Rare Mutations in CCDC7 Contribute to Early-Onset Preeclampsia by Inhibiting Trophoblast Migration and Invasion
Abstract
1. Introduction
2. Methods
2.1. Patients
2.2. Whole-Exome Sequencing and Data Analysis
2.3. Cell Culture
2.4. Generation of Stable CCDC7 Knockdown Cell Lines
2.5. RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction
2.6. Protein Extraction and Western Blot Analysis
2.7. Immunofluorescence
2.8. Wound-Healing Assay
2.9. Transwell Invasion Assay
2.10. Statistical Analysis
3. Results
3.1. Clinical Features and Rare Variants of CCDC7 Detected in Two EOPE Families
3.2. Whole-Exome Sequencing Identified Rare Variants in CCDC7 in Familial Preeclampsia
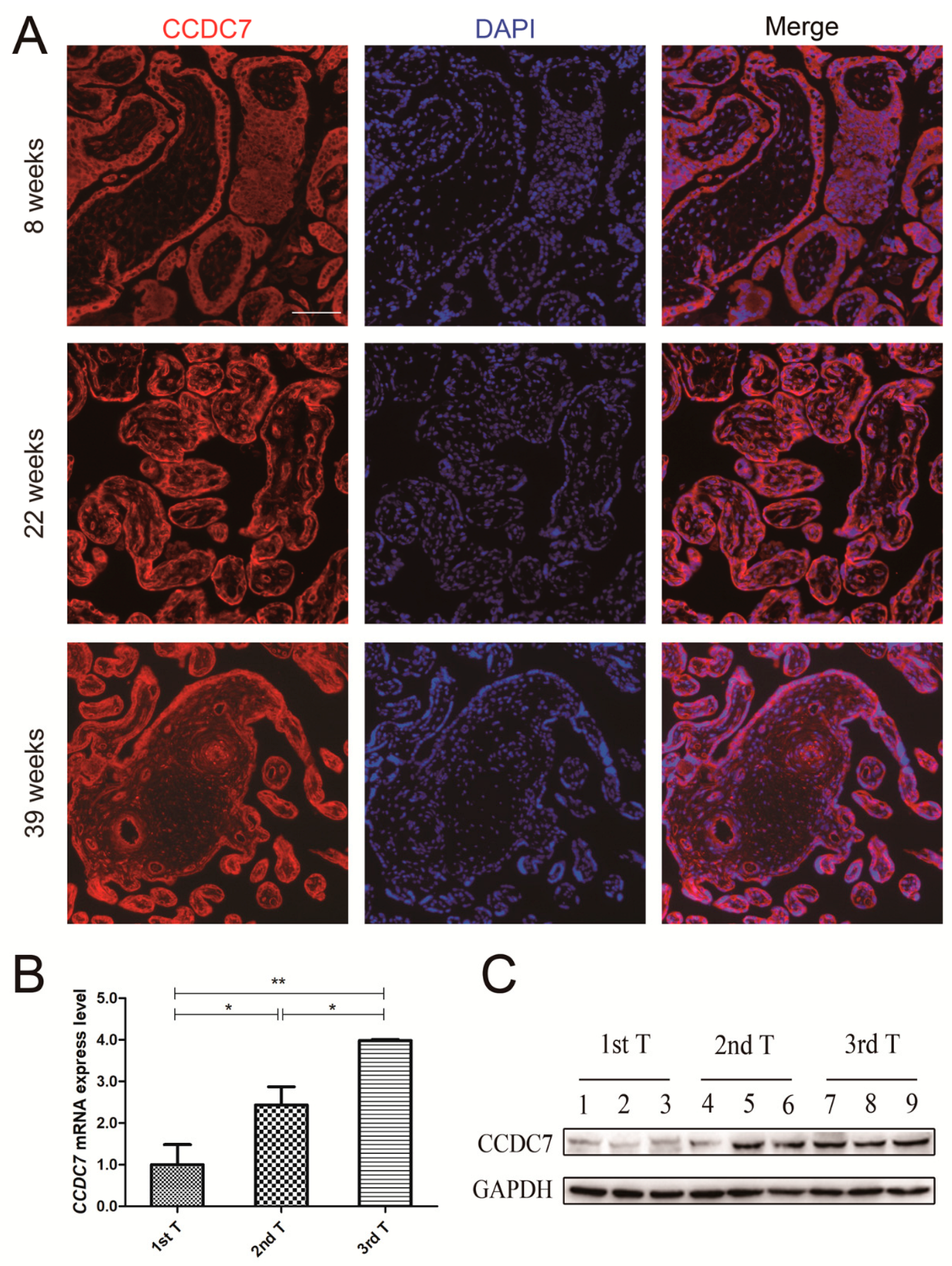
3.3. CCDC7 Spatial and Temporal Expression Patterns in the Human Placenta
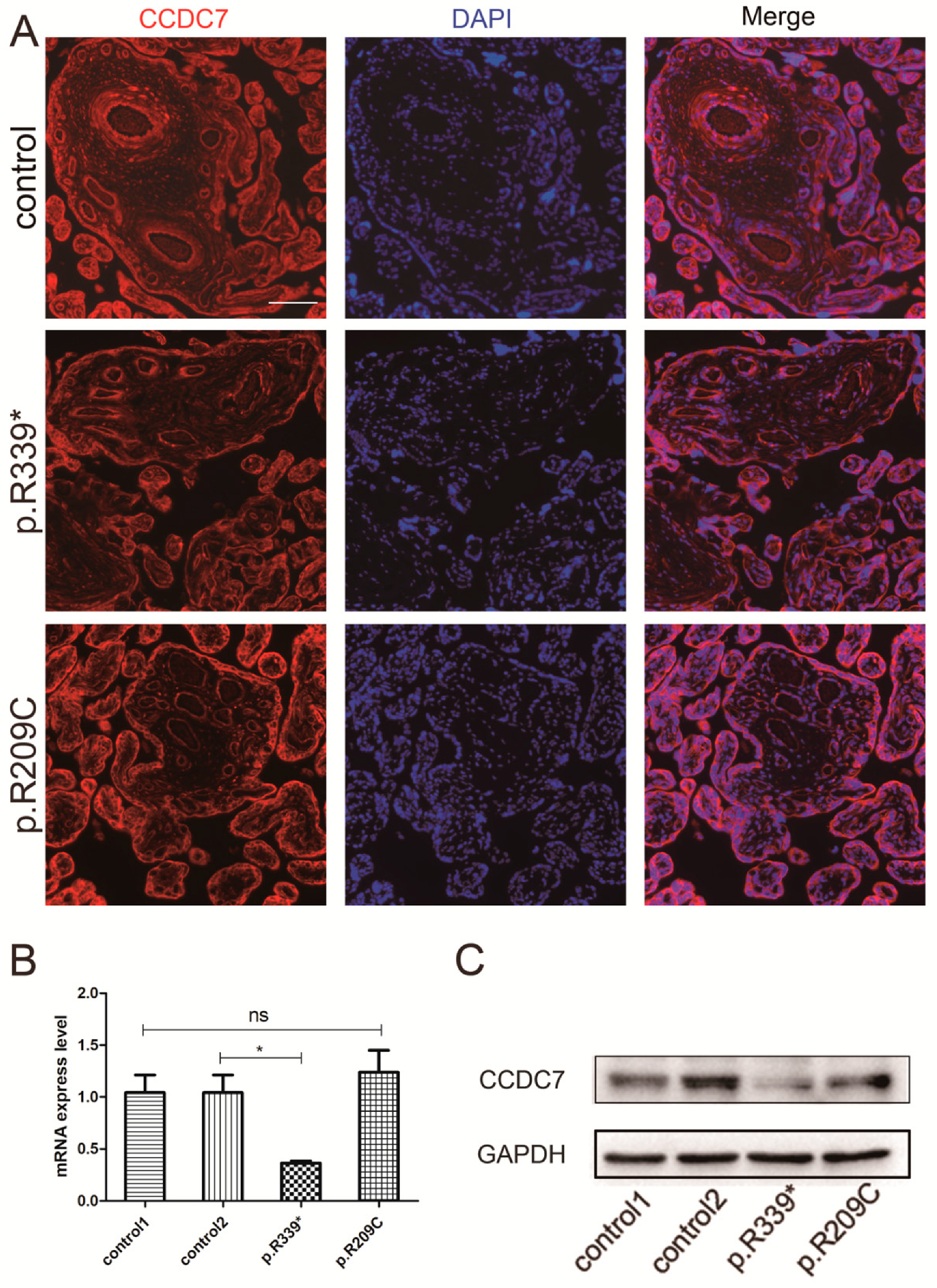
3.4. Expression and Distribution of Rare CCDC7 Variants in the Placenta
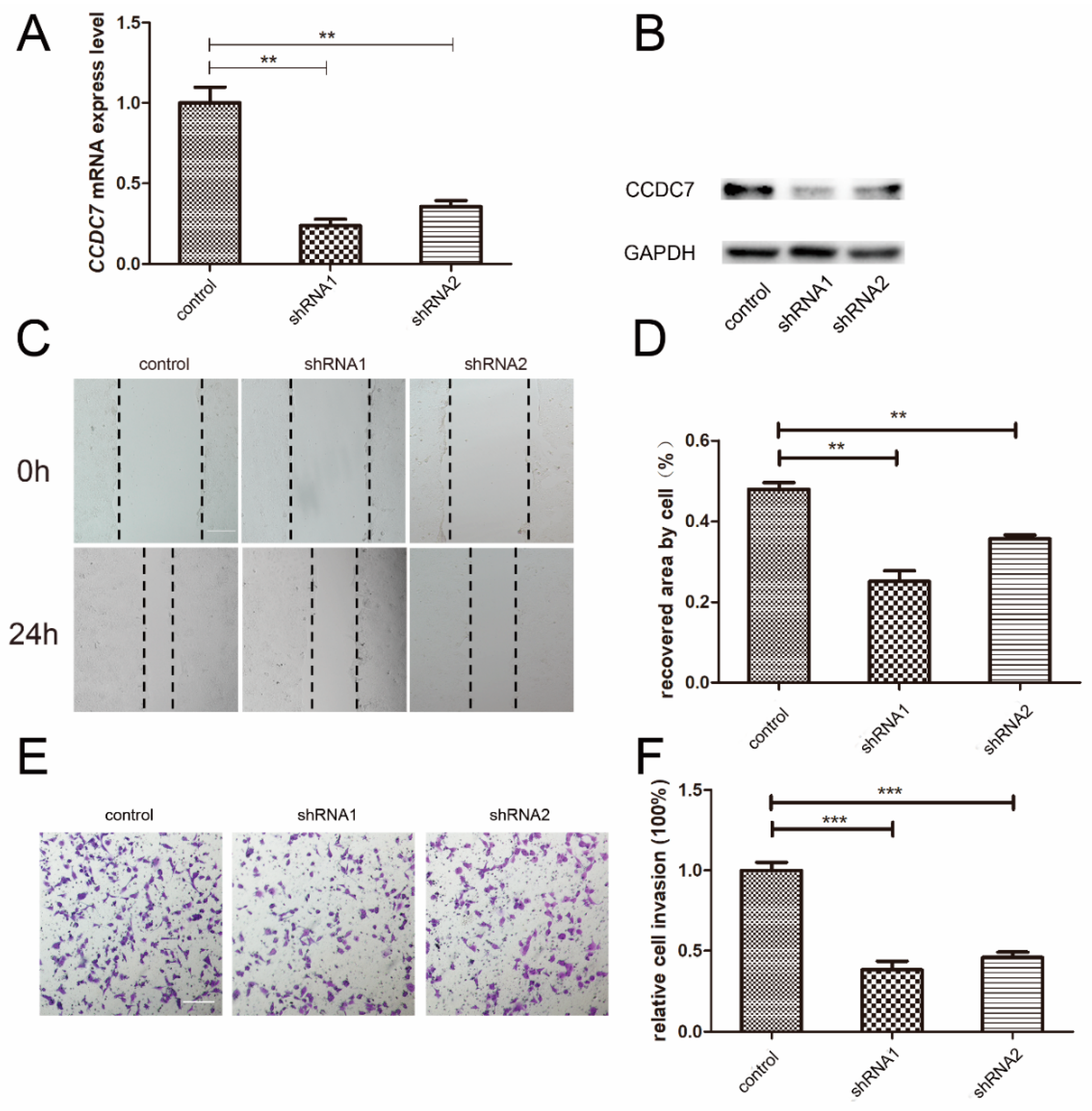
3.5. Effects of CCDC7 on JEG-3 Cell Migration and Invasion
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CCDC | Coiled-coil domain-containing |
| CTBs | cytotrophoblast cells |
| EOPE | Early-onset preeclampsia |
| GWAS | Genome-wide association studies |
| LOPE | Late-onset preeclampsia |
| MMPs | Matrix metalloproteinases |
| PVDF | Polyvinylidene fluoride |
| qRT-PCR | Quantitative real-time polymerase chain reaction |
| SDS-PAGE | Sodium dodecyl sulfate-polyacrylamide gel-electrophoresis |
| STBs | Syncytiotrophoblasts |
| WES | Whole-exome sequencing |
| WGS | Whole-genome sequencing |
References
- ACOG. Practice Bulletin No. 202: Gestational Hypertension and Preeclampsia. Obstet. Gynecol. 2019, 133, 1. [Google Scholar] [CrossRef]
- Dimitriadis, E.; Rolnik, D.L.; Zhou, W.; Estrada-Gutierrez, G.; Koga, K.; Francisco, R.P.V.; Whitehead, C.; Hyett, J.; Costa, F.d.S.; Nicolaides, K.; et al. Pre-eclampsia. Nat. Rev. Dis. Primers 2023, 9, 8. [Google Scholar] [CrossRef]
- Nilsson, E.; Ros, H.S.; Cnattingius, S.; Lichtenstein, P. The importance of genetic and environmental effects for pre-eclampsia and gestational hypertension: A family study. BJOG 2004, 111, 200. [Google Scholar] [CrossRef]
- Mogren, I.; Högberg, U.; Winkvist, A.; Stenlund, H. Familial occurrence of preeclampsia. Epidemiology 1999, 10, 518. [Google Scholar] [CrossRef]
- Parada-Niño, L.; Castillo-León, L.F.; Morel, A. Morel, Preeclampsia, Natural History, Genes, and miRNAs Associated with the Syndrome. J. Pregnancy 2022, 2022, 3851225. [Google Scholar] [CrossRef] [PubMed]
- Cnattingius, S.; Reilly, M.; Pawitan, Y.; Lichtenstein, P. Maternal and fetal genetic factors account for most of familial aggregation of preeclampsia: A population-based Swedish cohort study. Am. J. Med. Genet. A 2004, 130A, 365. [Google Scholar] [CrossRef] [PubMed]
- Gray, K.J.; Saxena, R.; Karumanchi, S.A. Genetic predisposition to preeclampsia is conferred by fetal DNA variants near FLT1, a gene involved in the regulation of angiogenesis. Am. J. Obstet. Gynecol. 2018, 218, 211. [Google Scholar] [CrossRef] [PubMed]
- van Dijk, M.; Mulders, J.; Poutsma, A.; Könst, A.A.M.; A Lachmeijer, A.M.; A Dekker, G.; A Blankenstein, M.; Oudejans, C.B.M. Maternal segregation of the Dutch preeclampsia locus at 10q22 with a new member of the winged helix gene family. Nat. Genet. 2005, 37, 514. [Google Scholar] [CrossRef] [PubMed]
- Dunk, C.E.; van Dijk, M.; Choudhury, R.; Wright, T.J.; Cox, B.; Leavey, K.; Harris, L.K.; Jones, R.L.; Lye, S.J. Functional Evaluation of STOX1 (STORKHEAD-BOX PROTEIN 1) in Placentation, Preeclampsia, and Preterm Birth. Hypertension 2021, 77, 475. [Google Scholar] [CrossRef] [PubMed]
- van Dijk, M.; van Bezu, J.; van Abel, D.; Dunk, C.; Blankenstein, M.A.; Oudejans, C.B.; Lye, S.J. The STOX1 genotype associated with pre-eclampsia leads to a reduction of trophoblast invasion by alpha-T-catenin upregulation. Hum. Mol. Genet. 2010, 19, 2658. [Google Scholar] [CrossRef] [PubMed]
- Iglesias-Platas, I.; Monk, D.; Jebbink, J.; Buimer, M.; Boer, K.; van der Post, J.; Hills, F.; Apostolidou, S.; Ris-Stalpers, C.; Stanier, P.; et al. STOX1 is not imprinted and is not likely to be involved in preeclampsia. Nat. Genet. 2007, 39, 279–280. [Google Scholar] [CrossRef]
- Kivinen, K.; Peterson, H.; Hiltunen, L.; Laivuori, H.; Heino, S.; Tiala, I.; Knuutila, S.; Rasi, V.; Kere, J. Evaluation of STOX1 as a preeclampsia candidate gene in a population-wide sample. Eur. J. Hum. Genet. 2007, 15, 494. [Google Scholar] [CrossRef]
- McGinnis, R.; The FINNPEC Consortium; Steinthorsdottir, V.; O Williams, N.; Thorleifsson, G.; Shooter, S.; Hjartardottir, S.; Bumpstead, S.; Stefansdottir, L.; Hildyard, L.; et al. Variants in the fetal genome near FLT1 are associated with risk of preeclampsia. Nat. Genet. 2017, 49, 1255. [Google Scholar] [CrossRef] [PubMed]
- Ghralaigh, F.N.; Gallagher, L.; Lopez, L.M. Autism spectrum disorder genomics: The progress and potential of genomic technologies. Genomics 2020, 112, 5136. [Google Scholar] [CrossRef] [PubMed]
- Hansen, R.D.; Christensen, A.F.; Olesen, J. Family studies to find rare high risk variants in migraine. J. Headache Pain. 2017, 18, 32. [Google Scholar] [CrossRef] [PubMed]
- Fritsche, L.G.; Igl, W.; Bailey, J.N.C.; Grassmann, F.; Sengupta, S.; Bragg-Gresham, J.L.; Burdon, K.P.; Hebbring, S.J.; Wen, C.; Gorski, M.; et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat. Genet. 2016, 48, 134. [Google Scholar] [CrossRef] [PubMed]
- Huusko, J.M.; Karjalainen, M.K.; Graham, B.E.; Zhang, G.; Farrow, E.G.; Miller, N.A.; Jacobsson, B.; Eidem, H.R.; Murray, J.C.; Bedell, B.; et al. Whole exome sequencing reveals HSPA1L as a genetic risk factor for spontaneous preterm birth. PLoS Genet. 2018, 14, e1007394. [Google Scholar] [CrossRef]
- Liu, X.; Lai, H.; Xin, S.; Li, Z.; Zeng, X.; Nie, L.; Liang, Z.; Wu, M.; Zheng, J.; Zou, Y. Whole-exome sequencing identifies novel mutations in ABC transporter genes associated with intrahepatic cholestasis of pregnancy disease: A case-control study. BMC Pregnancy Childbirth. 2021, 21, 110. [Google Scholar] [CrossRef] [PubMed]
- Canfield, J.; Arlier, S.; Mong, E.F.; Lockhart, J.; Van Wye, J.; Guzeloglu-Kayisli, O.; Schatz, F.; Magness, R.R.; Lockwood, C.J.; Tsibris, J.C.M.; et al. Decreased LIN28B in preeclampsia impairs human trophoblast differentiation and migration. Faseb. J. 2019, 33, 2759. [Google Scholar] [CrossRef] [PubMed]
- Phipps, E.A.; Thadhani, R.; Benzing, T.; Karumanchi, S.A. Pre-eclampsia: Pathogenesis, novel diagnostics and therapies. Nat. Rev. Nephrol. 2019, 15, 275. [Google Scholar] [CrossRef]
- Burton, G.J.; Redman, C.W.; Roberts, J.M.; Moffett, A. Pre-eclampsia: Pathophysiology and clinical implications. BMJ 2019, 366, l2381. [Google Scholar] [CrossRef] [PubMed]
- Melton, P.E.; Johnson, M.P.; Gokhale-Agashe, D.; Rea, A.J.; Ariff, A.; Cadby, G.; Peralta, J.M.; McNab, T.J.; Allcock, R.J.; Abraham, L.J.; et al. Whole-exome sequencing in multiplex preeclampsia families identifies novel candidate susceptibility genes. J. Hypertens. 2019, 37, 997. [Google Scholar] [CrossRef] [PubMed]
- Linhares, N.D.; Conceição, I.M.; Sandrim, V.C.; Luizon, M.R. Candidate genes identified by whole-exome sequencing in preeclampsia families: Insights into functional annotation and in-silico prediction of deleterious variants. J. Hypertens. 2020, 38, 372. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wu, X.; Dai, W.; Li, J.; Xiang, L.; Tang, W.; Lin, J.; Zhang, W.; Liu, G.; Yang, Q.; et al. The CCDC43-ADRM1 axis regulated by YY1, promotes proliferation and metastasis of gastric cancer. Cancer Lett. 2020, 482, 90. [Google Scholar] [CrossRef] [PubMed]
- Omari, S.; Lee, H.; Wang, J.; Zeng, S.X.; Lu, H. Extracellular and intracellular functions of coiled-coil domain containing 3. J. Mol. Cell Biol. 2023, 15, mjad037. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chen, S.; Jia, W.; Qian, Y.; Yang, X.; Zhang, M.; Fang, T.; Liu, H. Comprehensive analysis reveals CCDC60 as a potential biomarker correlated with prognosis and immune infiltration of head and neck squamous cell carcinoma. Front Oncol. 2023, 13, 1113781. [Google Scholar] [CrossRef]
- Shen, Y.-M.; He, X.; Deng, H.-X.; Xie, Y.-P.; Wang, C.-T.; Wei, Y.-Q.; Zhao, X. Overexpression of the hBiot2 gene is associated with development of human cervical cancer. Oncol. Rep. 2011, 25, 75. [Google Scholar]
- Sun, X.-F.; Shen, Y.-M.; Arbman, G.; Sandström, P.; Gullstrand, P.; Wei, Y.-Q.; Zhang, H.; Rosell, J.; Olsson, B.; Peng, F.; et al. Novel gene hBiot2 is an independent prognostic factor in colorectal cancer patients. Oncol. Rep. 2012, 27, 376. [Google Scholar] [CrossRef]
- Costanzo, V.; Costanzo, V.; Bardelli, A.; Bardelli, A.; Siena, S.; Siena, S.; Abrignani, S.; Abrignani, S. Exploring the links between cancer and placenta development. Open Biol. 2018, 8, 180081. [Google Scholar] [CrossRef]
- Zhang, X.-F.; An, M.-Z.; Ma, Y.-P.; Lu, Y.-M. Regulatory effects of CCDC3 on proliferation, migration, invasion and EMT of human cervical cancer cells. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 3217. [Google Scholar] [CrossRef]
- Feng, Y.; Gao, Y.; Yu, J.; Jiang, G.; Zhang, X.; Lin, X.; Han, Q.; Rong, X.; Xu, H.; Li, Q.; et al. CCDC85B promotes non-small cell lung cancer cell proliferation and invasion. Mol. Carcinog. 2019, 58, 126. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.L.; Pjanic, M.; Wang, T.; Nguyen, T.; Cohain, A.; Lee, J.D.; Perisic, L.; Hedin, U.; Kundu, R.K.; Majmudar, D.; et al. Integrative functional genomics identifies regulatory mechanisms at coronary artery disease loci. Nat. Commun. 2016, 7, 12092. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Deng, G.; Wei, R.; Wang, Q.; Zou, D.; Wei, J. Comprehensive Identification of Key Genes Involved in Development of Diabetes Mellitus-Related Atherogenesis Using Weighted Gene Correlation Network Analysis. Front. Cardiovasc. Med. 2020, 7, 580573. [Google Scholar] [CrossRef] [PubMed]
- Laulumaa, S.; Varjosalo, M. Commander Complex-A Multifaceted Operator in Intracellular Signaling and Cargo. Cells 2021, 10, 3447. [Google Scholar] [CrossRef]
- Zhang, T.; Shi, Q.; Gu, H.; Yu, B.; Yin, S.; Ge, Q.; Mo, X.; Liu, X.; Huang, J. CCDC134 facilitates T cell activation through the regulation of early T cell receptor signaling. Front. Immunol. 2023, 14, 1133111. [Google Scholar] [CrossRef]
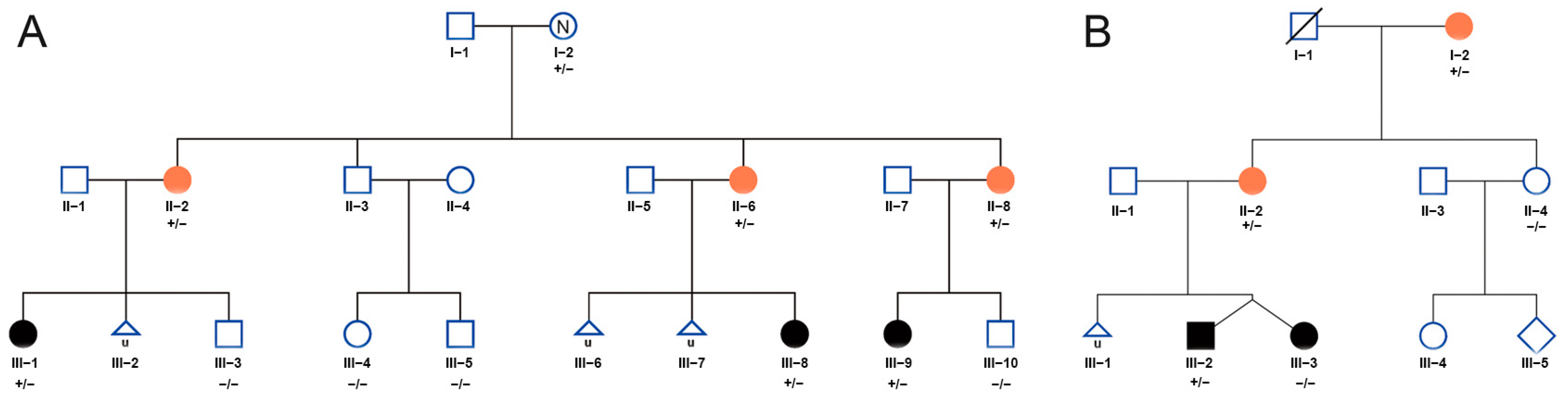

| Family No. | A | A | A | B | B | |
|---|---|---|---|---|---|---|
| Mother | II-2 | II-6 | II-8 | I-2 | II-2 | |
| Onset time at pregnancy (Week) | 24+1 | 27+0 | 32+0 | NA | 32+1 | |
| Gestational age at delivery (Week) | 34+4 | 27+4 | 32+0 | 36+4 | 36+4 | |
| High blood pressure * | Yes | Yes | Yes | Yes | Yes | |
| Proteinuria * | Yes | Yes | Yes | Yes | Yes | |
| Impaired liver function * | No | No | No | NA | No | |
| Renal insufficiency * | No | No | No | NA | No | |
| Thrombocytopenia * | No | No | No | NA | No | |
| Pulmonary edema * | No | No | No | NA | No | |
| Headache and/or visual symptoms * | No | No | Yes | NA | No | |
| Offspring | III-1 | III-8 | III-9 | II-2 | III-2 | IIII-3 |
| Sex | Female | Female | Female | Female | Male | Female |
| Birth weight (g) | 2100 | 750 | 1500 | NA | 2340 | 2470 |
| Apgar score 1 min | 10′ | 8′ | 4′ | NA | 10′ | 10′ |
| Apgar score 5 min | 10′ | 10′ | 7′ | NA | 10′ | 10′ |
| Apgar score 10 min | 10′ | 10′ | 10′ | NA | 10′ | 10′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, H.; Yu, L.; Chen, J.; Wang, X.; He, F.; Yu, L.; Du, L.; Chen, D. Rare Mutations in CCDC7 Contribute to Early-Onset Preeclampsia by Inhibiting Trophoblast Migration and Invasion. J. Pers. Med. 2024, 14, 253. https://doi.org/10.3390/jpm14030253
Tan H, Yu L, Chen J, Wang X, He F, Yu L, Du L, Chen D. Rare Mutations in CCDC7 Contribute to Early-Onset Preeclampsia by Inhibiting Trophoblast Migration and Invasion. Journal of Personalized Medicine. 2024; 14(3):253. https://doi.org/10.3390/jpm14030253
Chicago/Turabian StyleTan, Hu, Li Yu, Jingsi Chen, Xiaoyi Wang, Fang He, Lin Yu, Lili Du, and Dunjin Chen. 2024. "Rare Mutations in CCDC7 Contribute to Early-Onset Preeclampsia by Inhibiting Trophoblast Migration and Invasion" Journal of Personalized Medicine 14, no. 3: 253. https://doi.org/10.3390/jpm14030253
APA StyleTan, H., Yu, L., Chen, J., Wang, X., He, F., Yu, L., Du, L., & Chen, D. (2024). Rare Mutations in CCDC7 Contribute to Early-Onset Preeclampsia by Inhibiting Trophoblast Migration and Invasion. Journal of Personalized Medicine, 14(3), 253. https://doi.org/10.3390/jpm14030253







