melo, G.; Silva Guerra, E.N.; Warnakulasuriya, S.; Garnis, C.; Correa Rivero, E.R. Oral potentially malignant disorders: A scoping review of prognostic biomarkers. Crit. Rev. Oncol. Hematol. 2020, 153, 102986. [Google Scholar] [CrossRef]
- Oya, K.; Kokomoto, K.; Nozaki, K.; Toyosawa, S. Oral squamous cell carcinoma diagnosis in digitized histological images using convolutional neural network. J. Dent. Sci. 2023, 18, 322–329. [Google Scholar] [CrossRef]
- Rekow, E.D. Digital dentistry: The new state of the art—Is it disruptive or destructive. Dent. Mater. 2020, 36, 9–24. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, R.; Seitz, S. The Impact of Technological Innovation on Dentistry. In Biomedical Visualisation: Volume 15-Visualization in Teaching of Biomedical and Clinical Subjects: Anatomy, Advanced Microscopy and Radiology; Springer: Cham, Switzerland, 2023; pp. 79–102. [Google Scholar]
- Baker, M.J.; Trevisan, J.; Bassan, P.; Bhargava, R.; Butler, H.J.; Dorling, K.M.; Fielden, P.R.; Fogarty, S.W.; Fullwood, N.J.; Heys, K.A.; et al. Using Fourier transform IR spectroscopy to analyze biological materials. Nat. Protoc. 2014, 9, 1771–1791. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, R. Digital Histopathology by Infrared Spectroscopic Imaging. Annu. Rev. Anal. Chem. 2023, 16, 205–230. [Google Scholar] [CrossRef] [PubMed]
- Ellis, B.G.; Whitley, C.A.; Triantafyllou, A.; Gunning, P.J.; Smith, C.I.; Barrett, S.D.; Gardner, P.; Shaw, R.J.; Weightman, P.; Risk, J.M. Prediction of malignant transformation in oral epithelial dysplasia using infrared absorbance spectra. PLoS ONE 2022, 17, e0266043. [Google Scholar] [CrossRef]
- Wang, R.; Naidu, A.; Wang, Y. Oral Cancer Discrimination and Novel Oral Epithelial Dysplasia Stratification Using FTIR Imaging and Machine Learning. Diagnostics 2021, 11, 2133. [Google Scholar] [CrossRef]
- Townsend, D.; Milković, M.; Bird, B.; Lenau, K.; Old, O.; Almond, M.; Kendall, C.; Lloyd, G.; Shepherd, N.; Barr, H.; et al. Infrared micro-spectroscopy for cyto-pathological classification of esophageal cells. Analyst 2015, 140, 2215–2223. [Google Scholar] [CrossRef]
- Levin, I.W.; Bhargava, R. Fourier Transform Infrared Vibrational Spectroscopic Imaging: Integrating Microscopy and Molecular Recognition. Annu. Rev. Phys. Chem. 2005, 56, 429–474. [Google Scholar] [CrossRef]
- Petibois, C.; Déléris, G. Chemical mapping of tumor progression by FT-IR imaging: Towards molecular histopathology. Trends Biotechnol. 2006, 24, 455–462. [Google Scholar] [CrossRef]
- Kazarian, S.G.; Chan, K.L. Applications of ATR-FTIR spectroscopic imaging to biomedical samples. Biochim. Biophys. Acta Biomembr. 2006, 1758, 858–867. [Google Scholar] [CrossRef] [PubMed]
- Miller, L.M.; Dumas, P. From structure to cellular mechanism with infrared microspectroscopy. Curr. Opin. Struct. Biol. 2010, 20, 649–956. [Google Scholar] [CrossRef]
- Diem, M.; Mazur, A.; Lenau, K.; Schubert, J.; Bird, B.; Miljković, M.; Krafft, C.; Popp, J. Molecular pathology via IR and Raman spectral imaging. J. Biophotonics 2013, 6, 855–886. [Google Scholar] [CrossRef] [PubMed]
- Pilling, M.; Gardner, P. Fundamental developments in infrared spectroscopic imaging for biomedical applications. Chem. Soc. Rev. 2016, 45, 1935–1957. [Google Scholar] [CrossRef] [PubMed]
- Talari, A.C.; Martinez, M.A.; Movasaghi, Z.; Rehman, S.; Rehman, I.U. Advances in Fourier transform infrared (FTIR) spectroscopy of biological tissues. Appl. Spectrosc. Rev. 2017, 52, 456–506. [Google Scholar] [CrossRef]
- Kumar, S.; Srinivasan, A.; Nikolajeff, F. Role of infrared spectroscopy and imaging in cancer diagnosis. Curr. Med. Chem. 2018, 25, 1055–1072. [Google Scholar] [CrossRef]
- Hermes, M.; Morrish, R.B.; Huot, L.; Meng, L.; Junaid, S.; Tomko, J.; Lloyd, G.R.; Masselink, W.T.; Tidemand-Lichtenberg, P.; Pedersen, C.; et al. Mid-IR hyperspectral imaging for label-free histopathology and cytology. J. Opt. 2018, 20, 023002. [Google Scholar] [CrossRef]
- Fernandez, D.C.; Bhargava, R.; Hewitt, S.M.; Levin, I.W. Infrared spectroscopic imaging for histopathologic recognition. Nat. Biotechnol. 2005, 23, 469–474. [Google Scholar] [CrossRef]
- Yeh, K.; Lee, D.; Bhargava, R. Multicolor discrete frequency infrared spectroscopic imaging. Anal. Chem. 2019, 91, 2177–2185. [Google Scholar] [CrossRef] [PubMed]
- Yeh, K.; Sharma, I.; Falahkheirkhah, K.; Confer, M.P.; Orr, A.C.; Liu, Y.T.; Phal, Y.; Ho, R.J.; Mehta, M.; Bhargava, A.; et al. Infrared spectroscopic laser scanning confocal microscopy for whole-slide chemical imaging. Nat. Commun. 2023, 14, 5215. [Google Scholar] [CrossRef]
- Pounder, F.N.; Reddy, R.K.; Bhargava, R. Development of a practical spatial-spectral analysis protocol for breast histopathology using Fourier transform infrared spectroscopic imaging. Faraday Discuss. 2016, 187, 43–68. [Google Scholar] [CrossRef]
- Schnell, M.; Mittal, S.; Falahkheirkhah, K.; Mittal, A.; Yeh, K.; Kenkel, S.; Kajdacsy-Balla, A.; Carney, P.S.; Bhargava, R. All-digital histopathology by infrared-optical hybrid microscopy. Proc. Natl. Acad. Sci. USA 2020, 117, 3388–3396. [Google Scholar] [CrossRef]
- Li, M.; Razumtcev, A.; Yang, R.; Liu, Y.; Rong, J.; Geiger, A.C.; Blanchard, R.; Pfluegl, C.; Taylor, L.S.; Simpson, G.J. Fluorescence-Detected Mid-Infrared Photothermal Microscopy. J. Am. Chem. Soc. 2021, 143, 10809–10815. [Google Scholar] [CrossRef]
- Zhang, Y.; Zong, H.; Zong, C.; Tan, Y.; Zhang, M.; Zhan, Y.; Cheng, J.X. Fluorescence-Detected Mid-Infrared Photothermal Microscopy. J. Am. Chem. Soc. 2021, 143, 11490–11499. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Li, C.; Zhang, C.; Slipchenko, M.N.; Eakins, G.; Cheng, J.X. Depth-resolved mid-infrared photothermal imaging of living cells and organisms with submicrometer spatial resolution. Sci. Adv. 2016, 2, e1600521. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Yin, J.; Cheng, J.X. Bond-selective imaging by optically sensing the mid-infrared photothermal effect. Sci. Adv. 2021, 7, eabg1559. [Google Scholar] [CrossRef] [PubMed]
- Kenkel, S.; Gryka, M.; Chen, L.; Confer, M.P.; Rao, A.; Robinson, S.; Prasanth, K.V.; Bhargava, R. Chemical imaging of cellular ultrastructure by null-deflection infrared spectroscopic measurements. Proc. Natl. Acad. Sci. USA 2022, 119, e2210516119. [Google Scholar] [CrossRef]
- Dazzi, A.; Prater, C.B. AFM-IR: Technology and applications in nanoscale infrared spectroscopy and chemical imaging. Chem. Rev. 2017, 117, 5146–5173. [Google Scholar] [CrossRef]
- Mathurin, J.; Deniset-Besseau, A.; Bazin, D.; Dartois, E.; Wagner, M.; Dazzi, A. Photothermal AFM-IR spectroscopy and imaging: Status, challenges, and trends. J. Appl. Phys. 2022, 131, 010901. [Google Scholar] [CrossRef]
- Kurouski, D.; Dazzi, A.; Zenobi, R.; Centrone, A. Infrared and Raman chemical imaging and spectroscopy at the nanoscale. Chem. Soc. Rev. 2020, 49, 3315–3347. [Google Scholar] [CrossRef]
- Schwartz, J.J.; Jakob, D.S.; Centrone, A. A guide to nanoscale IR spectroscopy: Resonance enhanced transduction in contact and tapping mode AFM-IR. Chem. Soc. Rev. 2022, 51, 5248–5267. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep Learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Chen, L.-C.; Papandreou, G.; Schroff, F.; Adam, H. Rethinking Atrous Convolution for Sematic Image Segmentation. arXiv 2017, arXiv:1706.05587. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Networks. arXiv 2014, arXiv:1406.2661. [Google Scholar] [CrossRef]
- Echle, A.; Rindtorff, N.T.; Brinker, T.J.; Luedde, T.; Pearson, A.T.; Kather, J.N. Deep learning in cancer pathology: A new generation of clinical biomarkers. Br. J. Cancer 2020, 124, 686–696. [Google Scholar] [CrossRef] [PubMed]
- Mayerich, D.; Walsh, M.J.; Kadjacsy-Balla, A.; Ray, P.S.; Hewitt, S.M.; Bhargava, R. Stain-less staining for computed histopathology. Technology 2015, 3, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Falahkheirkhah, K.; Tiwari, S.; Yeh, K.; Gupta, S.; Herrera-Hernandez, L.; McCarthy, M.R.; Jimenez, R.E.; Cheville, J.C.; Bhargava, R. Deepfake Histologic Images for Enhancing Digital Pathology. Lab. Investig. 2023, 103, 100006. [Google Scholar] [CrossRef] [PubMed]
- Brunel, B.; Prada, P.; Slimano, F.; Boulagnon-Rombi, C.; Bouché, O.; Piot, O. Deep learning for the prediction of the chemotherapy response of metastatic colorectal cancer: Comparing and combining H&E staining histopathology and infrared spectral histopathology. Analyst 2023, 148, 3909–3917. [Google Scholar]
- Gerwert, K.; Schörner, S.; Broßerueschkamo, F.; Kraeft, A.-L.; Schuhmacher, D.; Sternemann, C.; Feder, I.S.; Wisser, S.; Lugnier, C.; Arnold, D.; et al. Fast and label-free automated detection of microsatellite status in early colon cancer using artificial intelligence integrated infrared imaging. Eur. J. Cancer 2023, 182, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Mittal, S.; Yeh, K.; Leslie, L.S.; Kenkel, S.; Kajdacsy-Balla, A.; Bhargava, R. Simultaneous Cancer and Tumor Microenvironment Subtyping using Confocal Infrared Microscopy for All-Digital Molecular Histopathology. Proc. Natl. Acad. Sci. USA 2018, 115, e5651–e5660. [Google Scholar] [CrossRef]
- Long, J.; Shelhamer, E.; Darrell, T. Fully Convolutional Networks for Semantic Segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. arXiv 2015, arXiv:1512.03385. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Cai, S.; Singh, B.R. Identification of β-turn and random coil amide III infrared bands for secondary structure estimation of proteins. Biophys. Chem. 1999, 80, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Anderle, G.; Mendelsohn, R. Thermal Denauturation of Globular Proteins Fourier Transform-Infrared Studies of the Amide III Spectral Region. Biophys. J. 1987, 52, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Matsui, T.; Tanaka, S. Quantiative Estimation of α-Helix Coil Content in Bovine Serum Albumin by Fourier Transform-Infrared Spectroscopy. Appl. Spectrosc. 1987, 41, 861–865. [Google Scholar] [CrossRef]
- Ataka, S.; Takeuchi, H.; Tasumi, M. Infrared studies of the less stable cis form of N-methylformamide and N-methylacetamide in low-temperature nitrogen matrices and vibrational analyses of the trans and cis forms of these molecules. J. Mol. Struct. 1984, 113, 147–160. [Google Scholar] [CrossRef]
- Gaigeot, M.P.; Vuilleumier, R.; Sprik, M.; Borgis, D. Infrared Spectroscopy of N-Methylacetamide Revisited by ab Initio Molecular Dynamics Simulations. J. Chem. Theory Comput. 2005, 1, 772–789. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Yang, X.; Ji, Z.; Zhu, L.; Ma, N.; Chen, D.; Jia, X.; Tang, J.; Cao, Y. DFT-Calculated IR Spectrum Amide I, II, and III Band Contributions of N-Methylacetamide Fine Components. ACS Omega 2020, 5, 8572–8578. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Mukamel, S. Two-dimensional vibrational lineshapes of amide III, II, I and A bands in a helical peptide. J. Mol. Liq. 2008, 141, 149–154. [Google Scholar] [CrossRef]
- Bradley, G.; Odell, E.W.; Raphael, S.; Ho, J.; Le, L.W.; Benchimol, S.; Kamel-Reid, S. Abnormal DNA Content in Oral Epithelial Dysplasia is Associated with Increased Risk of Progression to Carcinoma. Br. J. Cancer 2010, 103, 1432–1442. [Google Scholar] [CrossRef]
- Jara-Lazaro, A.R.; Thamboo, T.P.; Teh, M.; Tan, P.H. Digital pathology: Exploring its applications in diagnostic surgical pathology practice. Pathology 2010, 42, 512–518. [Google Scholar] [CrossRef]
- Remmerbach, T.W.; Meyer-Ebrecht, D.; Aach, T.; Würflinger, T.; Bell, A.A.; Schneider, T.E.; Nietzke, N.; Frerich, B.; Böcking, A. Toward a multimodal cell analysis of brush biopsies for the early detection of oral cancer. Cancer Cytopathol. 2009, 117, 228–235. [Google Scholar] [CrossRef]
- Tiwari, S.; Kajdacsy-Balla, A.; Whiteley, J.; Cheng, G.; Jirstrom, K.; Birgisson, H.; Hewitt, S.M.; Bhargava, R. INFORM: IN-Frared-Based Organizational Measurements of Tumor and its Microenvironmental to Predict Patient Survival. Sci. Adv. 2021, 7, eabb8292. [Google Scholar] [CrossRef] [PubMed]
- Shubhasini, A.R.; Praveen, B.N.; Usha, H.; Uma, K.; Shubha, G.; Keerthi, G.; Sil, S. Inter- and Intra-Observer Variability in Diagnosis of Oral Dysplasia. Asian Pac. J. Cancer Prev. 2017, 18, 3251–3254. [Google Scholar]
- Ranganathan, K.; Kavitha, L.; Sharada, P.; Bavle, R.M.; Rao, R.S.; Pattanshetty, S.M.; Hazarey, V.K.; Madhura, M.G.; Nagaraj, T.; Lingappa, A.; et al. Intra-Observer and Inter-Observer Variability in Two Grading Systems for Oral Epithelial Dysplasia: A Multi-Centre Study in India. J. Oral. Pathol. Med. 2020, 49, 948–955. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Confer, M.P.; Falahkheirkhah, K.; Surendran, S.; Sunny, S.P.; Yeh, K.; Liu, Y.-T.; Sharma, I.; Orr, A.C.; Lebovic, I.; Magner, W.J.; et al. Rapid and Label-Free Histopathology of Oral Lesions Using Deep Learning Applied to Optical and Infrared Spectroscopic Imaging Data. J. Pers. Med. 2024, 14, 304. https://doi.org/10.3390/jpm14030304
Confer MP, Falahkheirkhah K, Surendran S, Sunny SP, Yeh K, Liu Y-T, Sharma I, Orr AC, Lebovic I, Magner WJ, et al. Rapid and Label-Free Histopathology of Oral Lesions Using Deep Learning Applied to Optical and Infrared Spectroscopic Imaging Data. Journal of Personalized Medicine. 2024; 14(3):304. https://doi.org/10.3390/jpm14030304
Chicago/Turabian StyleConfer, Matthew P., Kianoush Falahkheirkhah, Subin Surendran, Sumsum P. Sunny, Kevin Yeh, Yen-Ting Liu, Ishaan Sharma, Andres C. Orr, Isabella Lebovic, William J. Magner, and et al. 2024. "Rapid and Label-Free Histopathology of Oral Lesions Using Deep Learning Applied to Optical and Infrared Spectroscopic Imaging Data" Journal of Personalized Medicine 14, no. 3: 304. https://doi.org/10.3390/jpm14030304
APA StyleConfer, M. P., Falahkheirkhah, K., Surendran, S., Sunny, S. P., Yeh, K., Liu, Y.-T., Sharma, I., Orr, A. C., Lebovic, I., Magner, W. J., Sigurdson, S. L., Aguirre, A., Markiewicz, M. R., Suresh, A., Hicks, W. L., Jr., Birur, P., Kuriakose, M. A., & Bhargava, R. (2024). Rapid and Label-Free Histopathology of Oral Lesions Using Deep Learning Applied to Optical and Infrared Spectroscopic Imaging Data. Journal of Personalized Medicine, 14(3), 304. https://doi.org/10.3390/jpm14030304




