Personalized Medicine in Cancer Pain Management
Abstract
1. Introduction
2. Materials and Methods
2.1. Data and Systematic Strategy
2.2. Bioinformatics Analyses
3. Results
3.1. Literature Review
3.2. Bioinformatics Analyses
4. Discussion
4.1. Cancer Pain Management Strategies
4.2. Personalized Medicine in Cancer-Pain Management
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Portenoy, R.K. Treatment of cancer pain. Lancet 2011, 377, 2236–2247. [Google Scholar] [CrossRef] [PubMed]
- Gorin, S.S.; Krebs, P.; Badr, H.; Janke, E.A.; Jim, H.S.; Spring, B.; Mohr, D.C.; Berendsen, M.A.; Jacobsen, P.B. Meta-analysis of psychosocial interventions to reduce pain in patients with cancer. J. Clin. Oncol. 2012, 30, 539. [Google Scholar] [CrossRef] [PubMed]
- Lalla, R.V.; Bowen, J.; Barasch, A.; Elting, L.; Epstein, J.; Keefe, D.M.; McGuire, D.B.; Migliorati, C.; Nicolatou-Galitis, O.; Peterson, D.E. MASCC/ISOO clinical practice guidelines for the management of mucositis secondary to cancer therapy. Cancer 2014, 120, 1453–1461. [Google Scholar] [CrossRef]
- Saibil, S.; Fitzgerald, B.; Freedman, O.; Amir, E.; Napolskikh, J.; Salvo, N.; Dranitsaris, G.; Clemons, M. Incidence of taxane-induced pain and distress in patients receiving chemotherapy for early-stage breast cancer: A retrospective, outcomes-based survey. Curr. Oncol. 2010, 17, 42–47. [Google Scholar] [CrossRef]
- Fassoulaki, A.; Melemeni, A.; Staikou, C.; Triga, A.; Sarantopoulos, C. Acute postoperative pain predicts chronic pain and long-term analgesic requirements after breast surgery for cancer. Acta Anaesthesiol. Belg. 2008, 59, 241. [Google Scholar]
- Bray, F.N.; Simmons, B.J.; Wolfson, A.H.; Nouri, K. Acute and chronic cutaneous reactions to ionizing radiation therapy. Dermatol. Ther. 2016, 6, 185–206. [Google Scholar] [CrossRef]
- Harb, A.H.; Fadel, C.A.; Sharara, A.I. Radiation enteritis. Curr. Gastroenterol. Rep. 2014, 16, 1–9. [Google Scholar] [CrossRef]
- Gewandter, J.S.; Freeman, R.; Kitt, R.A.; Cavaletti, G.; Gauthier, L.R.; McDermott, M.P.; Mohile, N.A.; Mohlie, S.G.; Smith, A.G.; Tejani, M.A. Chemotherapy-induced peripheral neuropathy clinical trials: Review and recommendations. Neurology 2017, 89, 859–869. [Google Scholar] [CrossRef] [PubMed]
- Paice, J.A.; Portenoy, R.; Lacchetti, C.; Campbell, T.; Cheville, A.; Citron, M.; Constine, L.S.; Cooper, A.; Glare, P.; Keefe, F. Management of chronic pain in survivors of adult cancers: American Society of Clinical Oncology clinical practice guideline. J. Clin. Oncol. 2016, 34, 3325–3345. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Tanco, K. How to measure pain. Curr. Oncol. Rep. 2021, 23, 1–6. [Google Scholar] [CrossRef]
- Villars, P.; Dodd, M.; West, C.; Koetters, T.; Paul, S.M.; Schumacher, K.; Tripathy, D.; Koo, P.; Miaskowski, C. Differences in the prevalence and severity of side effects based on type of analgesic prescription in patients with chronic cancer pain. J. Pain Symptom Manag. 2007, 33, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Hui, D.; Bruera, E. A personalized approach to assessing and managing pain in patients with cancer. J. Clin. Oncol. 2014, 32, 1640. [Google Scholar] [CrossRef] [PubMed]
- Chow, E. Genetic variants and biological markers of cancer-related pain sensitivity. J. Pain Manag. 2017, 10, 217–235. [Google Scholar]
- Kleine-Brueggeney, M.; Musshoff, F.; Stuber, F.; Stamer, U.M. Pharmacogenetics in palliative care. Forensic Sci. Int. 2010, 203, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Bruera, E.; Kuehn, N.; Miller, M.J.; Selmser, P.; Macmillan, K. The Edmonton Symptom Assessment System (ESAS): A simple method for the assessment of palliative care patients. J. Palliat. Care 1991, 7, 6–9. [Google Scholar] [CrossRef]
- Deplanque, G.; Demarchi, M.; Hebbar, M.; Flynn, P.; Melichar, B.; Atkins, J.; Nowara, E.; Moyé, L.; Piquemal, D.; Ritter, D. A randomized, placebo-controlled phase III trial of masitinib plus gemcitabine in the treatment of advanced pancreatic cancer. Ann. Oncol. 2015, 26, 1194–1200. [Google Scholar] [CrossRef]
- Griffith, K.A.; Zhu, S.; Johantgen, M.; Kessler, M.D.; Renn, C.; Beutler, A.S.; Kanwar, R.; Ambulos, N.; Cavaletti, G.; Bruna, J. Oxaliplatin-induced peripheral neuropathy and identification of unique severity groups in colorectal cancer. J. Pain Symptom Manag. 2017, 54, 701–706.e1. [Google Scholar] [CrossRef][Green Version]
- Okamoto, A.; Yamasaki, M.; Yokota, I.; Mori, M.; Matsuda, M.; Yamaguchi, Y.; Yamakita, S.; Ueno, H.; Sawa, T.; Taguchi, T. Classification of acute pain trajectory after breast cancer surgery identifies patients at risk for persistent pain: A prospective observational study. J. Pain Res. 2018, 11, 2197–2206. [Google Scholar] [CrossRef]
- Drewes, A.M.; Olesen, A.E.; Farmer, A.D.; Szigethy, E.; Rebours, V.; Olesen, S.S. Gastrointestinal pain. Nat. Rev. Dis. Primers 2020, 6, 1. [Google Scholar] [CrossRef]
- Coveler, A.L.; Mizrahi, J.; Eastman, B.; Apisarnthanarax, S.J.; Dalal, S.; McNearney, T.; Pant, S. Pancreas cancer-associated pain management. Oncologist 2021, 26, e971–e982. [Google Scholar] [CrossRef]
- de Sire, A.; Lippi, L.; Ammendolia, A.; Cisari, C.; Venetis, K.; Sajjadi, E.; Fusco, N.; Invernizzi, M. Physical Exercise with or without Whole-Body Vibration in Breast Cancer Patients Suffering from Aromatase Inhibitor—Induced Musculoskeletal Symptoms: A Pilot Randomized Clinical Study. J. Pers. Med. 2021, 11, 1369. [Google Scholar] [CrossRef] [PubMed]
- Parker, R.A.; Sande, T.A.; Laird, B.; Hoskin, P.; Fallon, M.; Colvin, L. Bayesian methods in palliative care research: Cancer-induced bone pain. BMJ Support. Palliat. Care 2022, 12, e5–e9. [Google Scholar] [CrossRef]
- Wilson, J.M.; Colebaugh, C.A.; Flowers, K.M.; Overstreet, D.; Edwards, R.R.; Maixner, W.; Smith, S.B.; Schreiber, K.L. Applying the Rapid OPPERA Algorithm to Predict Persistent Pain Outcomes Among a Cohort of Women Undergoing Breast Cancer Surgery. J. Pain 2022, 23, 2003–2012. [Google Scholar] [CrossRef]
- Lippi, L.; de Sire, A.; Folli, A.; Maconi, A.; Polverelli, M.; Vecchio, C.; Fusco, N.; Invernizzi, M. Effects of ultrasound-guided injection combined with a targeted therapeutic exercise in breast cancer women with subacromial pain syndrome: A randomized clinical study. J. Pers. Med. 2022, 12, 1833. [Google Scholar] [CrossRef] [PubMed]
- Bukkieva, T.; Pospelova, M.; Efimtsev, A.; Fionik, O.; Alekseeva, T.; Samochernykh, K.; Gorbunova, E.; Krasnikova, V.; Makhanova, A.; Nikolaeva, A. Microstructural Properties of Brain White Matter Tracts in Breast Cancer Survivors: A Diffusion Tensor Imaging Study. Pathophysiology 2022, 29, 595–609. [Google Scholar] [CrossRef]
- Manchikanti, L.; Knezevic, N.N.; Knezevic, E.; Abdi, S.; Sanapati, M.R.; Soin, A.; Wargo, B.W.; Navani, A.; Atluri, S.; Gharibo, C.G. A Systematic Review and Meta-analysis of the Effectiveness of Radiofrequency Neurotomy in Managing Chronic Neck Pain. Pain Ther. 2022, 12, 16–66. [Google Scholar] [CrossRef] [PubMed]
- Satkunananthan, S.E.; Suppiah, V.; Toh, G.-T.; Yow, H.-Y. Pharmacogenomics of Cancer Pain Treatment Outcomes in Asian Populations: A Review. J. Pers. Med. 2022, 12, 1927. [Google Scholar] [CrossRef]
- Lee, E.; Takita, C.; Wright, J.L.; Slifer, S.H.; Martin, E.R.; Urbanic, J.J.; Langefeld, C.D.; Lesser, G.J.; Shaw, E.G.; Hu, J.J. Genome-wide enriched pathway analysis of acute post-radiotherapy pain in breast cancer patients: A prospective cohort study. Hum. Genom. 2019, 13, 1–13. [Google Scholar] [CrossRef]
- Tang, P.C.-T.; Chung, J.Y.-F.; Liao, J.; Chan, M.K.-K.; Chan, A.S.-W.; Cheng, G.; Li, C.; Huang, X.-R.; Ng, C.S.-H.; Lam, E.W. Single-cell RNA sequencing uncovers a neuron-like macrophage subset associated with cancer pain. Sci. Adv. 2022, 8, eabn5535. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.S.; Barnes, N.M.; Lyon, D.E.; Dorsey, S.G. Genetic variants associated with cancer pain and response to opioid analgesics: Implications for precision pain management. Semin. Oncol. Nurs. 2019, 35, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Yennurajalingam, S.; Astolfi, A.; Indio, V.; Beccaro, M.; Schipani, A.; Yu, R.; Shete, S.; Reyes-Gibby, C.; Lu, Z.; Williams, J.L. Genetic factors associated with pain severity, daily opioid dose requirement, and pain response among advanced cancer patients receiving supportive care. J. Pain Symptom Manag. 2021, 62, 785–795. [Google Scholar] [CrossRef] [PubMed]
- Bortsov, A.V.; Devor, M.; Kaunisto, M.A.; Kalso, E.; Brufsky, A.; Kehlet, H.; Aasvang, E.; Bittner, R.; Diatchenko, L.; Belfer, I. CACNG2 polymorphisms associate with chronic pain following mastectomy. Pain 2019, 160, 561. [Google Scholar] [CrossRef] [PubMed]
- Bugada, D.; Lorini, L.F.; Fumagalli, R.; Allegri, M. Genetics and opioids: Towards more appropriate prescription in cancer pain. Cancers 2020, 12, 1951. [Google Scholar] [CrossRef] [PubMed]
- Rakvåg, T.T.; Ross, J.R.; Sato, H.; Skorpen, F.; Kaasa, S.; Klepstad, P. Genetic variation in the catechol-O-methyltransferase (COMT) gene and morphine requirements in cancer patients with pain. Mol. Pain 2008, 18, 1744–8069. [Google Scholar]
- Tchivileva, I.E.; Lim, P.F.; Smith, S.B.; Slade, G.D.; Diatchenko, L.; McLean, S.A.; Maixner, W. Effect of catechol-O-methyltransferase polymorphism on response to propranolol therapy in chronic musculoskeletal pain: A randomized, double–blind, placebo-controlled, crossover pilot study. Pharmacogenet. Genom. 2010, 20, 239. [Google Scholar] [CrossRef] [PubMed]
- Lopes, G.S.; Lopes, J.L.; Bielinski, S.J.; Armasu, S.M.; Zhu, Y.; Cavanaugh, D.C.; Moyer, A.M.; Jacobson, D.J.; Wang, L.; Jiang, R.; et al. Identification of sex-specific genetic associations in response to opioid analgesics in a White, non-Hispanic cohort from Southeast Minnesota. Pharmacogenom. J. 2022, 22, 117–123. [Google Scholar] [CrossRef]
- Mosley, S.A.; Hicks, J.K.; Portman, D.G.; Donovan, K.A.; Gopalan, P.; Schmit, J.; Starr, J.; Silver, N.; Gong, Y.; Langaee, T. Design and rational for the precision medicine guided treatment for cancer pain pragmatic clinical trial. Contemp. Clin. Trials 2018, 68, 7–13. [Google Scholar] [CrossRef]
- Rahmioglu, N.; Mortlock, S.; Ghiasi, M.; Møller, P.L.; Stefansdottir, L.; Galarneau, G.; Turman, C.; Danning, R.; Law, M.H.; Sapkota, Y. The genetic basis of endometriosis and comorbidity with other pain and inflammatory conditions. Nat. Genet. 2023, 55, 423–436. [Google Scholar] [CrossRef]
- Wang, J.; Lu, K.; Song, Y.; Xie, L.; Zhao, S.; Wang, Y.; Sun, W.; Liu, L.; Zhao, H.; Tang, D.; et al. Indications of clinical and genetic predictors for aromatase inhibitors related musculoskeletal adverse events in Chinese Han women with breast cancer. PLoS ONE 2013, 8, e68798. [Google Scholar] [CrossRef] [PubMed]
- Crescioli, G.; Lombardi, N.; Vagnoli, L.; Bettiol, A.; Giunti, L.; Cetica, V.; Coniglio, M.L.; Provenzano, A.; Giglio, S.; Bonaiuti, R. STOP Pain Project—Opioid Response in Pediatric Cancer Patients and Gene Polymorphisms of Cytokine Pathways. Pharmaceutics 2022, 14, 619. [Google Scholar] [CrossRef]
- Elens, L.; Norman, E.; Matic, M.; Rane, A.; Fellman, V.; Van Schaik, R.H. Genetic predisposition to poor opioid response in preterm infants: Impact of KCNJ6 and COMT polymorphisms on pain relief after endotracheal intubation. Ther. Drug Monit. 2016, 38, 525–533. [Google Scholar] [CrossRef]
- Skorpen, F.; von Hofacker, S.; Bjørngaard, M.; Skogholt, A.H.; Dale, O.; Kaasa, S.; Klepstad, P. The rare Arg181Cys mutation in the μ opioid receptor can abolish opioid responses. Acta Anaesthesiol. Scand. 2016, 60, 1084–1091. [Google Scholar] [CrossRef] [PubMed]
- Hajj, A.; Halepian, L.; Osta, N.E.; Chahine, G.; Kattan, J.; Khabbaz, L.R. OPRM1 c. 118A> G polymorphism and duration of morphine treatment associated with morphine doses and quality-of-life in palliative cancer pain settings. Int. J. Mol. Sci. 2017, 18, 669. [Google Scholar] [CrossRef]
- Lee, Y.S.; Kim, H.; Wu, T.X.; Wang, X.M.; Dionne, R.A. Genetically mediated interindividual variation in analgesic responses to cyclooxygenase inhibitory drugs. Clin. Pharmacol. Ther. 2006, 79, 407–418. [Google Scholar] [CrossRef] [PubMed]
- Galvan, A.; Skorpen, F.; Klepstad, P.; Knudsen, A.K.; Fladvad, T.; Falvella, F.S.; Pigni, A.; Brunelli, C.; Caraceni, A.; Kaasa, S. Multiple Loci Modulate Opioid Therapy Response for Cancer PainGenetic Control of Cancer Pain Response. Clin. Cancer Res. 2011, 17, 4581–4587. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.N.; Ho, I.K.; Tsou, H.H.; Fang, C.P.; Hsiao, C.F.; Chen, C.H.; Tan, H.K.; Lin, L.; Wu, C.S.; Su, L.W.; et al. UGT2B7 genetic polymorphisms are associated with the withdrawal symptoms in methadone maintenance patients. Pharmacogenomics 2012, 13, 879–888. [Google Scholar] [CrossRef] [PubMed]
- Genovese, T.J.; Mao, J.J. Genetic predictors of response to acupuncture for aromatase inhibitor–associated arthralgia among breast cancer survivors. Pain Med. 2019, 20, 191–194. [Google Scholar] [CrossRef] [PubMed]
- Bruera, E.; MacMillan, K.; Hanson, J.; MacDonald, R. The Edmonton staging system for cancer pain: Preliminary report. Pain 1989, 37, 203–209. [Google Scholar] [CrossRef]
- Nekolaichuk, C.L.; Fainsinger, R.L.; Lawlor, P.G. A validation study of a pain classification system for advanced cancer patients using content experts: The Edmonton Classification System for Cancer Pain. Palliat. Med. 2005, 19, 466–476. [Google Scholar] [CrossRef]
- Fainsinger, R.L.; Nekolaichuk, C.L.; Lawlor, P.G.; Neumann, C.M.; Hanson, J.; Vigano, A. A multicenter study of the revised Edmonton Staging System for classifying cancer pain in advanced cancer patients. J. Pain Symptom Manag. 2005, 29, 224–237. [Google Scholar] [CrossRef]
- Fainsinger, R.L.; Nekolaichuk, C.L. A “TNM” classification system for cancer pain: The Edmonton Classification System for Cancer Pain (ECS-CP). Support. Care Cancer 2008, 16, 547–555. [Google Scholar] [CrossRef] [PubMed]
- Bruera, E.; Schoeller, T.; Wenk, R.; MacEachern, T.; Marcelino, S.; Hanson, J.; Suarez-Almazor, M. A prospective multicenter assessment of the Edmonton staging system for cancer pain. J. Pain Symptom Manag. 1995, 10, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Hanks, G.; Cherny, N.I.; Christakis, N.A.; Kaasa, S. Oxford Textbook of Palliative Medicine; Oxford University Press: Oxford, UK, 2011. [Google Scholar]
- Comley, A.L.; DeMeyer, E. Assessing patient satisfaction with pain management through a continuous quality improvement effort. J. Pain Symptom Manag. 2001, 21, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Ward, S.E.; Gordon, D.B. Patient satisfaction and pain severity as outcomes in pain management: A longitudinal view of one setting’s experience. J. Pain Symptom Manag. 1996, 11, 242–251. [Google Scholar] [CrossRef]
- Cepeda, M.S.; Africano, J.M.; Polo, R.; Alcala, R.; Carr, D.B. Agreement between percentage pain reductions calculated from numeric rating scores of pain intensity and those reported by patients with acute or cancer pain. Pain 2003, 106, 439–442. [Google Scholar] [CrossRef]
- Musshoff, F.; Lachenmeier, K.; Trafkowski, J.; Madea, B.; Nauck, F.; Stamer, U. Determination of opioid analgesics in hair samples using liquid chromatography/tandem mass spectrometry and application to patients under palliative care. Ther. Drug Monit. 2007, 29, 655–661. [Google Scholar] [CrossRef]
- Dalal, S.; Bruera, E. Assessing cancer pain. Curr. Pain Headache Rep. 2012, 16, 314–324. [Google Scholar] [CrossRef]
- Sarzi-Puttini, P.; Vellucci, R.; Zuccaro, S.M.; Cherubino, P.; Labianca, R.; Fornasari, D. The appropriate treatment of chronic pain. Clin. Drug Investig. 2012, 32, 21–33. [Google Scholar] [CrossRef]
- Westerling, D. Pain and other symptoms in cancer survivors. J. Pain Palliat. Care Pharmacother. 2014, 28, 160–163. [Google Scholar] [CrossRef]
- Petersen, M.A.; Aaronson, N.K.; Chie, W.-C.; Conroy, T.; Costantini, A.; Hammerlid, E.; Hjermstad, M.J.; Kaasa, S.; Loge, J.H.; Velikova, G. Development of an item bank for computerized adaptive test (CAT) measurement of pain. Qual. Life Res. 2016, 25, 1–11. [Google Scholar] [CrossRef]
- Balducci, L.; Dolan, D. Palliative care of cancer in the older patient. Curr. Oncol. Rep. 2016, 18, 1–10. [Google Scholar] [CrossRef]
- Bhatnagar, S.; Gupta, M. Integrated pain and palliative medicine model. Ann. Palliat. Med. 2016, 5, 196–208. [Google Scholar] [CrossRef] [PubMed]
- Arthur, J.; Tanco, K.; Haider, A.; Maligi, C.; Park, M.; Liu, D.; Bruera, E. Assessing the prognostic features of a pain classification system in advanced cancer patients. Support. Care Cancer 2017, 25, 2863–2869. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.C.; Zheng, Z.X.; Tan, K.H.; Meredith, G.J. Multidimensional treatment of cancer pain. Curr. Oncol. Rep. 2017, 19, 1–9. [Google Scholar] [CrossRef]
- Colvin, L.A. Chemotherapy-induced peripheral neuropathy (CIPN): Where are we now? Pain 2019, 160, S1. [Google Scholar] [CrossRef]
- Vimalnath, K.; Rajeswari, A.; Sarma, H.D.; Dash, A.; Chakraborty, S. Ce-141-labeled DOTMP: A theranostic option in management of pain due to skeletal metastases. J. Label. Compd. Radiopharm. 2019, 62, 178–189. [Google Scholar] [CrossRef] [PubMed]
- Sica, A.; Casale, B.; Dato, M.T.D.; Calogero, A.; Spada, A.; Sagnelli, C.; Santagata, M.; Buonavolontà, P.; Fiorelli, A.; Salzano, A. Cancer-and non-cancer related chronic pain: From the physiopathological basics to management. Open Med. 2019, 14, 761–766. [Google Scholar] [CrossRef]
- Miller, K.R.; Patel, J.N.; Symanowski, J.T.; Edelen, C.A.; Walsh, D. Acupuncture for cancer pain and symptom management in a palliative medicine clinic. Am. J. Hosp. Palliat. Med. 2019, 36, 326–332. [Google Scholar] [CrossRef]
- Cuomo, A.; Bimonte, S.; Forte, C.A.; Botti, G.; Cascella, M. Multimodal approaches and tailored therapies for pain management: The trolley analgesic model. J. Pain Res. 2019, 12, 711–714. [Google Scholar] [CrossRef]
- Vitzthum, L.K.; Riviere, P.; Sheridan, P.; Nalawade, V.; Deka, R.; Furnish, T.; Mell, L.K.; Rose, B.; Wallace, M.; Murphy, J.D. Predicting persistent opioid use, abuse, and toxicity among cancer survivors. JNCI J. Natl. Cancer Inst. 2020, 112, 720–727. [Google Scholar] [CrossRef] [PubMed]
- LeBaron, V.; Hayes, J.; Gordon, K.; Alam, R.; Homdee, N.; Martinez, Y.; Ogunjirin, E.; Thomas, T.; Jones, R.; Blackhall, L. Leveraging smart health technology to empower patients and family caregivers in managing cancer pain: Protocol for a feasibility study. JMIR Res. Protoc. 2019, 8, e16178. [Google Scholar] [CrossRef] [PubMed]
- Chapman, C.R.; Beach, W.A. Patient-initiated pain expressions: Interactional asymmetries and consequences for cancer care. Health Commun. 2020, 35, 1643–1655. [Google Scholar] [CrossRef] [PubMed]
- Oldenmenger, W.H.; Lucas, A.; van der Werff, G.F.; Webber, K.; Visser, D.; van der Velden, A.W.; van der Rijt, C.C. Validation of the Dutch version of the Breakthrough Pain Assessment Tool in patients with cancer. J. Pain Symptom Manag. 2020, 59, 709–716.e2. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, L.; Hassett, A.; Sheridan, N. Pain assessment and registration in medical oncology clinics: Operationalised through the lens of health care professionals and patients. HRB Open Res. 2021, 4, 86. [Google Scholar] [CrossRef] [PubMed]
- Ben-Arye, E.; Elly, D.; Samuels, N.; Gressel, O.; Shulman, K.; Schiff, E.; Lavie, O.; Minerbi, A. Effects of a patient-tailored integrative oncology intervention in the relief of pain in palliative and supportive cancer care. J. Cancer Res. Clin. Oncol. 2021, 147, 2361–2372. [Google Scholar] [CrossRef]
- Mao, J.J.; Liou, K.T.; Baser, R.E.; Bao, T.; Panageas, K.S.; Romero, S.A.; Li, Q.S.; Gallagher, R.M.; Kantoff, P.W. Effectiveness of electroacupuncture or auricular acupuncture vs usual care for chronic musculoskeletal pain among cancer survivors: The PEACE randomized clinical trial. JAMA Oncol. 2021, 7, 720–727. [Google Scholar] [CrossRef] [PubMed]
- Soto-Perez-de-Celis, E.; Chavarri-Guerra, Y.; Ramos-Lopez, W.A.; Alcalde-Castro, J.; Covarrubias-Gomez, A.; Navarro-Lara, Á.; Quiroz-Friedman, P.; Sánchez-Román, S.; Alcocer-Castillejos, N.; Aguilar-Velazco, J.C. Patient navigation to improve early access to supportive care for patients with advanced cancer in resource-limited settings: A randomized controlled trial. Oncologist 2021, 26, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, M.; Luo, J.; Xie, J.; Chen, X.; Wang, H.; Li, S.; Yang, S.; Peng, C.; Yang, L. Practice, knowledge, and attitude of health care providers regarding cancer pain management: A national survey. Pain Res. Manag. 2021, 2021, 1247202. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Zhang, C.; Li, S.; Dai, R.; Deng, B.; Xu, Q.; Wang, J.; Shi, C.; Zhang, Y. Knowledge, attitudes, and practices toward cancer pain management amongst healthcare workers (physicians, pharmacists, and nurses): A cross-sectional study from first-tier cities in China. Support. Care Cancer 2022, 30, 7261–7269. [Google Scholar] [CrossRef]
- Batistaki, C.; Graczyk, M.; Janecki, M.; Lewandowska, A.A.; Moutinho, R.; Vagdatli, K. Relationship between breakthrough cancer pain, background cancer pain and analgesic treatment–case series and review of the literature. Drugs Context 2022, 11. [Google Scholar] [CrossRef]
- Masukawa, K.; Aoyama, M.; Yokota, S.; Nakamura, J.; Ishida, R.; Nakayama, M.; Miyashita, M. Machine learning models to detect social distress, spiritual pain, and severe physical psychological symptoms in terminally ill patients with cancer from unstructured text data in electronic medical records. Palliat. Med. 2022, 36, 1207–1216. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Baser, R.E.; Liou, K.T.; Li, S.Q.; Piulson, L.; Panageas, K.S.; Mao, J.J. Effect of acupuncture versus usual care on sleep quality in cancer survivors with chronic pain: Secondary analysis of a randomized clinical trial. Cancer 2023, 129, 2084–2094. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.; Haider, A.; Arthur, J.; Hui, D.; Dalal, S.; Dev, R.; Tanco, K.; Amaram-Davila, J.; Hernandez, F.; Chavez, P. Levorphanol as a Second Line Opioid in Cancer Patients Presenting to an Outpatient Supportive Care Center: An Open-label Study. J. Pain Symptom Manag. 2023, 65, e683–e690. [Google Scholar] [CrossRef]
- Aziz, M.B.; Cascella, M. Peripheral Neurolytic Blocks; StatPearls: Tampa, FL, USA, 2023. [Google Scholar]
- Dalal, S.; Hui, D.; Nguyen, L.; Chacko, R.; Scott, C.; Roberts, L.; Bruera, E. Achievement of personalized pain goal in cancer patients referred to a supportive care clinic at a comprehensive cancer center. Cancer 2012, 118, 3869–3877. [Google Scholar] [CrossRef] [PubMed]
- Ling, I.S.; Larsson, B. Individualized pharmacological treatment of oral mucositis pain in patients with head and neck cancer receiving radiotherapy. Support. Care Cancer 2011, 19, 1343–1350. [Google Scholar] [CrossRef]
- Khan, M.I.A.; Walsh, D.; Brito-Dellan, N. Opioid and adjuvant analgesics: Compared and contrasted. Am. J. Hosp. Palliat. Med. 2011, 28, 378–383. [Google Scholar] [CrossRef]
- Heintzelman, N.H.; Taylor, R.J.; Simonsen, L.; Lustig, R.; Anderko, D.; Haythornthwaite, J.A.; Childs, L.C.; Bova, G.S. Longitudinal analysis of pain in patients with metastatic prostate cancer using natural language processing of medical record text. J. Am. Med. Inform. Assoc. 2013, 20, 898–905. [Google Scholar] [CrossRef]
- Tverdohleb, T.; Dinc, B.; Knezevic, I.; Candido, K.D.; Knezevic, N.N. The role of cytochrome P450 pharmacogenomics in chronic non-cancer pain patients. Expert Opin. Drug Metab. Toxicol. 2016, 12, 1303–1311. [Google Scholar] [CrossRef]
- Sivanesan, E.; Gitlin, M.C. Desmoid tumors: A review of the literature and pharmacologic management. J. Pain Palliat. Care Pharmacother. 2016, 30, 99–105. [Google Scholar] [CrossRef]
- Obeng, A.O.; Hamadeh, I.; Smith, M. Review of opioid pharmacogenetics and considerations for pain management. Pharmacother. J. Hum. Pharmacol. Drug Ther. 2017, 37, 1105–1121. [Google Scholar] [CrossRef]
- Nissenbaum, J.; Devor, M.; Seltzer, Z.; Gebauer, M.; Michaelis, M.; Tal, M.; Dorfman, R.; Abitbul-Yarkoni, M.; Lu, Y.; Elahipanah, T. Susceptibility to chronic pain following nerve injury is genetically affected by CACNG2. Genome Res. 2010, 20, 1180–1190. [Google Scholar] [CrossRef]
- De Santis, S.; Simone, M.D.; Mercadante, S.; Mediati, R.D.; Vellucci, R.; Marchetti, P.; Tonini, G.; Cuomo, A.; Caraceni, A.; Natoli, S. Oxycodone/acetaminophen: The tailoring combination treatment for specific clinical profile of opioid well-responsive cancer pain. Cancer Manag. Res. 2021, 2021, 1747–1756. [Google Scholar] [CrossRef]
- Xu, X.; Yang, K.; Zhang, F.; Liu, W.; Wang, Y.; Yu, C.; Wang, J.; Zhang, K.; Zhang, C.; Nenadic, G. Identification of herbal categories active in pain disorder subtypes by machine learning help reveal novel molecular mechanisms of algesia. Pharmacol. Res. 2020, 156, 104797. [Google Scholar] [CrossRef]
- Hasuo, H.; Sakuma, H.; Fukunaga, M. Alexithymia in family caregivers of advanced cancer patients is associated with high personalized pain goal scores: A pilot study. J. Palliat. Med. 2020, 23, 930–936. [Google Scholar] [CrossRef]
- Di Lorenzo, G.; Autorino, R. Re: Olaparib for Metastatic Castration-resistant Prostate Cancer. Eur. Urol. 2020, 78, 767–768. [Google Scholar] [CrossRef]
- Rienzo, M.; Di Zazzo, E.; Casamassimi, A.; Gazzerro, P.; Perini, G.; Bifulco, M.; Abbondanza, C. PRDM12 in Health and Diseases. Int. J. Mol. Sci. 2021, 22, 12030. [Google Scholar] [CrossRef]
- Reizine, N.; Danahey, K.; Schierer, E.; Liu, P.; Middlestadt, M.; Ludwig, J.; Truong, T.M.; Wijk, X.M.; Yeo, K.-T.J.; Malec, M. Impact of CYP2D6 pharmacogenomic status on pain control among opioid-treated oncology patients. Oncologist 2021, 26, e2042–e2052. [Google Scholar] [CrossRef]
- Saloman, J.L.; Tang, G.; Stello, K.M.; Hall, K.E.; Wang, X.; AlKaade, S.; Banks, P.A.; Brand, R.E.; Conwell, D.L.; Coté, G.A. Serum biomarkers for chronic pancreatitis pain patterns. Pancreatology 2021, 21, 1411–1418. [Google Scholar] [CrossRef]
- Chang, C.Y.-Y.; Yang, L.; Tse, J.; Lo, L.-C.; Tseng, C.-C.; Sun, L.; Lai, M.-T.; Chen, P.-H.; Hwang, T.; Chen, C.-M. Genetic variations in UCA1, a lncRNA functioning as a miRNA sponge, determine endometriosis development and the potential associated infertility via regulating lipogenesis. PLoS ONE 2022, 17, e0271616. [Google Scholar] [CrossRef]
- Levran, O.; O’Hara, K.; Peles, E.; Li, D.; Barral, S.; Ray, B.; Borg, L.; Ott, J.; Adelson, M.; Kreek, M.J. ABCB1 (MDR1) genetic variants are associated with methadone doses required for effective treatment of heroin dependence. Hum. Mol. Genet. 2008, 17, 2219–2227. [Google Scholar] [CrossRef]
- Dennis, B.B.; Bawor, M.; Thabane, L.; Sohani, Z.; Samaan, Z. Impact of ABCB1 and CYP2B6 genetic polymorphisms on methadone metabolism, dose and treatment response in patients with opioid addiction: A systematic review and meta-analysis. PLoS ONE 2014, 9, e86114. [Google Scholar] [CrossRef] [PubMed]
- Tournier, N.; Decleves, X.; Saubamea, B.; Scherrmann, J.M.; Cisternino, S. Opioid Transport by ATP-Binding Cassette Transporters at the Blood-Brain Barrier: Implications for Neuropsychopharmacology. Curr. Pharm. Des. 2011, 17, 2829–2842. [Google Scholar] [CrossRef]
- Chaves, C.; Remiao, F.; Cisternino, S.; Decleves, X. Opioids and the Blood-Brain Barrier: A Dynamic Interaction with Consequences on Drug Disposition in Brain. Curr. Neuropharmacol. 2017, 15, 1156–1173. [Google Scholar] [CrossRef]
- Campa, D.; Gioia, A.; Tomei, A.; Poli, P.; Barale, R. Association of ABCB1/MDR1 and OPRM1 gene polymorphisms with morphine pain relief. Clin. Pharmacol. Ther. 2008, 83, 559–566. [Google Scholar] [CrossRef]
- Sia, A.T.; Sng, B.L.; Lim, E.C.; Law, H.; Tan, E.C. The influence of ATP-binding cassette sub-family B member-1 (ABCB1) genetic polymorphisms on acute and chronic pain after intrathecal morphine for caesarean section: A prospective cohort study. Int. J. Obstet. Anesth. 2010, 19, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Crews, K.R.; Monte, A.A.; Huddart, R.; Caudle, K.E.; Kharasch, E.D.; Gaedigk, A.; Dunnenberger, H.M.; Leeder, J.S.; Callaghan, J.T.; Samer, C.F.; et al. Clinical pharmacogenetics implementation consortium guideline for CYP2D6, OPRM1, and COMT genotypes and select opioid therapy. Clin. Pharmacol. Ther. 2021, 110, 888–896. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Luo, J.; Chen, X.; Hu, H.; Li, S.; Zhang, Y.; Shi, C. Clinical Observation of the Effects of Oral Opioid on Inflammatory Cytokines and Gut Microbiota in Patients with Moderate to Severe Cancer Pain: A Retrospective Cohort Study. Pain Ther. 2022, 11, 667–681. [Google Scholar] [CrossRef]
- Nishizawa, D.; Terui, T.; Ishitani, K.; Kasai, S.; Hasegawa, J.; Nakayama, K.; Ebata, Y.; Ikeda, K. Genome-Wide Association Study Identifies Candidate Loci Associated with Opioid Analgesic Requirements in the Treatment of Cancer Pain. Cancers 2022, 14, 4692. [Google Scholar] [CrossRef]
- Li, J.; Li, S.; Zhang, T.; Yu, L.; Wei, J.; Wu, M.; Xie, Y.; Tan, H. Label-free serum proteomics for the identification of the putative biomarkers of postoperative pain in patients with gastric cancer. Mol. Omics 2023, 19, 351–361. [Google Scholar] [CrossRef]
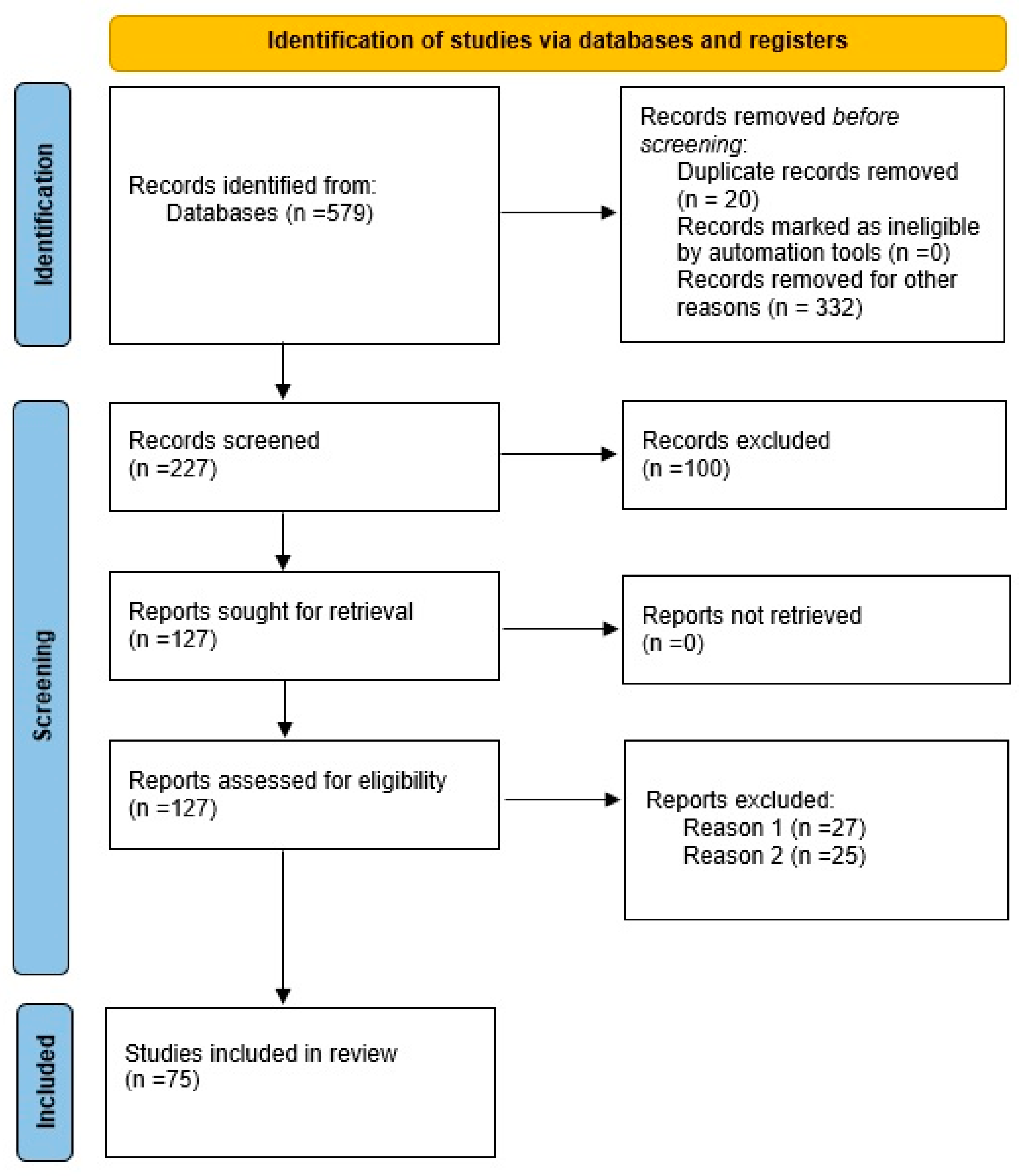

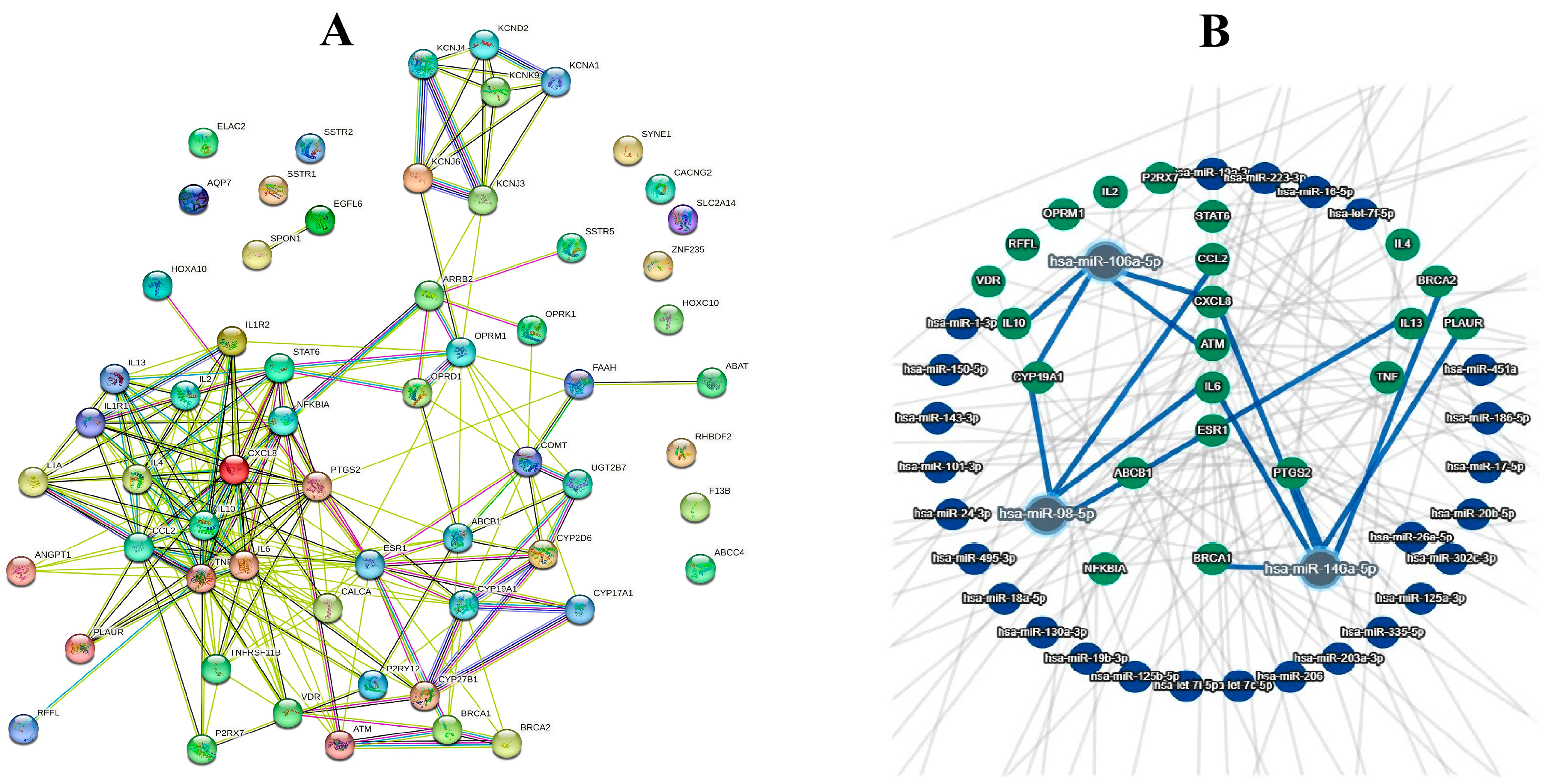
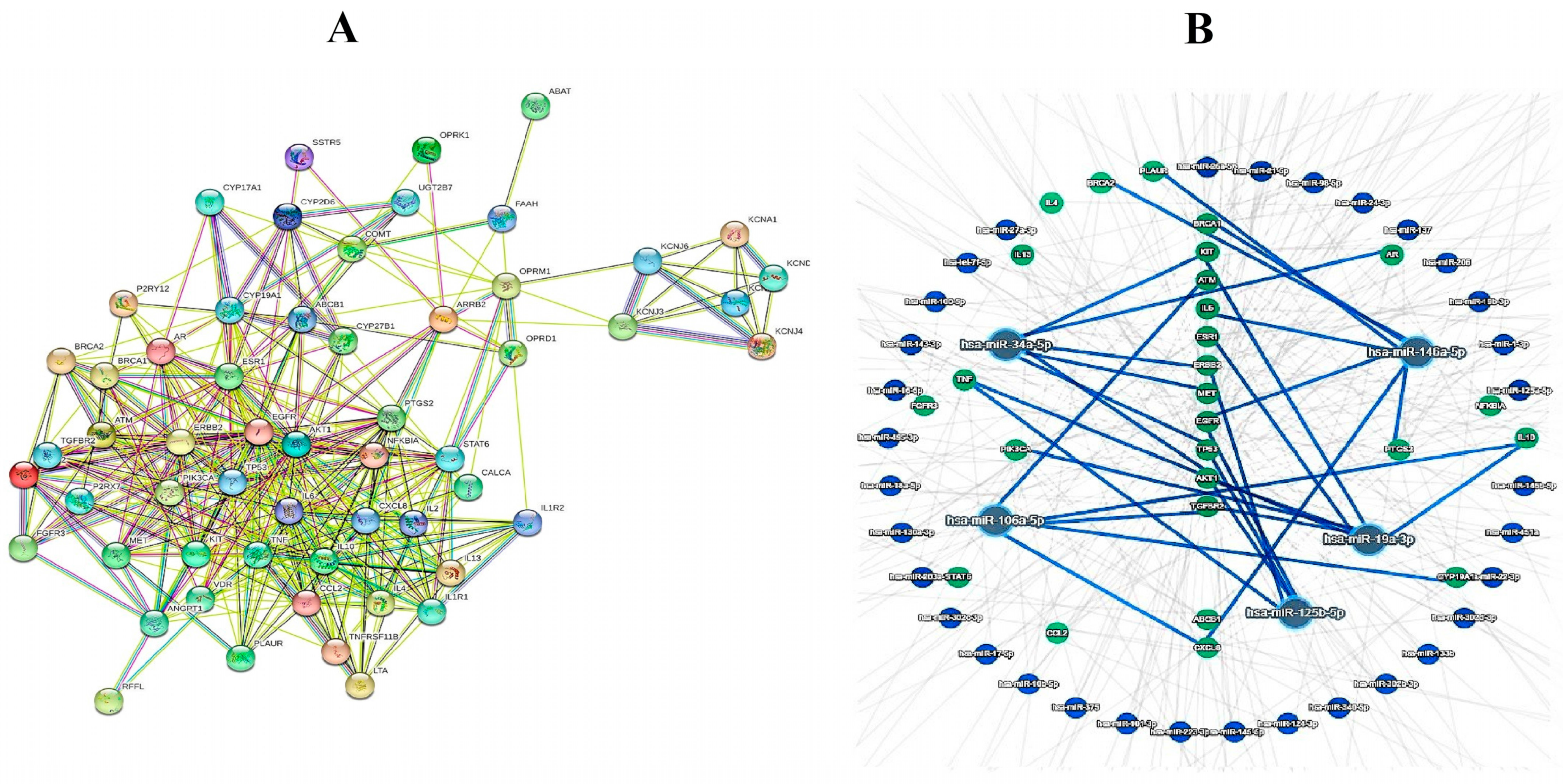
| Level 1 | Level 2 | Level 3 |
|---|---|---|
| ESR1 | ANGPT1 | ANGPT1 |
| EGFR | ATM | ATM |
| AR | CALCA | CALCA |
| ERBB2 | CCL2 | CCL2 |
| TGFBR2 | CXCL8 | CXCL8 |
| TP53 | CYP2D6, CYP19A1, CYP17A1, CYP27B1 | ESR1 |
| FGFR2 | ESR1 | IL10, IL13, IL2, IL4, IL6, IL1R1, IL1R2 |
| MET | IL2, IL4, IL6, IL10, IL13, IL1R1, IL1R2 | AKT1 |
| FGFR3 | LTA | LTA |
| KIT | NFKBIA | NFKBIA |
| PIK3CA | P2RX7 | P2RX7 |
| AKT1 | P2RY12 | P2RY12 |
| PLAUR | PLAUR | |
| PTGS2 | PTGS2 | |
| RFFL | RFFL | |
| STAT6 | STAT6 | |
| TNF | TNF, TNFRSF11B | |
| TNFRSF11B | VDR | |
| VDR | ARRB2 | |
| ARRB2 | KCNJ3, KCNJ4, KCNJ6, KCNK9, KCNA1, KCND2 | |
| KCNA1, KCND2, KCNJ3, KCNJ4, KCNJ6, KCNK9 | SSTR5 | |
| SSTR5 | ABAT | |
| ABAT | ABCB1 | |
| ABCB1 | BRCA1, BRCA2 | |
| BRCA1, BRCA2 | COMT | |
| COMT | CYP2D6, CYP17A1, CYP19A1, CYP27B1 | |
| FAAH | FAAH | |
| OPRD1, OPRK1, OPRM1 | OPRD1, OPRK1, OPRM1 | |
| UGT2B7 | UGT2B7 | |
| EGFR, FGFR2, FGFR3 | ||
| AR | ||
| ERBB2 | ||
| TGFBR2 | ||
| TP53 | ||
| KIT | ||
| PIK3CA | ||
| MET |
| Gene | Variant | Function | Author, Year | Country | Reference |
|---|---|---|---|---|---|
| ABAT | rs1641025 | Intronic | Satkunananthan et al., 2022 | Asian | [27] |
| ABCB1 | rs1045642 | Missense | Satkunananthan et al., 2022 | Asian | [27] |
| ABCB1 | rs1128503 | Synonymous | Satkunananthan et al., 2022 | Asian | [27] |
| ABCB1 | rs2032582 | Missense | Satkunananthan et al., 2022 | Asian | [27] |
| ABCC4/MPR4 | rs4584690 | Intronic | Lee et al., 2019 | European-Americans, Nigeria; Han Chinese; and Japanese | [28] |
| ABCC4/MPR4 | rs7335912 | Intergenic | Lee et al., 2019 | European-Americans, Nigeria; Han Chinese; and Japanese | [28] |
| ANGPT1 | rs1283671 | Upstream | Tang et al., 2022 | Cell line | [29] |
| ANGPT1 | rs1283720 | Upstream | Tang et al., 2022 | Cell line | [29] |
| AQP7 | rs76608797 | Missense | Yang et al., 2019 | NA | [30] |
| AQP7 | rs33386144 | Intergenic | Yang et al., 2019 | NA | [30] |
| ARRB2 | rs1045280 | Intronic | [31] | ||
| ATM | rs11212570 | Intronic | Yang et al., 2019 | NA | [30] |
| CACNG2 | rs2284017 | Upstream | [32] | ||
| CACNG2 | rs4820242 | Upstream | Yang et al., 2019 | NA | [30] |
| CACNG2 | rs2284015 | Upstream | Yang et al., 2019 | NA | [30] |
| CACNG2 | rs2284017 | Upstream | Yang et al., 2019 | NA | [30] |
| COMT | rs4680 (Val158Met) | Missense | Yang et al., 2019 | NA | [30,33] |
| COMT | rs165774 | Downstream | Yang et al., 2019 | NA | [30] |
| COMT | rs887200 | Intronic | Yang et al., 2019 | NA | [30] |
| COMT | rs4818 | Synonymous | Yang et al., 2019 | NA | [30] |
| COMT | rs9306234 | 3’UTR | Yennurajalingam et al., 2021 | USA | [31] |
| COMT | rs165728 | 3’UTR | Yennurajalingam et al., 2021 | USA | [31] |
| COMT | rs2020917 | Upstream | Yennurajalingam et al., 2021 | USA | [31] |
| COMT | rs2075507 | Upstream | Rakvåg et al., 2008 | Caucasian | [34] |
| COMT | rs4633 | Synonymous | Tchivileva et al., 2011 | Caucasian | [35] |
| COMT | rs6269 | 5’UTR | Tchivileva et al., 2011 | Caucasian | [35] |
| CXCL8 | rs4073 | Upstream | Yang et al., 2019 | NA | [30] |
| CYP17A1 | rs4919686 | Intronic | Yang et al., 2019 | NA | [30] |
| CYP17A1 | rs4919683 | Intronic | Yang et al., 2019 | NA | [30] |
| CYP17A1 | rs4919687 | Intronic | Yang et al., 2019 | NA | [30] |
| CYP17A1 | rs3781287 | Intronic | Yang et al., 2019 | NA | [30] |
| CYP17A1 | rs10786712 | Intronic | Yang et al., 2019 | NA | [30] |
| CYP17A1 | rs6163 | Synonymous | Yang et al., 2019 | NA | [30] |
| CYP17A1 | rs743572 | 5’UTR | Yang et al., 2019 | NA | [30] |
| CYP19A1 | rs4775936 | 5’UTR | Yang et al., 2019 | NA | [30] |
| CYP27B1 | rs4646536 | Intronic | Yang et al., 2019 | NA | [30] |
| CYP2D6 | rs35742686 | Frameshift | Lopes et al., 2022 | Non-Hispanic US | [36] |
| CYP2D6 | *2 (rs16947) (rs1135840) | Missense | Mosley et al., 2018 | USA | [37] |
| CYP2D6 | *3 (rs35742686) | Frameshift | Mosley et al., 2018 | USA | [37] |
| CYP2D6 | *4 (rs3892097 ) | Splicing | Mosley et al., 2018 | USA | [37] |
| CYP2D6 | *5 | Deletion | Mosley et al., 2018 | USA | [37] |
| CYP2D6 | *6 (rs5030655) | Frameshift | Mosley et al., 2018 | USA | [37] |
| CYP2D6 | *7 (rs5030867) | Missense | Mosley et al., 2018 | USA | [37] |
| CYP2D6 | *8 (rs5030865) | Missense | Mosley et al., 2018 | USA | [37] |
| CYP2D6 | *9 (rs5030656) | Deletion | Mosley et al., 2018 | USA | [37] |
| CYP2D6 | *10 (rs1065852) | Missense | Mosley et al., 2018; Satkunananthan et al., 2022 | USA; Asian | [27,37] |
| CYP2D6 | *11 (rs28399447) (rs28371685) | Missense | Mosley et al., 2018 | USA | [37] |
| CYP2D6 | *15 (rs5030867) | Missense | Mosley et al., 2018 | USA | [37] |
| CYP2D6 | *17 (rs28371706) (rs16947) | Missense | Mosley et al., 2018 | USA | [37] |
| CYP2D6 | *29 (rs61736512) (rs16947) (rs59421388) (rs1135840) | Missense | Mosley et al., 2018 | USA | [37] |
| CYP2D6 | *35 (rs769258) (rs1058164) (rs16947) (rs1135840) | Missense | Mosley et al., 2018 | USA | [37] |
| CYP2D6 | *41 (rs28371725) | Intronic | Mosley et al., 2018 | USA | [37] |
| EGFL6 | rs73633565 | Intronic | Lee et al., 2019 | European-Americans, Nigeria; Han Chinese; and Japanese | [28] |
| ELAC2 | rs11545302 | Synonymous | Yang et al., 2019 | NA | [30] |
| ESR1 | rs73625113 | Intronic | Rahmioglu et al., 2023 | European and East Asian | [38] |
| ESR1 | rs2234693 | Upstream | Wang et al., 2013 | China | [39] |
| ESR1 | rs9340799 | Intronic | Wang et al., 2013 | China | [39] |
| FAAH | rs324420 | Missense | Yang et al., 2019 | NA | [30] |
| FAAH | rs1571138 | Intronic | Yang et al., 2019 | NA | [30] |
| FAAH | rs3766246 | Intronic | Yang et al., 2019 | NA | [30] |
| FAAH | rs4660928 | TF binding site | Yang et al., 2019 | NA | [30] |
| HOXA10 | rs6970537 | Intronic | Rahmioglu et al., 2023 | European and East Asian | [38] |
| HOXC10 | rs3803042 | Non-coding exon | Rahmioglu et al., 2023 | European and East Asian | [38] |
| IL-10 | rs1800871 | Upstream | Yang et al., 2019 | NA | [30] |
| IL-10 | rs3024505 | Intergenic | Yang et al., 2019 | NA | [30] |
| IL-10 | rs3024498 | 3’UTR | Yang et al., 2019 | NA | [30] |
| IL-10 | rs3024496 | 3’UTR | Yang et al., 2019 | NA | [30] |
| IL-10 | rs1878672 | Intronic | Yang et al., 2019 | NA | [30] |
| IL-10 | rs1518111 | Upstream | Yang et al., 2019 | NA | [30] |
| IL-10 | rs1518110 | Upstream | Yang et al., 2019 | NA | [30] |
| IL-10 | rs3024491 | Intronic | Yang et al., 2019 | NA | [30] |
| IL-13 | rs1295686 | Intronic | Yang et al., 2019 | NA | [30] |
| IL1R1 | rs2110726 | 3’UTR | Yang et al., 2019 | NA | [30] |
| IL1R2 | rs11674595 | Intronic | Yang et al., 2019 | NA | [30] |
| IL-6 | rs2006984 | 5’UTR | Yang et al., 2019 | NA | [30] |
| IL-6 | rs1800797 | Upstream | Crescioli et al. 2022 | Italy | [40] |
| KCNA1 | rs4766311 | 3’UTR | Yang et al., 2019 | NA | [30] |
| KCND2 | rs1072198 | Intronic | Yang et al., 2019 | NA | [30] |
| KCNJ3 | rs12995382 | Intronic | Yang et al., 2019 | NA | [30] |
| KCNJ4 | rs17641121 | Intronic | Yang et al., 2019 | NA | [30] |
| KCNJ6 | rs858003 | Intronic | Yang et al., 2019 | NA | [30] |
| KCNJ6 | rs6517442 | Upstream | Elens et al., 2016 | Sweden | [41] |
| KCNK9 | rs2542424 | Intronic | Yang et al., 2019 | NA | [30] |
| KCNK9 | rs2545457 | Intronic | Yang et al., 2019 | NA | [30] |
| LINC00629 | rs73241342 | Intronic | Rahmioglu et al., 2023 | European and East Asian | [38] |
| LNC-LBCS | rs6456259 | Intronic | Rahmioglu et al., 2023 | European and East Asian | [38] |
| LTA | rs1799964 | Upstream | Yang et al., 2019 | NA | [30] |
| NFKBIA | rs8904 | 3’UTR | Yang et al., 2019 | NA | [30] |
| NFKBIA | rs2233419 | Intronic | Yennurajalingam et al., 2021 | USA | [31] |
| NFKBIA | rs2233417 | Intronic | Yennurajalingam et al., 2021 | USA | [31] |
| NFKBIA | rs3138054 | Intronic | Yennurajalingam et al., 2021 | USA | [31] |
| NFKBIA | rs1050851 | Synonymous | Yennurajalingam et al., 2021 | USA | [31] |
| NF-κB | rs230493 | Intronic | Yang et al., 2019 | NA | [30] |
| OPG | rs2073618 | Upstream | Yang et al., 2019 | NA | [30] |
| OPRM1 | rs79910351 (Arg181Cys) | Missense | Skorpen et al., 2016 | European | [42] |
| OPRM1 | rs1799971 (Asn40Asp) | Missense | Bugada, 2020; Hajj et al., 2017; Satkunananthan et al., 2022; Yang et al., 2019 | NA; Lebanon; Asian; NA | [27,30,33,43] |
| OPRM1 | rs9479759 | Intronic | Yennurajalingam et al., 2021 | USA | [31] |
| OPRM1 | rs2003185 | Intronic | Yennurajalingam et al., 2021 | USA | [31] |
| OPRM1 | rs636433 | 3’UTR | Yennurajalingam et al., 2021 | USA | [31] |
| P2RX7 | rs1718125 | Intronic | Satkunananthan et al., 2022 | Asian | [27] |
| P2RY12 | rs3732765 | Missense | Yang et al., 2019 | NA | [30] |
| P2RY12 | rs9859538 | Intronic | Yang et al., 2019 | NA | [30] |
| P2RY12 | rs17283010 | Intronic | Yang et al., 2019 | NA | [30] |
| P2RY12 | rs11713504 | Intronic | Yang et al., 2019 | NA | [30] |
| P2RY12 | rs10935840 | Intronic | Yang et al., 2019 | NA | [30] |
| PLAUR | rs4760 | Missense | Yang et al., 2019 | NA | [30] |
| PTGS2 | rs5275 | 3’UTR | Yang et al., 2019 | NA | [30] |
| PTGS2 | rs20417 | Upstream | Lee et al., 2006 | USA | [44] |
| RFFL | rs16970540 | 3’UTR | Lee et al., 2019 | European-Americans, Nigeria; Han Chinese; and Japanese | [28] |
| RHBDF2 | rs12948783 | Upstream | Galvan et al., 2011 | European | [45] |
| SPON1 | rs13421094 | Intergenic | Galvan et al., 2011 | European | [45] |
| SPON1 | rs12211463 | Intergenic | Galvan et al., 2011 | European | [45] |
| SPON1 | rs7757130 | Intronic | Galvan et al., 2011 | European | [45] |
| SPON1 | rs2473967 | Intronic | Galvan et al., 2011 | European | [45] |
| SPON1 | rs2884129 | Intergenic | Galvan et al., 2011 | European | [45] |
| SPON1 | rs7104613 | Intronic | Galvan et al., 2011 | European | [45] |
| SYNE1 | rs71575922 | Intronic | Rahmioglu et al., 2023 | European and East Asian | [38] |
| TNF | rs1800629 | Upstream | Yang et al., 2019 | NA | [30] |
| TNF | rs1800610 | Intronic | Yang et al., 2019 | NA | [30] |
| TNF | rs1800469 | Upstream | Yang et al., 2019 | NA | [30] |
| TNF | rs2241716 | Intronic | Yang et al., 2019 | NA | [30] |
| TNF | rs1800629 | Upstream | Crescioli et al. 2022 | Italy | [40] |
| UGT2B7 | rs7439366 | Missense | Satkunananthan et al., 2022 | Asian | [27] |
| UGT2B7 | rs7438135 | Upstream | Tian et al., 2012 | Italy | [46] |
| VDR | rs11568820 | Intronic | Yang et al., 2019 | NA | [30] |
| ZNF235 | rs10413396 | 5’UTR | Galvan et al., 2011 | European | [45] |
| * | rs2369049 | Intergenic | Genovese and Mao, 2019 | USA | [47] |
| Study | Tools Used to Collect Pain Information from Patients | Findings | Ref. |
|---|---|---|---|
| Cepeda et al. | CPPR and PRPPR | Pain decreased from initial pain following analgesic administration. | [56] |
| Musshoff et al. | Hair analysis | Hair analysis can be a valuable and supplementary method for isolating patients who take opioid analgesics for pain relief. | [57] |
| Dalal and Bruera | Personalized therapy and education of patient/family in decision-making | Cancer-related pain is a multidimensional construct resulting from a complicated combination of physiological, socio-cultural, psychological, behavioral, sensory, and cognitive factors. | [58] |
| Sarzi-Puttini et al. | Selecting an effective, suitable, personalized analgesic prescription for individuals with chronic pain is feasible | Personalized analgesic prescription will improve compliance, general functioning, and QoL. | [59] |
| Petersen et al. | Prepared an item bank of 16 pain-measurement items appropriate for CAT | The pain CAT is now available for “experimental” uses by the EORTC. | [61] |
| Balducci and Dolan | Investigated palliative care for disease in elderly patients | Target planning, symptom control, and caregiver attention are the three foundations of effective palliative care. | [62] |
| Bhatnagar and Gupta | Integrating cancer pain and symptom management into present pain management | Simultaneously collect information and develop guidelines and suggestions for accurate symptom management across a wide range of patients and diseases to provide a personalized strategy for patient care. | [63] |
| Arthur et al. | Relationship between ECS-CP characteristics and pain treatment results among outpatients | Neuropathy was a poor predictive factor in the treatment of advanced cancer pain. | [64] |
| Colvin | Repurposing drugs such as metformin | During oncological treatment, any new therapies adopted must not conflict with the tumoricidal impacts of chemotherapy. | [66] |
| Vimalnath et al. | Production and investigation of Ce-141 as an effective theragnostic agent for metastatic skeletal lesions | Potential value of 141Ce-DOTMP as a theragnostic component proved for tailored patient treatment of cancer patients. | [67] |
| Sica et al. | Studied the efficiency of the intrathecal pump in 140 patients | Intrathecal is safer than systemic opioids, which frequently require greater dosages to be efficient leading to the probable major side effects. | [68] |
| Cuomo et al. | Trolley analgesic model | The employment of personalized therapies with dynamic multimodal methods for pain management found. | [70] |
| LeBaron et al. | BE-SI-C | BE-SI-C has the ability to track and predict pain while also improving self-efficacy, safety, communication, and QoL in cancer patients. | [72] |
| Oldenmenger et al. | Examined nine BAT questions, such as “How often do you get breakthrough pain?” | BAT is a legitimate and precise questionnaire that may be used in everyday practice to measure breakthrough pain in Dutch cancer patients. | [74] |
| Ben-Arye et al. | IO therapy in 815 eligible patients receiving cancer treatment in adjuvant, neo-adjuvant, and palliative care settings. | An initial consultation with an integrative clinician and follow-up visits and receiving adjuvant and neo-adjuvant were the benefits. | [76] |
| Mao et al. | Electroacupuncture or auricular acupuncture | Electroacupuncture and auricular acupuncture reduced pain. | [77] |
| Batistaki et al. | Investigated the relationship between BTcP, background cancer pain, and analgesic therapy | A multimodal analgesic approach is proposed. | [81] |
| Masukawa et al. | Established machine learning models in CPM | They predicted social pain, spiritual pain, and severe signs in terminally suffering cancer patients using text data from electronic healthcare records. | [82] |
| Reddy et al. | Transition from opioids to levorphanol utilizing an ORR of 8.5 | Levorphanol was well tolerated and linked to better pain and symptom management. | [84] |
| Aziz and Cascella | Peripheral neurolytic blocks | Some forms of painful diseases must be handled by administration of less-invasive analgesic procedures. | [85] |
| Dalal et al. | Utilized level of pain reduction on a scale from 0 to 10 and median PPG | Regular PPG recording may help with personalized pain management. | [86] |
| Study | Genes | Drugs/Methods | Ref. |
|---|---|---|---|
| Galvan et al. | SPON1, RHBDF2, ZNF235 | Opioids | [45] |
| Skorpen et al. | OPRM1 | Opioids | [42] |
| Tverdohleb et al. | CYP2D6 expression | Opioids | [90] |
| Sivanesan and Gitlin | OPRM1 | Tramadol and ziconotide | [91] |
| Obeng et al. | OPRM1, CYP2D6 | Morphine, oxycodone, and hydrocodone | [92] |
| Haji et al. | OPRM1 | Morphine | [43] |
| Mosley et al. | CYP2D6 | Oxycodone | [37] |
| Yang et al. | OPRM1, COMT, CYP2D6, and ILs | Opioid analgesics | [30] |
| Nissenbaum et al. | CACNG2 | - | [93] |
| Bortsov et al. | CACNG2 | Anti-epileptics | [32] |
| Lee et al. | RFFL/LIG3, ABCC4/MPR4, EGFL6 | - | [28] |
| Genovese and Mao | COMT | Acupuncture | [47] |
| Xu et al. | OPRM1, OPRK1, OPRD1, SSTR1, SSTR2, and SSTR5 | Herbal drugs | [95] |
| De Bono et al. | BRCA1, BRCA2, ATM | Olaparib | [97] |
| Bugada et al. | COMT, OPRM1 | Opioids | [33] |
| Rienzo et al. | PRDM12 | - | [98] |
| Reizine et al. | CYP2D6 | Codeine, tramadol, hydrocodone | [99] |
| Saloman et al. | IL1β, IL6, IL2, TNF, MCP1, IL-4, IL-8, CGRP | Pain biomarkers in serum | [100] |
| Crescioli et al. | IL6, TNF | Opioids | [40] |
| Satkunananthan et al. | CYP2D6, OPRM1, COMT, ABCB1 | Tramadol | [27] |
| Wang et al. | IL-6, TNF-α | Oxycodone | [109] |
| Nishizawa et al. | ANGPT1, SLC2A14 | Opioid analgesic | [110] |
| Li et al. | F13B | Sufentanil | [111] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raad, M.; López, W.O.C.; Sharafshah, A.; Assefi, M.; Lewandrowski, K.-U. Personalized Medicine in Cancer Pain Management. J. Pers. Med. 2023, 13, 1201. https://doi.org/10.3390/jpm13081201
Raad M, López WOC, Sharafshah A, Assefi M, Lewandrowski K-U. Personalized Medicine in Cancer Pain Management. Journal of Personalized Medicine. 2023; 13(8):1201. https://doi.org/10.3390/jpm13081201
Chicago/Turabian StyleRaad, Mohammad, William Omar Contreras López, Alireza Sharafshah, Marjan Assefi, and Kai-Uwe Lewandrowski. 2023. "Personalized Medicine in Cancer Pain Management" Journal of Personalized Medicine 13, no. 8: 1201. https://doi.org/10.3390/jpm13081201
APA StyleRaad, M., López, W. O. C., Sharafshah, A., Assefi, M., & Lewandrowski, K.-U. (2023). Personalized Medicine in Cancer Pain Management. Journal of Personalized Medicine, 13(8), 1201. https://doi.org/10.3390/jpm13081201







