Blood Plasma Circulating DNA-Protein Complexes: Involvement in Carcinogenesis and Prospects for Liquid Biopsy of Breast Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Blood Samples
- -
- Patients with metabolic obesity syndrome;
- -
- Patients with the presence of hematogenous metastases at the time of surgery (M1);
- -
- Patients who received neoadjuvant therapy.
2.2. Histone-Contained NPC Isolation by Affinity Chromatography
2.3. Characterization of Blood NPC Proteins
2.4. Bioinformatics Analysis of NPC Proteins
3. Results
3.1. Characterization of Blood Plasma NPCs
3.2. Annotation of Proteins from Blood Plasma NPCs
3.3. Involvement of NPC Proteins in Tumor Dissemination
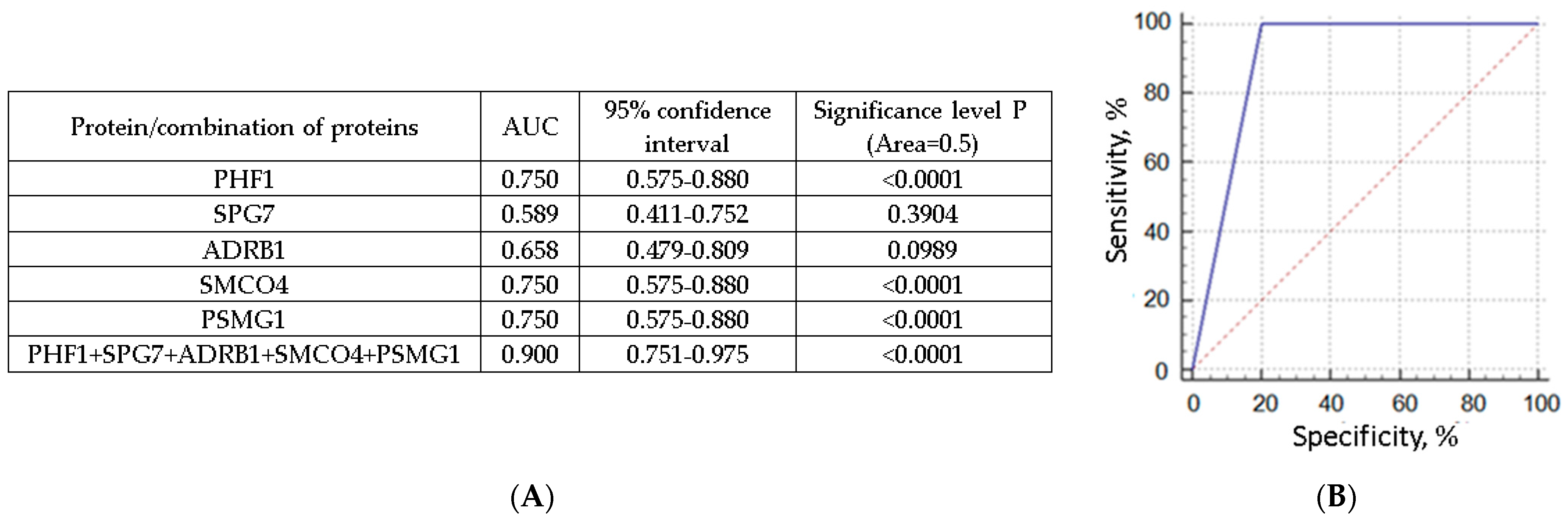
3.4. NPC Proteins as Potential Markers for Liquid Biopsy of Breast Cancer
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, M.; Xie, S.; Lu, C.; Zhu, L.; Zhu, L. Application of data science in circulating tumor DNA detection: A promising avenue towards liquid biopsy. Front. Oncol. 2021, 11, 692322. [Google Scholar] [CrossRef] [PubMed]
- Duque, G.; Manterola, C.; Otzen, T.; Arias, C.; Galindo, B.; Mora, M.; Guerrero, E.; García, N. Clinical utility of liquid biopsy in breast cancer: A systematic review. Clin. Genet. 2022, 101, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Stejskal, P.; Goodarzi, H.; Srovnal, J.; Hajdúch, M.; van’t Veer, L.J.; Magbanua, M.J.M. Circulating tumor nucleic acids: Biology, release mechanisms, and clinical relevance. Mol. Cancer 2023, 22, 15. [Google Scholar] [CrossRef] [PubMed]
- Mouliere, F.; Thierry, A.R. The importance of examining the proportion of circulating DNA originating from tumor, microenvironment and normal cells in colorectal cancer patients. Expert Opin. Biol. Ther. 2012, 12, 209–215. [Google Scholar] [CrossRef]
- Tutanov, O.; Tamkovich, S. The influence of proteins on fate and biological role of circulating DNA. Int. J. Mol. Sci. 2022, 23, 7224. [Google Scholar] [CrossRef]
- Gould, T.J.; Lysov, Z.; Liaw, P.C. Extracellular DNA and histones: Double-edged swords in immunothrombosis. J. Thromb. Haemost. 2015, 13, S82–S91. [Google Scholar] [CrossRef]
- Bryzgunova, O.E.; Tamkovich, S.N.; Cherepanova, A.V.; Yarmoshchuk, S.V.; Permyakova, V.I.; Anykeeva, O.Y.; Laktionov, P.P. Redistribution of free- and cell-surface-bound DNA in blood of benign and malignant prostate tumor patients. Acta Naturae 2015, 7, 115–118. [Google Scholar] [CrossRef]
- Shaw, J.-P.; Kent, K.; Bird, J.; Fishback, J.; Froehler, B. Modified deoxyoligonucleotides stable to exsonuclease degradation in serum. Nucleic Acids Res. 1991, 19, 747–750. [Google Scholar] [CrossRef]
- Tsoneva, D.K.; Ivanov, M.N.; Conev, N.V.; Manev, R.; Stoyanov, D.S.; Vinciguerra, M. Circulating histones to detect and monitor the progression of cancer. Int. J. Mol. Sci. 2023, 24, 942. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, X.; Pelayo, R.; Monestier, M.; Ammollo, C.T.; Semeraro, F.; Taylor, F.B.; Esmon, N.L.; Lupu, F.; Esmon, C.T. Extracellular histones are major mediators of death in sepsis. Nat. Med. 2009, 15, 1318–1321. [Google Scholar] [CrossRef]
- Tian, J.; Avalos, A.M.; Mao, S.; Chen, B.; Senthil, K.; Wu, H.; Parroche, P.; Drabic, S.; Golenbock, D.; Sirois, C.; et al. Toll-like receptor 9- dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE. Nat. Immunol. 2007, 8, 487–496. [Google Scholar] [CrossRef]
- Scaffidi, P.; Misteli, T.; Bianchi, M.E. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature 2002, 418, 191–195. [Google Scholar] [CrossRef]
- Chen, Z.; Fadiel, A.; Naftolin, F.; Eichenbaum, K.D.; Xia, Y. Circulation DNA: Biological implications for cancer metastasis and immunology. Med. Hypotheses 2005, 65, 956–961. [Google Scholar] [CrossRef]
- Wang, K.; Shan, S.; Wang, S.; Gu, X.; Zhou, X.; Ren, T. HMGB1-containing nucleosome mediates chemotherapy-induced metastasis of human lung cancer. Biochem. Biophys. Res. Commun. 2018, 500, 758–764. [Google Scholar] [CrossRef]
- Tanner, J.E.; Forte, A.; Panchal, C. Nucleosomes bind fibroblast growth factor-2 for increased angiogenesis in vitro and in vivo. Mol. Cancer Res. 2004, 2, 281–288. [Google Scholar] [CrossRef]
- Tanner, J.E. Nucleosomes activate NF-kappaB in endothelial cells for induction of the proangiogenic cytokine IL-8. Int. J. Cancer 2004, 112, 155–160. [Google Scholar] [CrossRef]
- Holdenrieder, S.; Stieber, P. Clinical use of circulating nucleosomes. Crit. Rev. Clin. Lab. Sci. 2009, 46, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Kuroi, K.; Tanaka, C.; Toi, M. Plasma nucleosome levels in node-negative breast cancer patients. Breast Cancer 1999, 6, 361–364. [Google Scholar] [CrossRef] [PubMed]
- Barrea, L.; Muscogiuri, G.; Pugliese, G.; de Alteriis, G.; Colao, A.; Savastano, S. Metabolically Healthy Obesity (MHO) vs. Metabolically Unhealthy Obesity (MUO) Phenotypes in PCOS: Association with endocrine-metabolic profile, adherence to the mediterranean diet, and body composition. Nutrients 2021, 13, 3925. [Google Scholar] [CrossRef]
- Tutanov, O.; Shefer, A.; Tsentalovich, Y.; Tamkovich, S. Comparative Analysis of Molecular Functions and Biological Role of Proteins from Cell-Free DNA-Protein Complexes Circulating in Plasma of Healthy Females and Breast Cancer Patients. Int. J. Mol. Sci. 2023, 24, 7279. [Google Scholar] [CrossRef]
- Bryzgunova, O.; Bondar, A.; Morozkin, E.; Mileyko, V.; Vlassov, V.; Laktionov, P. A reliable method to concentrate circulating DNA. Anal. Biochem. 2011, 408, 354–356. [Google Scholar] [CrossRef] [PubMed]
- Gaal, O.; Medgyesi, G.A.; Vereczkey, L. Electrophoresis in the Separation of Biological Macromolecules; John Wiley: New York, NY, USA, 1980. [Google Scholar]
- Rosenfeld, J.; Capdevielle, J.; Guillemot, J.C.; Ferrara, P. In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal. Biochem. 1992, 203, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Villar-Garea, A.; Israel, L.; Imhof, A. Analysis of histone modifications by mass spectrometry. Curr. Protoc. Protein Sci. 2008, 51, 14.10.1–14.10.14. [Google Scholar] [CrossRef] [PubMed]
- Binns, D.; Dimmer, E.; Huntley, R.; Barrell, D.; O’Donovan, C.; Apweiler, R. QuickGO: A web-based tool for Gene Ontology searching. Bioinformatics 2009, 25, 3045–3046. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- Finn, R.D.; Attwood, T.K.; Babbitt, P.C. InterPro in 2017—Beyond protein family and domain annotations. Nucleic Acids Res. 2017, 45, D190–D199. [Google Scholar] [CrossRef]
- Bryzgunova, O.E.; Konoshenko, M.Y.; Laktionov, P.P. Concentration of cell-free DNA in different tumor types. Expert Rev. Mol. Diagn. 2021, 21, 63–75. [Google Scholar] [CrossRef]
- Hudečková, M.; Koucký, V.; Rottenberg, J.; Gál, B. Gene mutations in circulating tumour DNA as a diagnostic and prognostic marker in head and neck cancer-a systematic review. Biomedicines 2021, 9, 1548. [Google Scholar] [CrossRef]
- Arisi, M.F.; Dotan, E.; Fernandez, S.V. Circulating tumor DNA in precision oncology and its applications in colorectal cancer. Int. J. Mol. Sci. 2022, 23, 4441. [Google Scholar] [CrossRef]
- Telekes, A.; Horváth, A. The role of cell-free DNA in cancer treatment decision making. Cancers 2022, 14, 6115. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Pantel, K. Circulating DNA as biomarker in breast cancer. Breast Cancer Res. 2015, 17, 136. [Google Scholar] [CrossRef]
- Panagopoulou, M.; Esteller, M.; Chatzaki, E. Circulating cell-free DNA in breast cancer: Searching for hidden information towards precision medicine. Cancers 2021, 13, 728. [Google Scholar] [CrossRef] [PubMed]
- Dhayat, S.A.; Yang, Z. Impact of circulating tumor DNA in hepatocellular and pancreatic carcinomas. J. Cancer Res. Clin. Oncol. 2020, 146, 1625–1645. [Google Scholar] [CrossRef] [PubMed]
- Bronkhorst, A.J.; Ungerer, V.; Oberhofer, A.; Gabriel, S.; Polatoglou, E.; Randeu, H.; Uhlig, C.; Pfister, H.; Mayer, Z.; Holdenrieder, S. New perspectives on the importance of cell-free DNA biology. Diagnostics 2022, 12, 2147. [Google Scholar] [CrossRef]
- Settasatian, C.; Whitmore, S.; Crawford, J.; Bilton, R.L.; Cleton-Jansen, A.-M.; Sutherland, G.R.; Callen, D.F. Genomic structure and expression analysis of the spastic paraplegia gene, SPG7. Hum. Genet. 1999, 105, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Boyenle, I.D.; Oyedele, A.K.; Ogunlana, A.T.; Adeyemo, A.F.; Oyelere, F.S.; Akinola, O.B.; Adelusi, T.I.; Ehigie, L.O.; Ehigie, A.F. Targeting the mitochondrial permeability transition pore for drug discovery: Challenges and opportunities. Mitochondrion 2022, 63, 57–71. [Google Scholar] [CrossRef]
- Xu, K.; Wu, C.L.; Wang, Z.X.; Wang, H.J.; Yin, F.J.; Li, W.D.; Li, C.C.; Fan, H.N. Family gene expression as prognostic biomarkers for Alzheimer’s disease and primary liver cancer. Comput. Math. Methods Med. 2021, 2021, 3422393. [Google Scholar] [CrossRef]
- Deng, C.; Guo, H.; Yan, D.; Liang, T.; Ye, X.; Li, Z. Pancancer analysis of neurovascular-related NRP family genes as potential prognostic biomarkers of bladder urothelial carcinoma. BioMed Res. Int. 2021, 2021, 5546612. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, X.; Li, J.; Ma, X.; Feng, F.; Liu, L.; Wu, J.; Sun, C. ADRB1 was identified as a potential biomarker for breast cancer by the co-analysis of tumor mutational burden and immune infiltration. Aging 2020, 13, 351–363. [Google Scholar] [CrossRef]
- Montoya, A.; Amaya, C.N.; Belmont, A.; Diab, N.; Trevino, R.; Villanueva, G.; Rains, S.; Sanchez, L.A.; Badri, N.; Otoukesh, S.; et al. Use of non-selective β-blockers is associated with decreased tumor proliferative indices in early stage breast cancer. Oncotarget 2017, 8, 6446–6460. [Google Scholar] [CrossRef] [PubMed]
- Chang, A.; Yeung, S.; Thakkar, A.; Huang, K.M.; Liu, M.M.; Kanassatega, R.-S.; Parsa, C.; Orlando, R.; Jackson, E.K.; Andresen, B.T.; et al. Prevention of skin carcinogenesis by the β-blocker carvedilol. Cancer Prev. Res. 2015, 8, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.-Y.; Huang, W.-Y.; Lin, C.-L.; Huang, T.-C.; Wu, Y.-Y.; Chen, J.-H.; Kao, C.H. Propranolol reduces cancer risk: A population-based cohort study. Medicine 2015, 94, e1097. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; Wang, H.; He, J.; Erdjument-Bromage, H.; Tempst, P.; Zhang, Y. Role of hPHF1 in H3K27 methylation and Hox gene silencing. Mol. Cell. Biol. 2008, 28, 1862–1872. [Google Scholar] [CrossRef] [PubMed]
- Sarma, K.; Margueron, R.; Ivanov, A.; Pirrotta, V.; Reinberg, D. Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Mol. Cell. Biol. 2008, 28, 2718–2731. [Google Scholar] [CrossRef] [PubMed]
- Hong, Z.; Jiang, J.; Lan, L.; Nakajima, S.; Kanno, S.; Koseki, H.; Yasui, A. A polycomb group protein, PHF1, is involved in the response to DNA double-strand breaks in human cell. Nucleic Acids Res. 2008, 36, 2939–2947. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, C.; Zhang, P.; Gao, K.; Wang, D.; Yu, H.; Zhang, T.; Jiang, S.; Hexige, S.; Hong, Z.; et al. Polycomb group protein PHF1 regulates p53-dependent cell growth arrest and apoptosis. J. Biol. Chem. 2013, 288, 529–539. [Google Scholar] [CrossRef]
- Yagin, B.; Yagin, F.H.; Colak, C.; Inceoglu, F.; Kadry, S.; Kim, J. Cancer Metastasis Prediction and Genomic Biomarker Identification through Machine Learning and eXplainable Artificial Intelligence in Breast Cancer Research. Diagnostics 2023, 13, 3314. [Google Scholar] [CrossRef]
- Waterman, M.; Xu, W.; Stempak, J.M.; Milgrom, R.; Bernstein, C.N.; Griffiths, A.M.; Greenberg, G.R.; Steinhart, H.; Silverberg, M.S. Distinct and overlapping genetic loci in Crohn’s disease and ulcerative colitis: Correlations with pathogenesis. Inflamm. Bowel Dis. 2011, 17, 1936–1942. [Google Scholar] [CrossRef]
- Latiano, A.; Palmieri, O.; Latiano, T.; Corritore, G.; Bossa, F.; Martino, G.; Biscaglia, G.; Scimeca, D.; Valvano, M.R.; Pastore, M.; et al. Investigation of multiple susceptibility loci for inflammatory bowel disease in an Italian cohort of patients. PLoS ONE 2011, 6, e22688. [Google Scholar] [CrossRef]
- Li, K.; Liu, T. Evaluation of Oncogene NUP37 as a Potential Novel Biomarker in Breast Cancer. Front. Oncol. 2021, 11, 669655. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Tang, W.; Dorsey, T.H.; Ambs, S. miR-484 is associated with disease recurrence and promotes migration in prostate cancer. Biosci. Rep. 2020, 40, BSR20191028. [Google Scholar] [CrossRef] [PubMed]
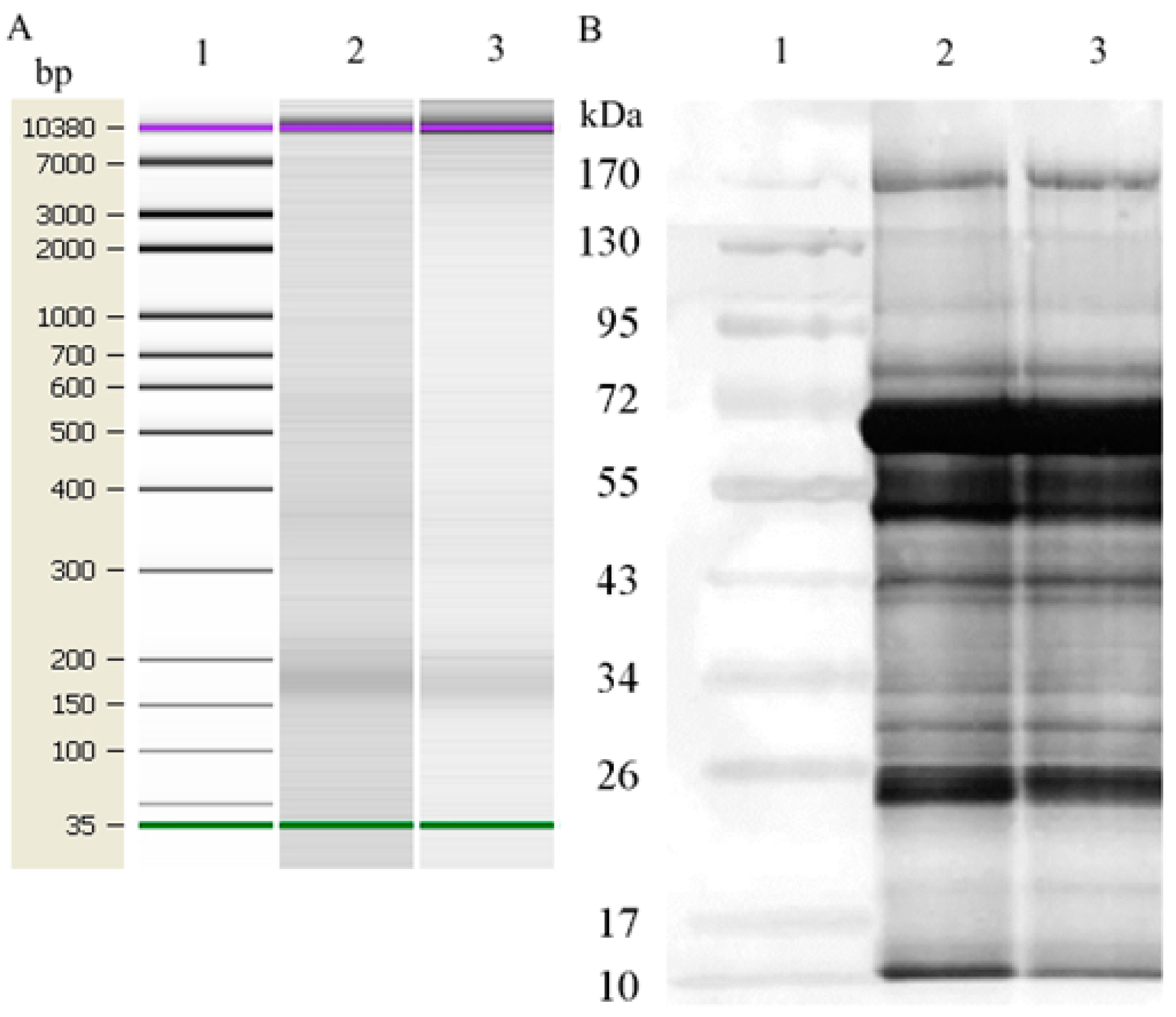

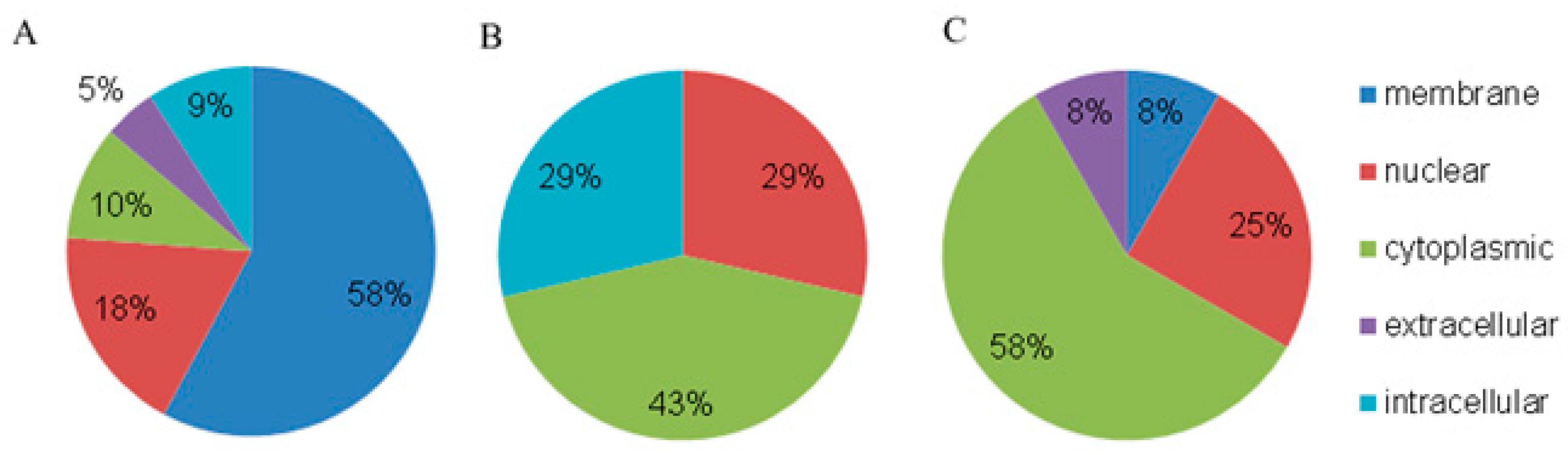
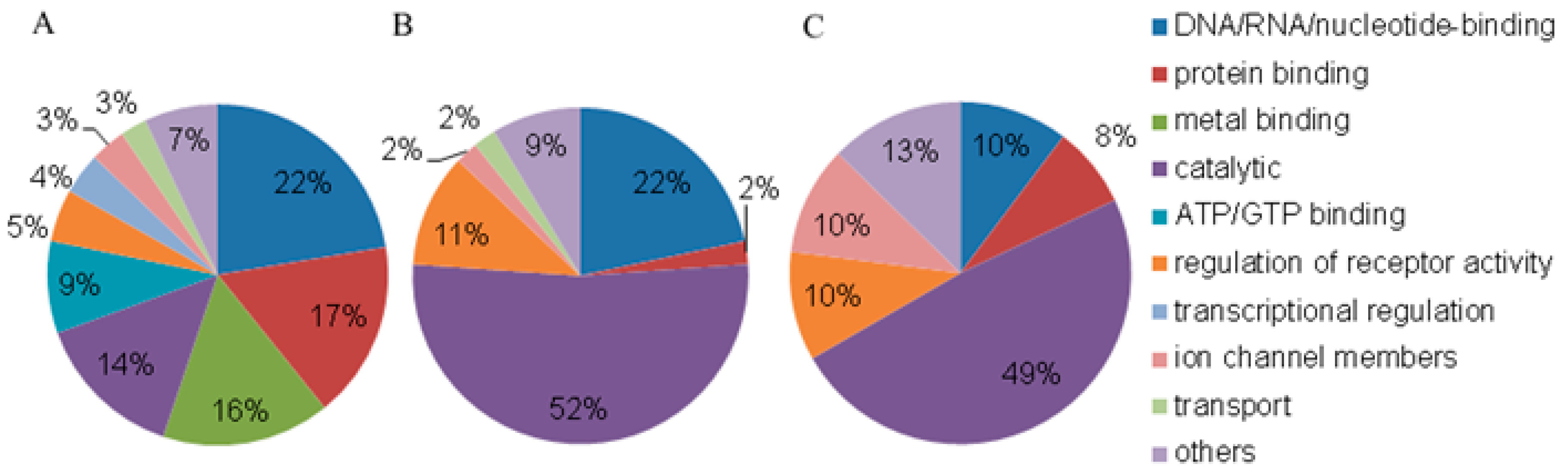
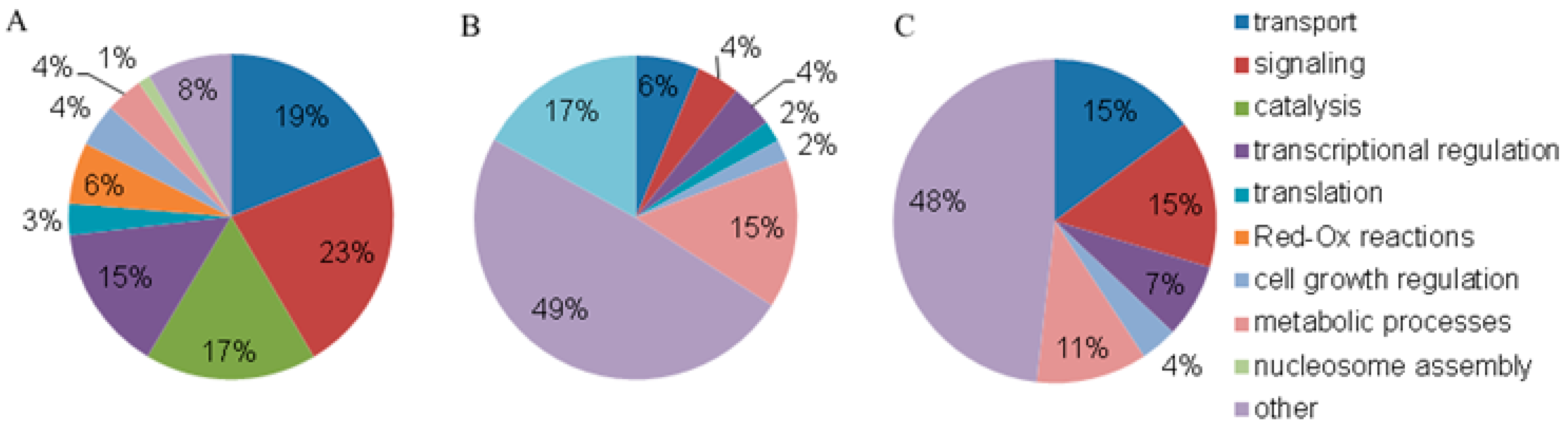
| N (%) | ||
|---|---|---|
| Tumor stage | T1 | 20 (100%) |
| Lymph node status | N0 | 20 (100%) |
| Distant metastasis | M0 | 20 (100%) |
| Histologic grade | II III | 19 (95%) 1 (5%) |
| Histological type | Invasive ductal carcinoma | 20 (100%) |
| Uniprot ID | Protein Name | Gene Name | Score | Detected in More Than 75% of Samples |
|---|---|---|---|---|
| Q7Z5R6 | Amyloid beta A4 precursor protein-binding family B member 1-interacting protein | APBB1IP | 63 | |
| P08588 | Beta-1 adrenergic receptor | ADRB1 | 60 | + |
| Q6Y288 | Beta-1,3-glucosyltransferase | B3GALTL | 94 | |
| Q03060 | cAMP-responsive element modulator | CREM | 56 | + |
| A2IDD5 | Coiled-coil domain-containing protein 78 | CCD78 | 70 | |
| Q7Z7J5 | Developmental pluripotency-associated protein 2 | DPPA2 | 71 | |
| A8MZ26 | EF-hand calcium-binding domain-containing protein 9 | EFCAB9 | 65 | + |
| Q9BY07 | Electrogenic sodium bicarbonate cotransporter 4 | SLC4A5 | 78 | |
| Q02108 | Guanylate cyclase soluble subunit alpha-3 | GUCY1A3 | 60 | |
| Q00444 | Homeobox protein Hox-C5 | HOXC5 | 92 | + |
| Q8NBZ0 | INO80 complex subunit E | INO80E | 89 | |
| P14735 | Insulin-degrading enzyme | IDE | 77 | + |
| O00522 | Krev interaction trapped protein 1 | KRIT1 | 70 | |
| Q96AQ8 | Mitochondrial calcium uniporter regulator 1 | CCDC90A | 60 | |
| Q99558 | Mitogen-activated protein kinase kinase kinase 14 | MAP3K14 | 61 | |
| P19105 | Myosin regulatory light chain 12A | MYL12A | 60 | + |
| P24844 | Myosin regulatory light polypeptide 9 | MYL9 | 57 | |
| P48163 | NADP-dependent malic enzyme | MAOX | 73 | |
| P48745 | NOV homolog | NOV | 57 | |
| Q9UQ90 | Paraplegin | SPG7 | 61 | |
| O75570 | Peptide chain release factor 1, mitochondrial | MTRF1 | 70 | |
| Q99680 | Probable G-protein coupled receptor 22 | GPR22 | 76 | + |
| Q5JUK9 | Putative G antigen family D member 1 | PAGE3 | 56 | + |
| A8MU76 | Putative UPF0607 protein ENSP00000381418 | N/A | 60 | |
| A8MX80 | Putative UPF0607 protein ENSP00000383144 | YM017 | 68 | |
| P11233 | Ras-related protein Ral-A | RALA | 59 | |
| Q8WU08 | Serine/threonine-protein kinase 32A | STK32A | 68 | |
| P62314 | Small nuclear ribonucleoprotein Sm D1 | SNRPD1 | 65 | |
| Q8NHX4 | Spermatogenesis-associated protein 3 | SPATA3 | 67 | + |
| Q01081 | Splicing factor U2AF 35 kDa subunit | U2AF1 | 112 | + |
| Q13033 | Striatin-3 | STRN3 | 66 | |
| Q7Z422 | SUZ domain-containing protein 1 | SZRD1 | 58 | |
| Q9H2G4 | Testis-specific Y-encoded-like protein 2 | TSPYL2 | 79 | |
| P17535 | Transcription factor jun-D | JUND | 68 | + |
| Q15629 | Translocating chain-associated membrane protein 1 | TRAM1 | 73 | |
| Q8N609 | Translocating chain-associated membrane protein 1-like 1 | TRAM1L1 | 62 | + |
| Q9UPT9 | Ubiquitin carboxyl-terminal hydrolase 22 | UBP22 | 68 | |
| Q16763 | Ubiquitin-conjugating enzyme E2 | UBE2S | 63 |
| Protein | EMT | Cell Proliferation | Invasion | Cell Migration | Vasculature Development | Immune Response |
|---|---|---|---|---|---|---|
| A0JLT2 | ||||||
| A6NJT0 | ||||||
| ACOT11 | ||||||
| O00159 | ||||||
| O00401 | ||||||
| O00522 | ||||||
| O14929 | ||||||
| O15392 | ||||||
| O43307 | ||||||
| O43918 | ||||||
| O75084 | ||||||
| O75190 | ||||||
| O75317 | ||||||
| O75912 | ||||||
| O95236 | ||||||
| O95260 | ||||||
| O95837 | ||||||
| O96004 | ||||||
| O96020 | ||||||
| P02795 | ||||||
| P06858 | ||||||
| P08311 | ||||||
| P08588 | ||||||
| P11233 | ||||||
| P14316 | ||||||
| P15692 | ||||||
| P17482 | ||||||
| P17483 | ||||||
| P17535 | ||||||
| P17812 | ||||||
| P17987 | ||||||
| P20592 | ||||||
| P23443 | ||||||
| P23942 | ||||||
| P24844 | ||||||
| P29371 | ||||||
| P29466 | ||||||
| P31271 | ||||||
| P34896 | ||||||
| P43487 | ||||||
| P46777 | ||||||
| P48163 | ||||||
| P48995 | ||||||
| P49137 | ||||||
| P49757 | ||||||
| P50454 | ||||||
| P51665 | ||||||
| P55010 | ||||||
| P60059 | ||||||
| P60608 | ||||||
| P62314 | ||||||
| Q01081 | ||||||
| Q02108 | ||||||
| Q03060 | ||||||
| Q03252 | ||||||
| Q05215 | ||||||
| Q06416 | ||||||
| Q13033 | ||||||
| Q13111 | ||||||
| Q13114 | ||||||
| Q13118 | ||||||
| Q13243 | ||||||
| Q13454 | ||||||
| Q14332 | ||||||
| Q15391 | ||||||
| Q15404 | ||||||
| Q15629 | ||||||
| Q15645 | ||||||
| Q16206 | ||||||
| Q3YEC7 | ||||||
| Q5T447 | ||||||
| Q5T4B2 | ||||||
| Q5VIR6 | ||||||
| Q69YI7 | ||||||
| Q6NXG1 | ||||||
| Q6PCT2 | ||||||
| Q6STE5 | ||||||
| Q75V66 | ||||||
| Q7LGA3 | ||||||
| Q7Z422 | ||||||
| Q7Z5R6 | ||||||
| Q7Z6I6 | ||||||
| Q7Z7J5 | ||||||
| Q8IV76 | ||||||
| Q8IX30 | ||||||
| Q8N1L4 | ||||||
| Q8NEC5 | ||||||
| Q8WU08 | ||||||
| Q92623 | ||||||
| Q96AQ8 | ||||||
| Q96HJ3 | ||||||
| Q99442 | ||||||
| Q99558 | ||||||
| Q99608 | ||||||
| Q99626 | ||||||
| Q99683 | ||||||
| Q99784 | ||||||
| Q99999 | ||||||
| Q9BT49 | ||||||
| Q9BY07 | ||||||
| Q9BYE3 | ||||||
| Q9H000 | ||||||
| Q9H239 | ||||||
| Q9H2G4 | ||||||
| Q9H6E4 | ||||||
| Q9H6Y7 | ||||||
| Q9HCP0 | ||||||
| Q9NS84 | ||||||
| Q9NYL4 | ||||||
| Q9NZE8 | ||||||
| Q9NZU5 | ||||||
| Q9P126 | ||||||
| Q9UKG4 | ||||||
| Q9ULL5 | ||||||
| Q9UPG8 | ||||||
| Q9UQ90 | ||||||
| Q9Y3C8 | ||||||
| Q9Y3M8 |
 HF;
HF;  BCP;
BCP;  universal.
universal.| Protein Name | Gene Name | Protein Description |
|---|---|---|
| PHD finger protein 1 | PHF1 | Zinc-binding protein is a component of a methyltransferase complex specific for Lys-27 of histone H3 (H3K27); it is involved in the repression of homeotic gene transcription. The protein is also recruited to double-strand breaks, and decreased levels of the protein result in sensitivity to X-rays and increased homologous recombination. |
| Beta-1 adrenergic receptor | ADRB1 | An integral membrane protein that mediates catecholamine-induced activation of adenylate cyclase through the action of G-proteins. This receptor binds adrenaline and noradrenaline with approximately equal affinity. Mediates the activation of Ras through G(s)-α- and cAMP-mediated signaling. Also present in the early endosome. |
| Proteasome assembly chaperone 1 | PSMG1 | A cytoplasmic/nuclear chaperone protein that promotes assembly of the 20S proteasome as part of a heterodimer with PSMG2. The PSMG1-PSMG2 heterodimer binds to PSMA5 and PSMA7 proteasome subunits, promotes assembly of the α-subunits of the proteasome into a heteroheptameric α-ring, and prevents dimerization of the α-ring. |
| Single-pass membrane and coiled-coil domain-containing protein 4 | SMCO4 | The transmembrane protein. 17-β-estradiol decreases and cisplatin increases SMCO4 mRNA expression. |
| Paraplegin | SPG7 | Transmembrane ATP-dependent zinc metalloprotease that is involved in protein folding and proteolysis. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shefer, A.; Tutanov, O.; Belenikin, M.; Tsentalovich, Y.P.; Tamkovich, S. Blood Plasma Circulating DNA-Protein Complexes: Involvement in Carcinogenesis and Prospects for Liquid Biopsy of Breast Cancer. J. Pers. Med. 2023, 13, 1691. https://doi.org/10.3390/jpm13121691
Shefer A, Tutanov O, Belenikin M, Tsentalovich YP, Tamkovich S. Blood Plasma Circulating DNA-Protein Complexes: Involvement in Carcinogenesis and Prospects for Liquid Biopsy of Breast Cancer. Journal of Personalized Medicine. 2023; 13(12):1691. https://doi.org/10.3390/jpm13121691
Chicago/Turabian StyleShefer, Aleksei, Oleg Tutanov, Maxim Belenikin, Yuri P. Tsentalovich, and Svetlana Tamkovich. 2023. "Blood Plasma Circulating DNA-Protein Complexes: Involvement in Carcinogenesis and Prospects for Liquid Biopsy of Breast Cancer" Journal of Personalized Medicine 13, no. 12: 1691. https://doi.org/10.3390/jpm13121691
APA StyleShefer, A., Tutanov, O., Belenikin, M., Tsentalovich, Y. P., & Tamkovich, S. (2023). Blood Plasma Circulating DNA-Protein Complexes: Involvement in Carcinogenesis and Prospects for Liquid Biopsy of Breast Cancer. Journal of Personalized Medicine, 13(12), 1691. https://doi.org/10.3390/jpm13121691







