A Statistical Approach to Assess the Robustness of Radiomics Features in the Discrimination of Mammographic Lesions
Abstract
1. Introduction
2. Methods
2.1. Dataset
2.2. Methodological Workflow
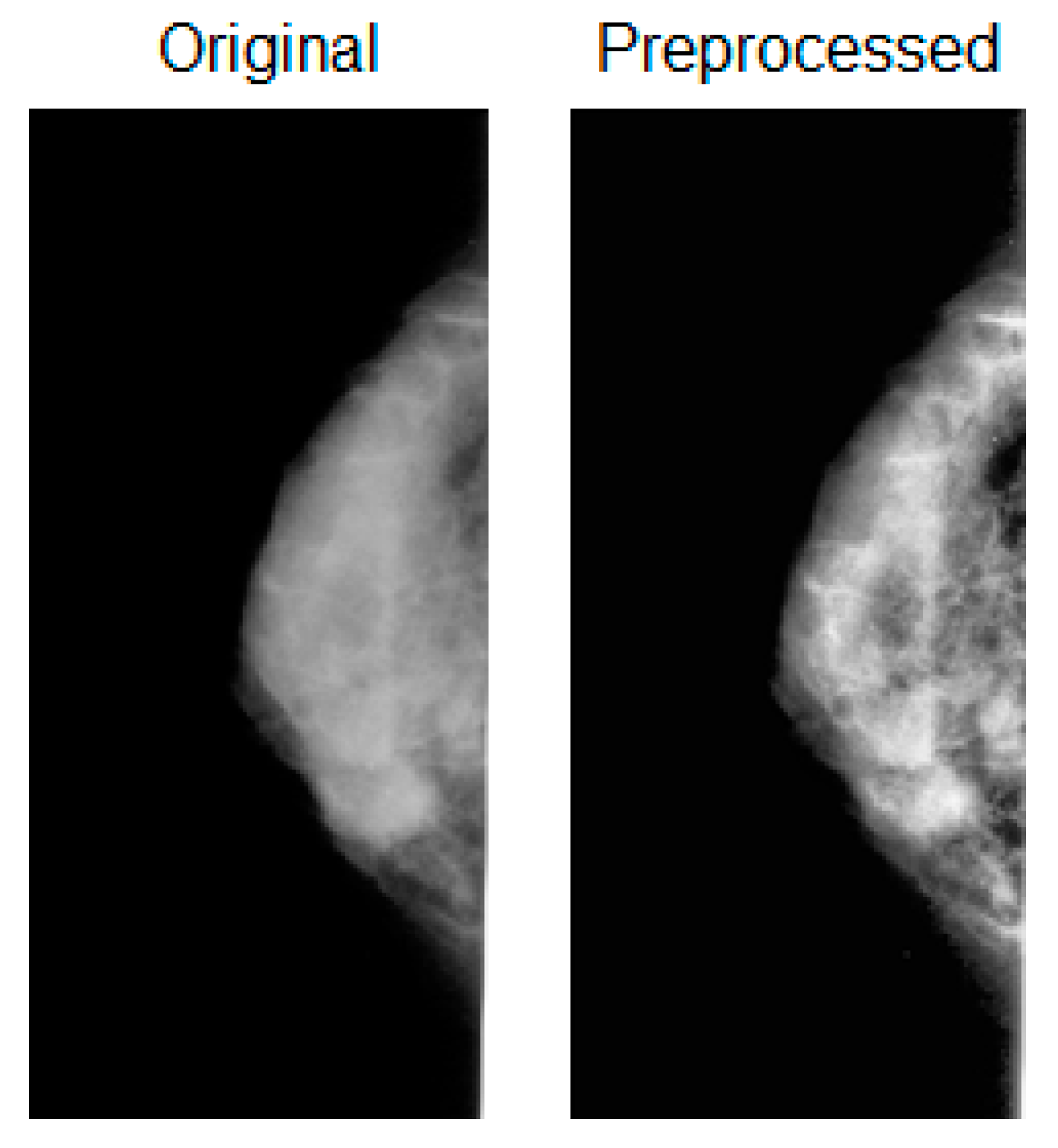
2.3. Preprocessing
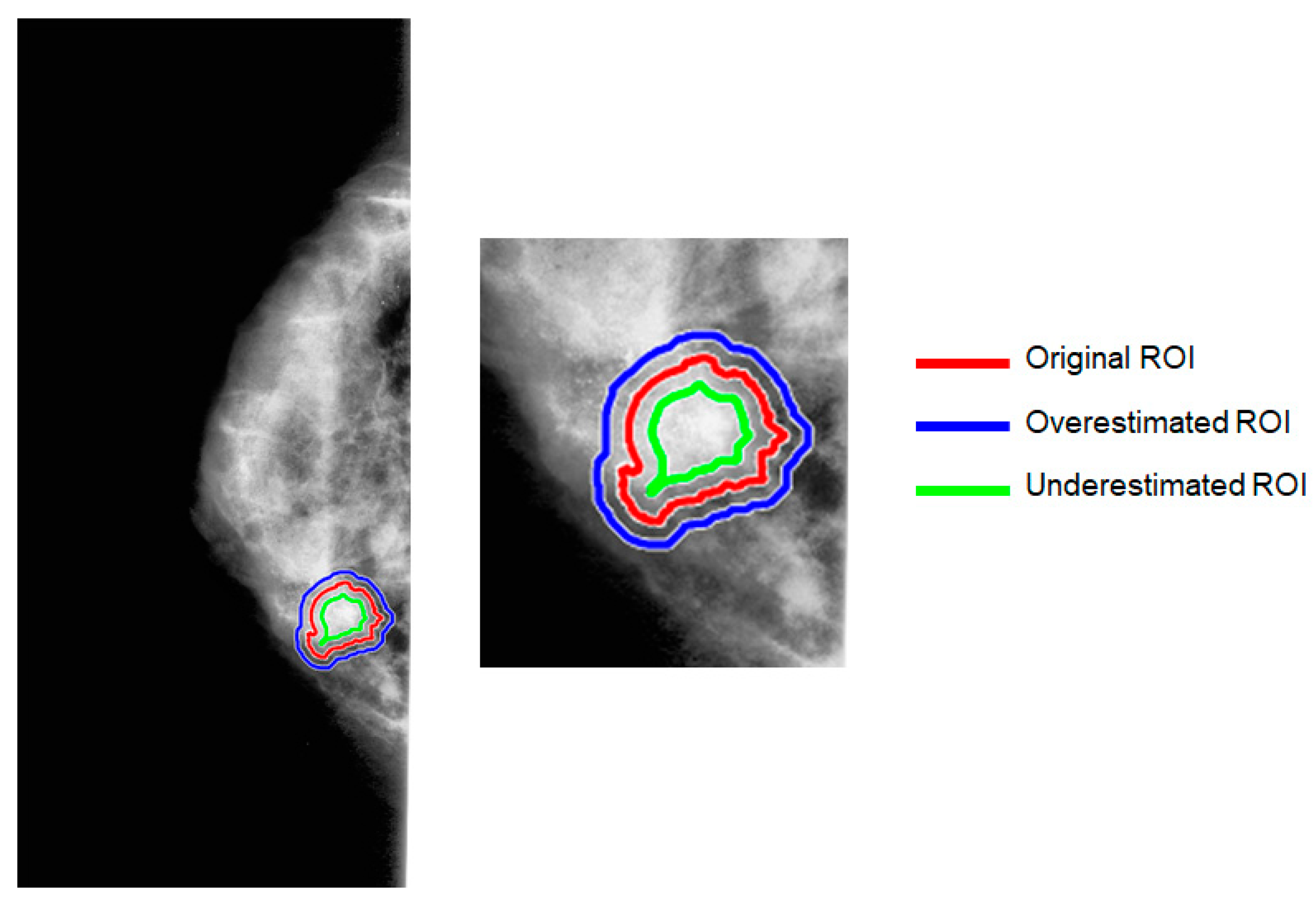
2.4. Generation of Artificial ROI
2.5. Radiomics Feature Extraction
2.6. Statistical Analysis and Radiomics Robustness Score
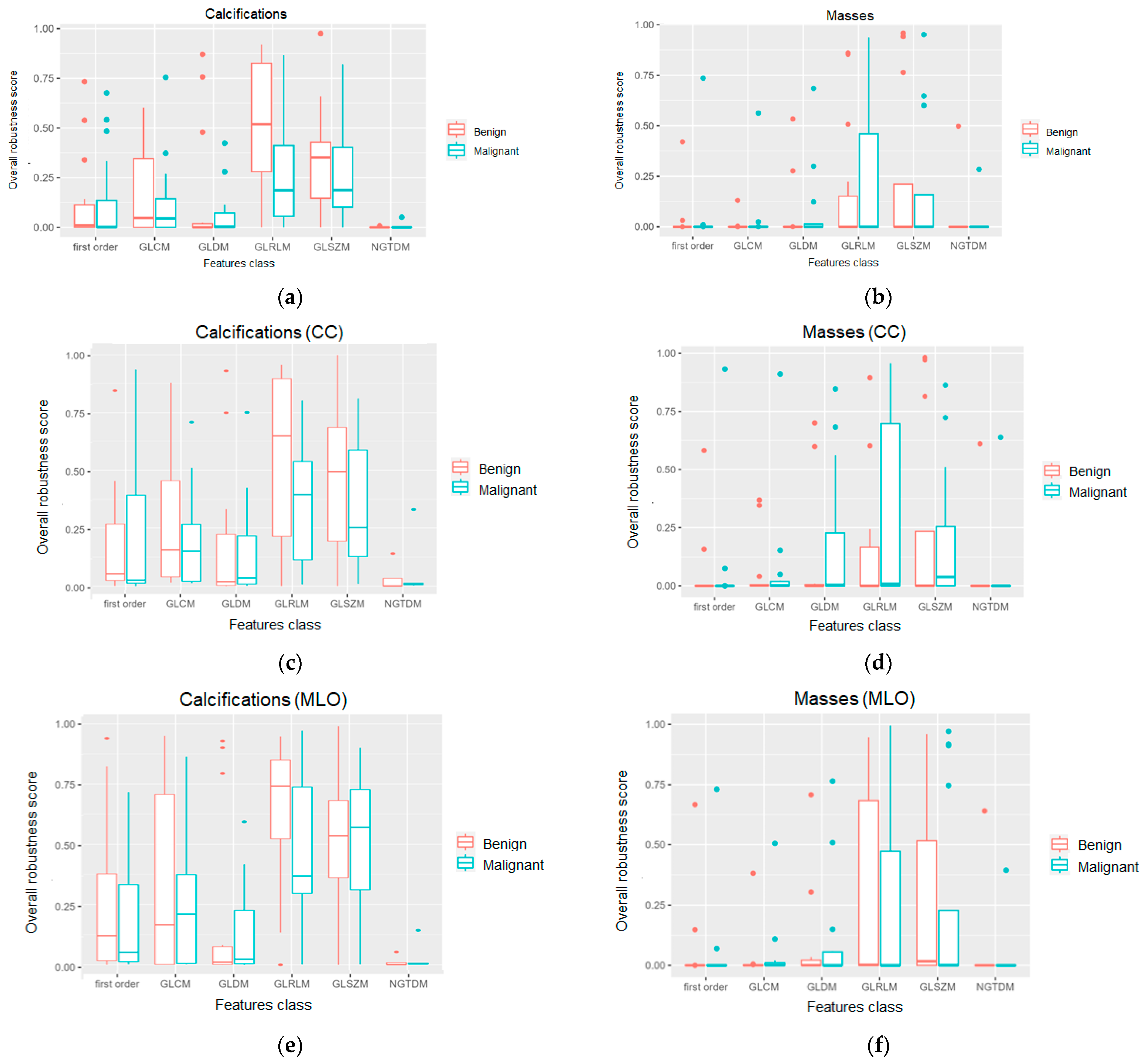
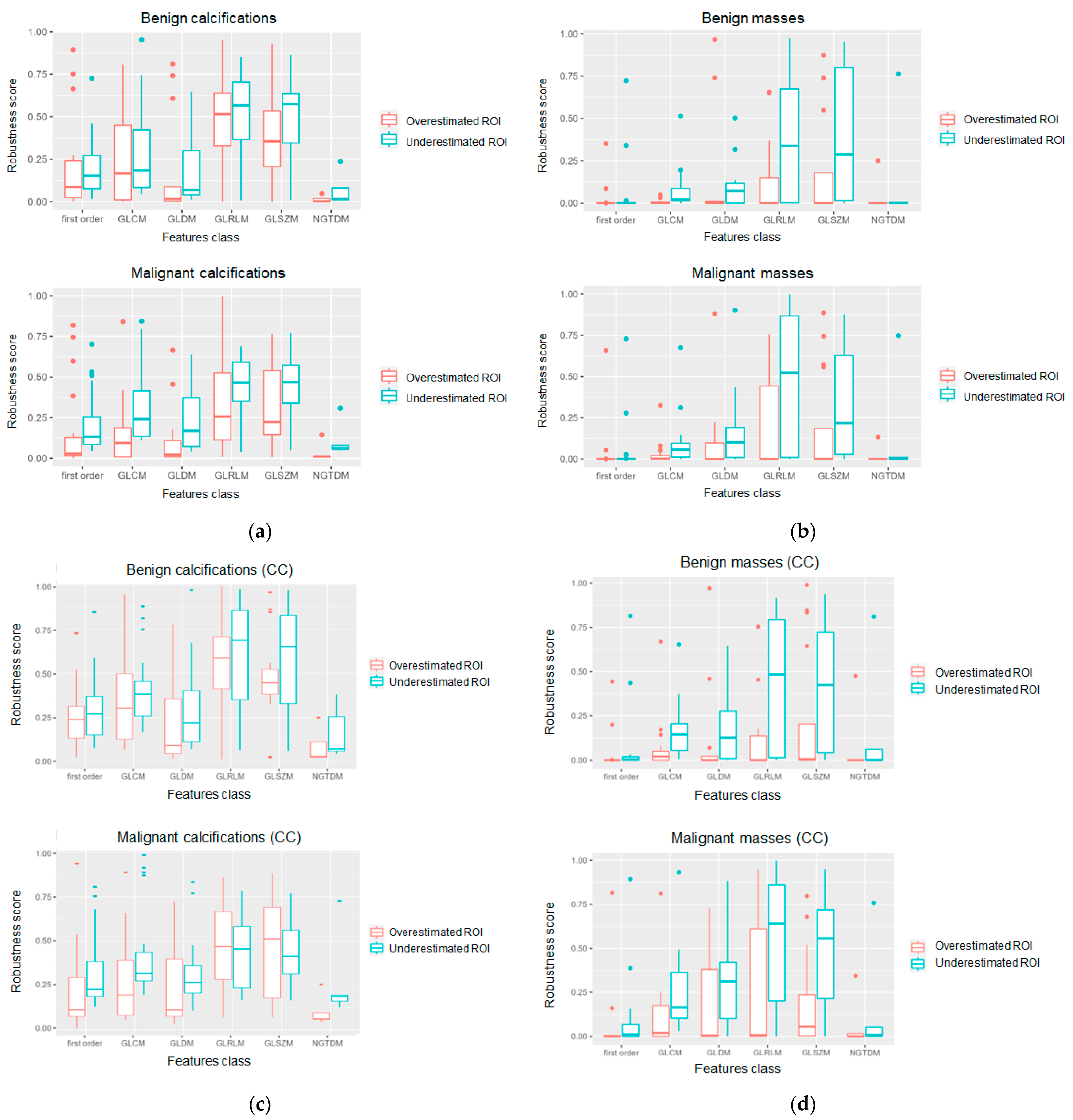
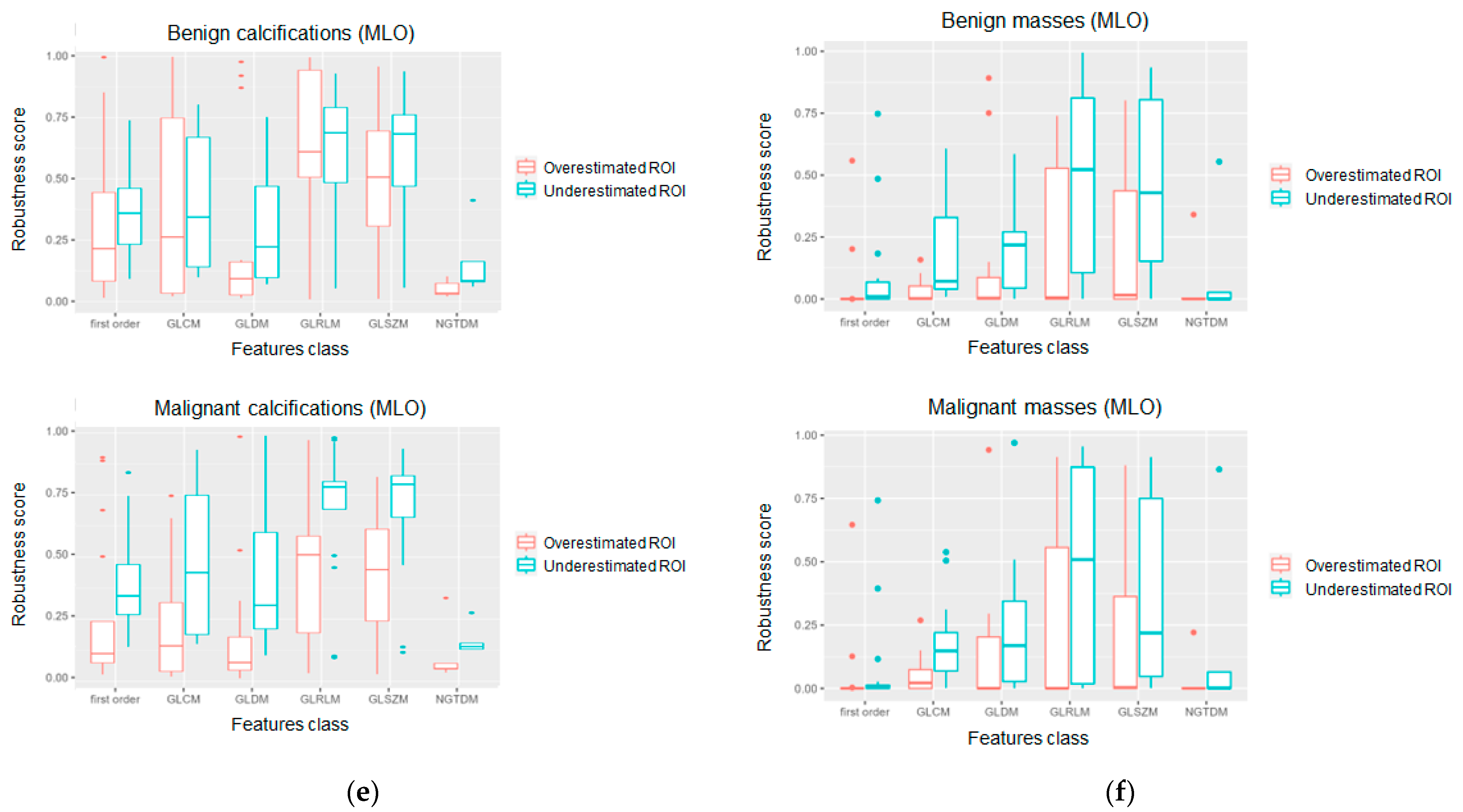
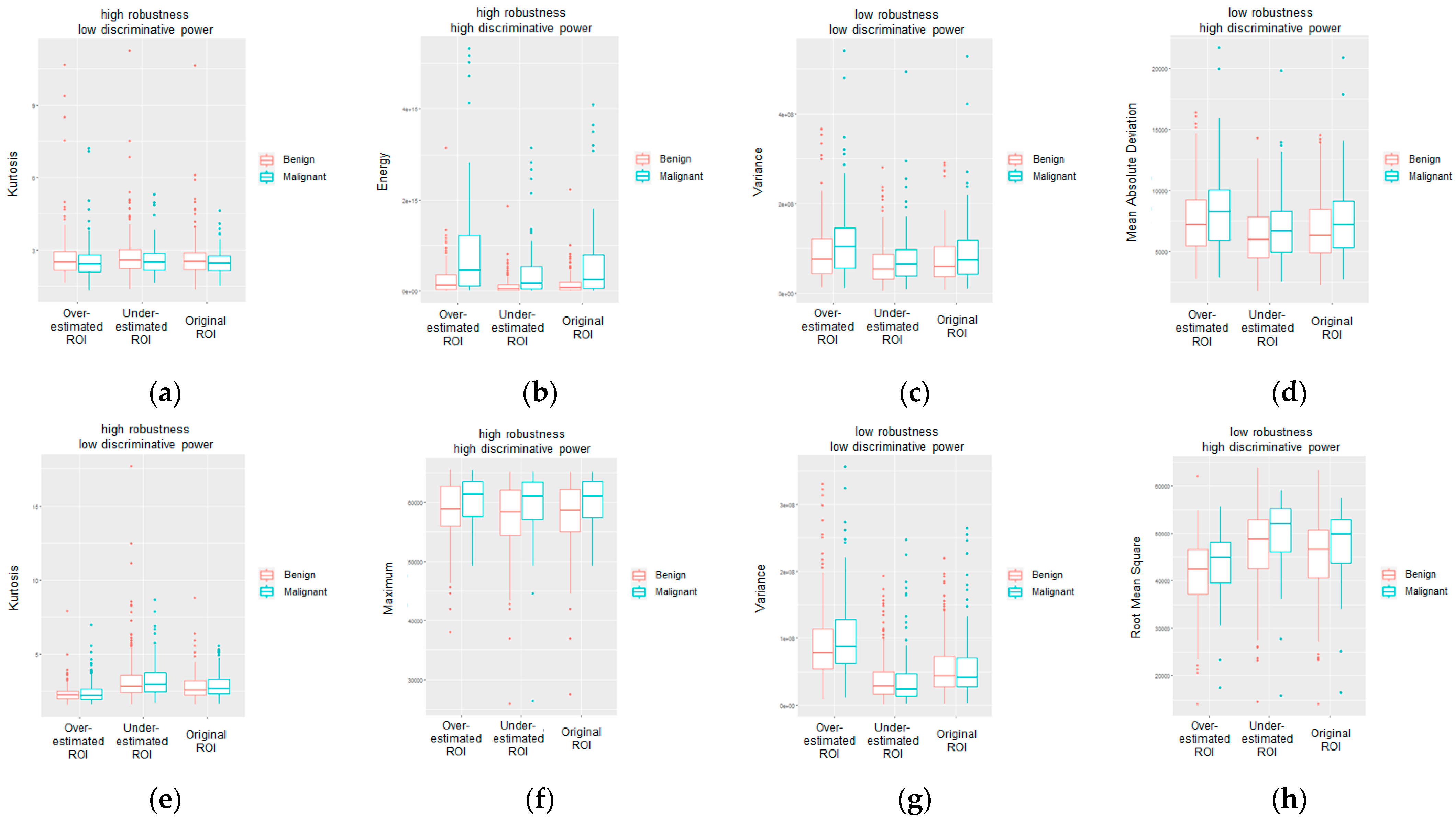
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nindrea, R.D.; Aryandono, T.; Lazuardi, L.; Dwiprahasto, I. Diagnostic Accuracy of Different Machine Learning Algorithms for Breast Cancer Risk Calculation: A Meta-Analysis. Asian Pac. J. Cancer Prev. APJCP 2018, 19, 1747–1752. [Google Scholar] [CrossRef]
- Davis, J.; Liang, J.; Petterson, M.B.; Roh, A.T.; Chundu, N.; Kang, P.; Matz, S.L.; Connell, M.J.; Gridley, D.G. Risk Factors for Late Screening Mammography. Curr. Probl. Diagn. Radiol. 2019, 48, 40–44. [Google Scholar] [CrossRef] [PubMed]
- Takkar, N.; Kochhar, S.; Garg, P.; Pandey, A.K.; Dalal, U.R.; Handa, U. Screening methods (clinical breast examination and mammography) to detect breast cancer in women aged 40–49 years. J. Life Health 2017, 8, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Ho, P.J.; Lau, H.S.H.; Ho, W.K.; Wong, F.Y.; Yang, Q.; Tan, K.W.; Tan, M.-H.; Chay, W.Y.; Chia, K.S.; Hartman, M.; et al. Incidence of breast cancer attributable to breast density, modifiable and non-modifiable breast cancer risk factors in Singapore. Sci. Rep. 2020, 10, 503. [Google Scholar] [CrossRef]
- Tumori, A.I.R. I numeri del cancro in italia 2021. Intermedia Ed. 2021, 64–66. Available online: https://www.aiom.it/wp-content/uploads/2021/10/2021_NumeriCancro_web.pdf (accessed on 1 May 2023).
- Tubiana-Hulin, M.; Stevens, D.; Lasry, S.; Guinebretière, J.M.; Bouita, L.; Cohen-Solal, C.; Cherel, P.; Rouëssé, J. Response to neoadjuvant chemotherapy in lobular and ductal breast carcinomas: A retrospective study on 860 patients from one institution. Ann. Oncol. 2006, 17, 1228–1233. [Google Scholar] [CrossRef] [PubMed]
- Swanick, C.W.; Smith, B.D. Indications for adjuvant radiation therapy in breast cancer: A review of the evidence and recommendations for clinical practice. Chin. Clin. Oncol. 2016, 5, 38. [Google Scholar] [CrossRef]
- Riaz, N.; Jeen, T.; Whelan, T.J.; Nielsen, T.O. Recent Advances in Optimizing Radiation Therapy Decisions in Early Invasive Breast Cancer. Cancers 2023, 15, 1260. [Google Scholar] [CrossRef]
- Gøtzsche, P.C.; Jørgensen, K.J. Screening for breast cancer with mammography. Cochrane Database Syst. Rev. 2013, 2013, CD001877. [Google Scholar] [CrossRef]
- Burrell, H.C.; Sibbering, D.M.; Wilson, A.R.; Pinder, S.E.; Evans, A.J.; Yeoman, L.J.; Elston, C.W.; Ellis, I.O.; Blamey, R.W.; Robertson, J.F. Screening interval breast cancers: Mammographic features and prognosis factors. Radiology 1996, 199, 811–817. [Google Scholar] [CrossRef]
- Mann, R.M.; Hooley, R.; Barr, R.G.; Moy, L. Novel Approaches to Screening for Breast Cancer. Radiology 2020, 297, 266–285. [Google Scholar] [CrossRef] [PubMed]
- Dang, L.-A.; Chazard, E.; Poncelet, E.; Serb, T.; Rusu, A.; Pauwels, X.; Parsy, C.; Poclet, T.; Cauliez, H.; Engelaere, C.; et al. Impact of artificial intelligence in breast cancer screening with mammography. Breast Cancer 2022, 29, 967–977. [Google Scholar] [CrossRef]
- Tari, D.U.; Pinto, F. Mammography in Breast Disease Screening and Diagnosis. J. Pers. Med. 2023, 13, 228. [Google Scholar] [CrossRef] [PubMed]
- Houn, F.; Brown, M.L. Current practice of screening mammography in the United States: Data from the National Survey of Mammography Facilities. Radiology 1994, 190, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Melekoodappattu, J.G.; Subbian, P.S. A Hybridized ELM for Automatic Micro Calcification Detection in Mammogram Images Based on Multi-Scale Features. J. Med. Syst. 2019, 43, 183. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Ruiz, A.; Krupinski, E.; Mordang, J.-J.; Schilling, K.; Heywang-Köbrunner, S.H.; Sechopoulos, I.; Mann, R.M. Detection of Breast Cancer with Mammography: Effect of an Artificial Intelligence Support System. Radiology 2019, 290, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Korte, J.C.; Cardenas, C.; Hardcastle, N.; Kron, T.; Wang, J.; Bahig, H.; Elgohari, B.; Ger, R.; Court, L.; Fuller, C.D.; et al. Radiomics feature stability of open-source software evaluated on apparent diffusion coefficient maps in head and neck cancer. Sci. Rep. 2021, 11, 17633. [Google Scholar] [CrossRef]
- Dinnes, J.; Moss, S.; Melia, J.; Blanks, R.; Song, F.; Kleijnen, J. Effectiveness and cost-effectiveness of double reading of mammograms in breast cancer screening: Findings of a systematic review. In Database of Abstracts of Reviews of Effects (DARE): Quality-assessed Reviews [Internet]; Centre for Reviews and Dissemination: Edinburgh, UK, 2001. [Google Scholar]
- Posso, M.; Puig, T.; Carles, M.; Rué, M.; Canelo-Aybar, C.; Bonfill, X. Effectiveness and cost-effectiveness of double reading in digital mammography screening: A systematic review and meta-analysis. Eur. J. Radiol. 2017, 96, 40–49. [Google Scholar] [CrossRef]
- Taplin, S.; Abraham, L.; Barlow, W.E.; Fenton, J.J.; Berns, E.A.; Carney, P.A.; Cutter, G.R.; Sickles, E.A.; Carl, D.; Elmore, J.G. Mammography Facility Characteristics Associated with Interpretive Accuracy of Screening Mammography. JNCI J. Natl. Cancer Inst. 2008, 100, 876–887. [Google Scholar] [CrossRef]
- Taplin, S.H.; Rutter, C.M.; Elmore, J.G.; Seger, D.; White, D.; Brenner, R.J. Accuracy of Screening Mammography Using Single Versus Independent Double Interpretation. Am. J. Roentgenol. 2000, 174, 1257–1262. [Google Scholar] [CrossRef]
- Andreadis, I.; Nikita, K.; Spyrou, G. Investigating the performance of a CADx scheme for mammography in specific BIRADS categories. In Proceedings of the 2014 IEEE International Conference on Imaging Systems and Techniques (IST) Proceedings, Santorini, Greece, 14–17 October 2014; pp. 335–339. [Google Scholar]
- Li, L.; Zheng, Y.; Zhang, L.; Clark, R.A. Anisotropic diffusion filtering of mammographic image for CAD in digital mammography. In Proceedings of the WCC 2000—ICSP 2000. 2000 5th International Conference on Signal Processing Proceedings. 16th World Computer Congress 2000, Beijing, China, 21–25 August 2000; Volume 2, pp. 1159–1162. [Google Scholar]
- Baker, J.A.; Rosen, E.L.; Lo, J.Y.; Gimenez, E.I.; Walsh, R.; Soo, M.S. Computer-Aided Detection (CAD) in Screening Mammography: Sensitivity of Commercial CAD Systems for Detecting Architectural Distortion. Am. J. Roentgenol. 2003, 181, 1083–1088. [Google Scholar] [CrossRef] [PubMed]
- Warren Burhenne, L.J.; Wood, S.A.; D’Orsi, C.J.; Feig, S.A.; Kopans, D.B.; O’Shaughnessy, K.F.; Sickles, E.A.; Tabar, L.; Vyborny, C.J.; Castellino, R.A. Potential Contribution of Computer-aided Detection to the Sensitivity of Screening Mammography. Radiology 2000, 215, 554–562. [Google Scholar] [CrossRef] [PubMed]
- Malich, A.; Fischer, D.R.; Böttcher, J. CAD for mammography: The technique, results, current role and further developments. Eur. Radiol. 2006, 16, 1449–1460. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Lin, K.; Yan, W.; Guo, Y.; Wang, Y.; Li, J.; Zhu, J. Computer-Aided Diagnosis of Pancreas Serous Cystic Neoplasms: A Radiomics Method on Preoperative MDCT Images. Technol. Cancer Res. Treat. 2019, 18, 1533033818824339. [Google Scholar] [CrossRef]
- Yang, S.K.; Moon, W.K.; Cho, N.; Park, J.S.; Cha, J.H.; Kim, S.M.; Kim, S.J.; Im, J.-G. Screening Mammography–detected Cancers: Sensitivity of a Computer-aided Detection System Applied to Full-Field Digital Mammograms. Radiology 2007, 244, 104–111. [Google Scholar] [CrossRef]
- Chen, W.; Liu, B.; Peng, S.; Sun, J.; Qiao, X. Computer-Aided Grading of Gliomas Combining Automatic Segmentation and Radiomics. Int. J. Biomed. Imaging 2018, 2018, e2512037. [Google Scholar] [CrossRef] [PubMed]
- Lo, C.-M.; Weng, R.-C.; Cheng, S.-J.; Wang, H.-J.; Hsieh, K.L.-C. Computer-aided diagnosis of isocitrate dehydrogenase genotypes in glioblastomas from radiomic patterns. Medicine 2020, 99, e19123. [Google Scholar] [CrossRef]
- Suter, Y.; Knecht, U.; Alão, M.; Valenzuela, W.; Hewer, E.; Schucht, P.; Wiest, R.; Reyes, M. Radiomics for glioblastoma survival analysis in pre-operative MRI: Exploring feature robustness, class boundaries, and machine learning techniques. Cancer Imaging 2020, 20, 55. [Google Scholar] [CrossRef]
- Belfiore, M.P.; Sansone, M.; Monti, R.; Marrone, S.; Fusco, R.; Nardone, V.; Grassi, R.; Reginelli, A. Robustness of Radiomics in Pre-Surgical Computer Tomography of Non-Small-Cell Lung Cancer. J. Pers. Med. 2023, 13, 83. [Google Scholar] [CrossRef]
- Fusco, R.; Piccirillo, A.; Sansone, M.; Granata, V.; Rubulotta, M.R.; Petrosino, T.; Barretta, M.L.; Vallone, P.; Di Giacomo, R.; Esposito, E.; et al. Radiomics and Artificial Intelligence Analysis with Textural Metrics Extracted by Contrast-Enhanced Mammography in the Breast Lesions Classification. Diagnostics 2021, 11, 815. [Google Scholar] [CrossRef]
- Angelone, F.; Ricciardi, C.; Gatta, G.; Sansone, M.; Ponsiglione, A.M.; Belfiore, M.P.; Amato, F.; Grassi, R. Breast Density Analysis on Mammograms: Application of Machine Learning with Textural Features. In Proceedings of the 2022 IEEE International Conference on Metrology for Extended Reality, Artificial Intelligence and Neural Engineering (MetroXRAINE), Rome, Italy, 26–28 October 2022; pp. 295–300. [Google Scholar]
- Sansone, M.; Ponsiglione, A.M.; Angelone, F.; Amato, F.; Grassi, R. Effect of X-ray scatter correction on the estimation of attenuation coefficient in mammography: A simulation study. In Proceedings of the 2022 IEEE International Conference on Metrology for Extended Reality, Artificial Intelligence and Neural Engineering (MetroXRAINE), Rome, Italy, 26–28 October 2022; pp. 323–328. [Google Scholar]
- Sansone, M.; Fusco, R.; Grassi, F.; Gatta, G.; Belfiore, M.P.; Angelone, F.; Ricciardi, C.; Ponsiglione, A.M.; Amato, F.; Galdiero, R.; et al. Machine Learning Approaches with Textural Features to Calculate Breast Density on Mammography. Curr. Oncol. Tor. Ont 2023, 30, 839–853. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.G.F.; Campos, J.P.M.; de Aquino e Aquino, G.; de Albuquerque Pereira, W.C.; Costa Filho, C.F.F. Evaluating the performance of convolutional neural networks with direct acyclic graph architectures in automatic segmentation of breast lesion in US images. BMC Med. Imaging 2019, 19, 85. [Google Scholar] [CrossRef] [PubMed]
- Fleury, E.; Marcomini, K. Performance of machine learning software to classify breast lesions using BI-RADS radiomic features on ultrasound images. Eur. Radiol. Exp. 2019, 3, 34. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, T.; Li, J.; Pei, Y.; Akhtar, F.; Imran, A.; Yaqub, M. An Automatic Detection and Localization of Mammographic Microcalcifications ROI with Multi-Scale Features Using the Radiomics Analysis Approach. Cancers 2021, 13, 5916. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Margolies, L.R.; Rothstein, J.H.; Fluder, E.; McBride, R.; Sieh, W. Deep Learning to Improve Breast Cancer Detection on Screening Mammography. Sci. Rep. 2019, 9, 12495. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Aghaei, F.; Zarafshani, A.; Qiu, Y.; Qian, W.; Zheng, B. Computer-aided Classification of Mammographic Masses Using Visually Sensitive Image Features. J. X-ray Sci. Technol. 2017, 25, 171–186. [Google Scholar] [CrossRef]
- Sawyer-Lee, R.; Gimenez, F.; Hoogi, A.; Rubin, D. Curated Breast Imaging Subset of Digital Database for Screening Mammography (CBIS-DDSM). 2016. Available online: https://wiki.cancerimagingarchive.net/x/lZNXAQ (accessed on 1 May 2023).
- Lee, R.S.; Gimenez, F.; Hoogi, A.; Miyake, K.K.; Gorovoy, M.; Rubin, D.L. A curated mammography data set for use in computer-aided detection and diagnosis research. Sci. Data 2017, 4, 170177. [Google Scholar] [CrossRef]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef]
- Tripathy, S.; Swarnkar, T. Unified Preprocessing and Enhancement Technique for Mammogram Images. Procedia Comput. Sci. 2020, 167, 285–292. [Google Scholar] [CrossRef]
- Zuiderveld, K. Contrast Limited Adaptive Histogram Equalization; Elsevier: Amsterdam, The Netherlands, 1994; pp. 474–485. [Google Scholar]
- van den Boomgaard, R.; van Balen, R. Methods for fast morphological image transforms using bitmapped binary images. CVGIP Graph. Model. Image Process. 1992, 54, 252–258. [Google Scholar] [CrossRef]
- Gonzalez, R.C.; Woods, R.E.; Eddins, S.L. Digital Image Processing Using MATLAB. 3; Gatesmark Publishing: Knoxville, Tennessee, 2020. [Google Scholar]
- Mendel, K.R.; Li, H.; Lan, L.; Cahill, C.M.; Rael, V.; Abe, H.; Giger, M.L. Quantitative texture analysis: Robustness of radiomics across two digital mammography manufacturers’ systems. J. Med. Imaging 2018, 5, 011002. [Google Scholar] [CrossRef] [PubMed]
- Robinson, K.; Li, H.; Lan, L.; Schacht, D.; Giger, M. Radiomics robustness assessment and classification evaluation: A two-stage method demonstrated on multivendor FFDM. Med. Phys. 2019, 46, 2145–2156. [Google Scholar] [CrossRef] [PubMed]
- Sansone, M.; Grassi, R.; Belfiore, M.P.; Gatta, G.; Grassi, F.; Pinto, F.; La Casella, G.V.; Fusco, R.; Cappabianca, S.; Granata, V.; et al. Radiomic features of breast parenchyma: Assessing differences between FOR PROCESSING and FOR PRESENTATION digital mammography. Insights Imaging 2021, 12, 147. [Google Scholar] [CrossRef]
- Rashid, H.U.; Ibrikci, T.; Paydaş, S.; Binokay, F.; Çevik, U. Analysis of breast cancer classification robustness with radiomics feature extraction and deep learning techniques. Expert Syst. 2022, 39, e13018. [Google Scholar] [CrossRef]
- Niu, S.; Jiang, W.; Zhao, N.; Jiang, T.; Dong, Y.; Luo, Y.; Yu, T.; Jiang, X. Intra- and peritumoral radiomics on assessment of breast cancer molecular subtypes based on mammography and MRI. J. Cancer Res. Clin. Oncol. 2022, 148, 97–106. [Google Scholar] [CrossRef]
- Militello, C.; Rundo, L.; Dimarco, M.; Orlando, A.; D’Angelo, I.; Conti, V.; Bartolotta, T.V. Robustness Analysis of DCE-MRI-Derived Radiomic Features in Breast Masses: Assessing Quantization Levels and Segmentation Agreement. Appl. Sci. 2022, 12, 5512. [Google Scholar] [CrossRef]
- Caballo, M.; Pangallo, D.R.; Mann, R.M.; Sechopoulos, I. Deep learning-based segmentation of breast masses in dedicated breast CT imaging: Radiomic feature stability between radiologists and artificial intelligence. Comput. Biol. Med. 2020, 118, 103629. [Google Scholar] [CrossRef]
- Losurdo, L.; Fanizzi, A.; Basile, T.M.A.; Bellotti, R.; Bottigli, U.; Dentamaro, R.; Didonna, V.; Lorusso, V.; Massafra, R.; Tamborra, P.; et al. Radiomics Analysis on Contrast-Enhanced Spectral Mammography Images for Breast Cancer Diagnosis: A Pilot Study. Entropy 2019, 21, 1110. [Google Scholar] [CrossRef]
- Wang, S.; Sun, Y.; Mao, N.; Duan, S.; Li, Q.; Li, R.; Jiang, T.; Wang, Z.; Xie, H.; Gu, Y. Incorporating the clinical and radiomics features of contrast-enhanced mammography to classify breast lesions: A retrospective study. Quant. Imaging Med. Surg. 2021, 11, 4418–4430. [Google Scholar] [CrossRef]
- Li, H.; Mendel, K.R.; Lan, L.; Sheth, D.; Giger, M.L. Digital Mammography in Breast Cancer: Additive Value of Radiomics of Breast Parenchyma. Radiology 2019, 291, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Acciavatti, R.J.; Cohen, E.A.; Maghsoudi, O.H.; Gastounioti, A.; Pantalone, L.; Hsieh, M.-K.; Conant, E.F.; Scott, C.G.; Winham, S.J.; Kerlikowske, K.; et al. Robust radiomic feature selection in digital mammography: Understanding the effect of imaging acquisition physics using phantom and clinical data analysis. In Proceedings of the Medical Imaging 2020: Computer-Aided Diagnosis, Houston, TX, USA, 16–19 February 2020; Volume 11314, p. 113140W. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ponsiglione, A.M.; Angelone, F.; Amato, F.; Sansone, M. A Statistical Approach to Assess the Robustness of Radiomics Features in the Discrimination of Mammographic Lesions. J. Pers. Med. 2023, 13, 1104. https://doi.org/10.3390/jpm13071104
Ponsiglione AM, Angelone F, Amato F, Sansone M. A Statistical Approach to Assess the Robustness of Radiomics Features in the Discrimination of Mammographic Lesions. Journal of Personalized Medicine. 2023; 13(7):1104. https://doi.org/10.3390/jpm13071104
Chicago/Turabian StylePonsiglione, Alfonso Maria, Francesca Angelone, Francesco Amato, and Mario Sansone. 2023. "A Statistical Approach to Assess the Robustness of Radiomics Features in the Discrimination of Mammographic Lesions" Journal of Personalized Medicine 13, no. 7: 1104. https://doi.org/10.3390/jpm13071104
APA StylePonsiglione, A. M., Angelone, F., Amato, F., & Sansone, M. (2023). A Statistical Approach to Assess the Robustness of Radiomics Features in the Discrimination of Mammographic Lesions. Journal of Personalized Medicine, 13(7), 1104. https://doi.org/10.3390/jpm13071104







