Exome Sequencing Reveals Novel Germline Variants in Breast Cancer Patients in the Southernmost Region of Thailand
Abstract
1. Introduction
2. Materials and Methods
2.1. Population
2.2. Whole Exome Sequencing (WES)
2.3. Bioinformatics Analysis
2.4. Sanger Sequencing
3. Results
3.1. Patient Characteristics
3.2. Germline Variant
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hodgson, A.; Turashvili, G. Pathology of Hereditary Breast and Ovarian Cancer. Front. Oncol. 2020, 10, 531790. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, F.C.; van Overeem Hansen, T.; Sorensen, C.S. Hereditary Breast and Ovarian Cancer: New Genes in Confined Pathways. Nat. Rev. Cancer 2016, 16, 599–612. [Google Scholar] [CrossRef]
- Lee, A.; Moon, B.I.; Kim, T.H. Brca1/Brca2 Pathogenic Variant Breast Cancer: Treatment and Prevention Strategies. Ann. Lab. Med. 2020, 40, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Robson, M.; Im, S.A.; Senkus, E.; Xu, B.; Domchek, S.M.; Masuda, N.; Delaloge, S.; Li, W.; Tung, N.; Armstrong, A.; et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline Brca Mutation. N. Engl. J. Med. 2017, 377, 523–533. [Google Scholar] [CrossRef]
- Choi, M.; Kipps, T.; Kurzrock, R. Atm Mutations in Cancer: Therapeutic Implications. Mol. Cancer Ther. 2016, 15, 1781–1791. [Google Scholar] [CrossRef]
- Jerzak, K.J.; Mancuso, T.; Eisen, A. Ataxia-Telangiectasia Gene (Atm) Mutation Heterozygosity in Breast Cancer: A Narrative Review. Curr. Oncol. 2018, 25, e176–e180. [Google Scholar] [CrossRef] [PubMed]
- Evans, D.G.; Woodward, E.R.; Bajalica-Lagercrantz, S.; Oliveira, C.; Frebourg, T. Germline Tp53 Testing in Breast Cancers: Why, When and How? Cancers 2020, 12, 3762. [Google Scholar] [CrossRef] [PubMed]
- Haffty, B.G.; Euhus, D.M.; Pierce, L.J. Genetic Factors in the Locoregional Management of Breast Cancer. J. Clin. Oncol. 2020, 38, 2220–2229. [Google Scholar] [CrossRef] [PubMed]
- Stolarova, L.; Kleiblova, P.; Janatova, M.; Soukupova, J.; Zemankova, P.; Macurek, L.; Kleibl, Z. Chek2 Germline Variants in Cancer Predisposition: Stalemate Rather Than Checkmate. Cells 2020, 9, 2675. [Google Scholar] [CrossRef]
- Wu, S.; Zhou, J.; Zhang, K.; Chen, H.; Luo, M.; Lu, Y.; Sun, Y.; Chen, Y. Molecular Mechanisms of Palb2 Function and Its Role in Breast Cancer Management. Front. Oncol. 2020, 10, 301. [Google Scholar] [CrossRef]
- Desmedt, C.; Yates, L.; Kulka, J. Catalog of Genetic Progression of Human Cancers: Breast Cancer. Cancer Metastasis Rev. 2016, 35, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Manchanda, R.; Gaba, F. Population Based Testing for Primary Prevention: A Systematic Review. Cancers 2018, 10, 424. [Google Scholar] [CrossRef]
- Asphaug, L.; Melberg, H.O. The Cost-Effectiveness of Multigene Panel Testing for Hereditary Breast and Ovarian Cancer in Norway. MDM Policy Pract. 2019, 4, 2381468318821103. [Google Scholar] [CrossRef] [PubMed]
- Manahan, E.R.; Kuerer, H.M.; Sebastian, M.; Hughes, K.S.; Boughey, J.C.; Euhus, D.M.; Boolbol, S.K.; Taylor, W.A. Consensus Guidelines on Genetic; Testing for Hereditary Breast Cancer from the American Society of Breast Surgeons. Ann. Surg. Oncol. 2019, 26, 3025–3031. [Google Scholar] [CrossRef] [PubMed]
- Kurian, A.W.; Ward, K.C.; Abrahamse, P.; Bondarenko, I.; Hamilton, A.S.; Deapen, D.; Morrow, M.; Berek, J.S.; Hofer, T.P.; Katz, S.J. Time Trends in Receipt of Germline Genetic Testing and Results for Women Diagnosed with Breast Cancer or Ovarian Cancer, 2012–2019. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2021, 39, 1631–1640. [Google Scholar] [CrossRef]
- Yip, C.H.; Evans, D.G.; Agarwal, G.; Buccimazza, I.; Kwong, A.; Morant, R.; Prakash, I.; Song, C.Y.; Taib, N.A.; Tausch, C.; et al. Global Disparities in Breast Cancer Genetics Testing, Counselling and Management. World J. Surg. 2019, 43, 1264–1270. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, R. Hereditary Breast and Ovarian Cancer (Hboc): Review of Its Molecular Characteristics, Screening, Treatment, and Prognosis. Breast Cancer 2021, 28, 1167–1180. [Google Scholar] [CrossRef] [PubMed]
- Albritton, R.B. Political Diversity among Muslims in Thailand. Asian Stud. Rev. 1999, 23, 233–246. [Google Scholar] [CrossRef]
- Sriplung, H.; Bilheem, S.; Kuntipundee, T.; Geater, S.L. Differences in Cancer Incidence among Predominantly Muslim and Buddhist Subpopulations in Songkhla. Asian Pac. J. Cancer Prev. 2014, 15, 9979–9983. [Google Scholar] [CrossRef]
- Virani, S.; Wetzel, E.C.; Laohawiriyakamol, S.; Boonyaphiphat, P.; Geater, A.; Kleer, C.G.; Pang, J.; Rentschler, K.M.; Colacino, J.A.; de Leon, C.F.M.; et al. Ethnic Disparity in Breast Cancer Survival in Southern Thai Women. Cancer Epidemiol. 2018, 54, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Lertwilaiwittaya, P.; Roothumnong, E.; Nakthong, P.; Dungort, P.; Meesamarnpong, C.; Tansa-Nga, W.; Pongsuktavorn, K.; Wiboonthanasarn, S.; Tititumjariya, W.; Thongnoppakhun, W.; et al. Thai Patients Who Fulfilled Nccn Criteria for Breast/Ovarian Cancer Genetic Assessment Demonstrated High Prevalence of Germline Mutations in Cancer Susceptibility Genes: Implication to Asian Population Testing. Breast Cancer Res. Treat. 2021, 188, 237–248. [Google Scholar] [CrossRef]
- AS ISO 15189-2012; Medical laboratories Requirements for Quality and Competence. Bureau of Laboratory Quality Standards, Department of Medical Sciences: Nonthaburi, Thailand, 2012.
- AS ISO 15190-2020; Medical Laboratories Requirements for Quality and Competence. Bureau of Laboratory Quality Standards, Department of Medical Sciences: Nonthaburi, Thailand, 2020.
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A. Primer3plus, an Enhanced Web Interface to Primer3. Nucleic Acids Res. 2007, 35, W71–W74. [Google Scholar] [CrossRef]
- Sukpan, P.; Kanokwiroon, K.; Sriplung, H.; Laochareonsuk, W.; Choochuen, P.; Auseng, N.; Wanawanakorn, K.; Sangkhathat, S. Prevalence of Pathogenic Germline Mutations in 13 Hereditary Cancer-Related Genes in Breast Cancer Patients in Narathiwat Province, Thailand. Asian Pac. J. Cancer Prev. 2023, 24, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Ferrell, S.D., Jr.; Ahmad, I.; Nguyen, C.; Petrova, S.C.; Wilhelm, S.R.; Ye, Y.; Barsky, S.H. Why Is Cancer So Common a Disease in People yet So Rare at a Cellular Level? Med. Hypotheses 2020, 144, 110171. [Google Scholar] [CrossRef] [PubMed]
- Knudson, A.G. Hereditary Cancer: Two Hits Revisited. J. Cancer Res. Clin. Oncol. 1995, 122, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Knudson, A.G. Hereditary predisposition to cancer. Ann. N. Y. Acad. Sci. 1997, 833, 58–67. [Google Scholar] [CrossRef]
- Tomlinson, I.P.M.; Roylance, R.; Houlston, R.S. Two Hits Revisited Again. J. Med. Genet. 2001, 38, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Abbass, M.A.; Leach, B.; Church, J.M. The Second Allele: A Key to Understanding the Timing of Sporadic and Hereditary Colorectal Tumorigenesis. Genes 2021, 12, 1515. [Google Scholar] [CrossRef]
- Mastrodomenico, L.; Piombino, C.; Ricco, B.; Barbieri, E.; Venturelli, M.; Piacentini, F.; Dominici, M.; Cortesi, L.; Toss, A. Personalized Systemic Therapies in Hereditary Cancer Syndromes. Genes 2023, 14, 684. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.A.; Jian, J.W.; Hung, C.F.; Peng, H.P.; Yang, C.F.; Cheng, H.S.; Yang, A.S. Germline Breast Cancer Susceptibility Gene Mutations and Breast Cancer Outcomes. BMC Cancer 2018, 18, 315. [Google Scholar] [CrossRef]
- Sun, J.; Meng, H.; Yao, L.; Lv, M.; Bai, J.; Zhang, J.; Wang, L.; Ouyang, T.; Li, J.; Wang, T.; et al. Germline Mutations in Cancer Susceptibility Genes in a Large Series of Unselected Breast Cancer Patients. Clin. Cancer Res. 2017, 23, 6113–6119. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.R.; Devi, B.C.R.; Sung, H.; Guida, J.; Mucaki, E.J.; Xiao, Y.; Best, A.; Garland, L.; Xie, Y.; Hu, N.; et al. Prevalence and Spectrum of Germline Rare Variants in Brca1/2 and Palb2 among Breast Cancer Cases in Sarawak, Malaysia. Breast Cancer Res. Treat. 2017, 165, 687–697. [Google Scholar] [CrossRef]
- Thorat, M.A.; Balasubramanian, R. Breast Cancer Prevention in High-Risk Women. Best Pract. Res. Clin. Obstet. Gynaecol. 2020, 65, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.; Paul, S. The Breast Cancer Susceptibility Genes (Brca) in Breast and Ovarian Cancers. Front. Biosci. Landmark Ed. 2014, 19, 605–618. [Google Scholar] [CrossRef]
- Kaur, R.P.; Shafi, G.; Benipal, R.P.S.; Munshi, A. Frequency of Pathogenic Germline Mutations in Cancer Susceptibility Genes in Breast Cancer Patients. Med. Oncol. 2018, 35, 81. [Google Scholar] [CrossRef] [PubMed]
- Couch, F.J.; Shimelis, H.; Hu, C.; Hart, S.N.; Polley, E.C.; Na, J.; Hallberg, E.; Moore, R.; Thomas, A.; Lilyquist, J.; et al. Associations between Cancer Predisposition Testing Panel Genes and Breast Cancer. JAMA Oncol. 2017, 3, 1190–1196. [Google Scholar] [CrossRef] [PubMed]
- Piombino, C.; Cortesi, L.; Lambertini, M.; Punie, K.; Grandi, G.; Toss, A. Secondary Prevention in Hereditary Breast and/or Ovarian Cancer Syndromes Other Than Brca. J. Oncol. 2020, 2020, 6384190. [Google Scholar] [CrossRef]
- Mohamad, S.; Isa, N.M.; Muhammad, R.; Emran, N.A.; Kitan, N.M.; Kang, P.; Kang, I.N.; Taib, N.A.; Teo, S.H.; Akmal, S.N. Low Prevalence of Chek2 Gene Mutations in Multiethnic Cohorts of Breast Cancer Patients in Malaysia. PLoS ONE 2015, 10, e0117104. [Google Scholar] [CrossRef] [PubMed]
- Sehgal, R.; Sheahan, K.; O’Connell, P.R.; Hanly, A.M.; Martin, S.T.; Winter, D.C. Lynch Syndrome: An Updated Review. Genes 2014, 5, 497–507. [Google Scholar] [CrossRef]
- Antoniou, A.C.; Casadei, S.; Heikkinen, T.; Barrowdale, D.; Pylkas, K.; Roberts, J.; Lee, A.; Subramanian, D.; De Leeneer, K.; Fostira, F.; et al. Breast-Cancer Risk in Families with Mutations in Palb2. N. Engl. J. Med. 2014, 371, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Phuah, S.Y.; Lee, S.Y.; Kang, P.; Kang, I.N.; Yoon, S.Y.; Thong, M.K.; Hartman, M.; Sng, J.H.; Yip, C.H.; Taib, N.A.; et al. Prevalence of Palb2 Mutations in Breast Cancer Patients in Multi-Ethnic Asian Population in Malaysia and Singapore. PLoS ONE 2013, 8, e73638. [Google Scholar] [CrossRef][Green Version]
- Kechagioglou, P.; Papi, R.M.; Provatopoulou, X.; Kalogera, E.; Papadimitriou, E.; Grigoropoulos, P.; Nonni, A.; Zografos, G.; Kyriakidis, D.A.; Gounaris, A. Tumor Suppressor Pten in Breast Cancer: Heterozygosity, Mutations and Protein Expression. Anticancer Res. 2014, 34, 1387–1400. [Google Scholar] [PubMed]
- Ngeow, J.; Sesock, K.; Eng, C. Breast Cancer Risk and Clinical Implications for Germline Pten Mutation Carriers. Breast Cancer Res. Treat. 2017, 165, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Hoxhaj, A.; Drissen, M.; Vos, J.R.; Bult, P.; Mann, R.M.; Hoogerbrugge, N. The Yield and Effectiveness of Breast Cancer Surveillance in Women with Pten Hamartoma Tumor Syndrome. Cancer 2022, 128, 2883–2891. [Google Scholar] [CrossRef]
- Oranratnachai, S.; Yamkaew, W.; Tunteeratum, A.; Sukarayothin, T.; Iemwimangsa, N.; Panvichien, R. Characteristics of Breast Cancer Patients Tested for Germline Brca1/2 Mutations by Next-Generation Sequencing in Ramathibodi Hospital, Mahidol University. Cancer Rep. 2023, 6, e1664. [Google Scholar] [CrossRef]
- Manchana, T.; Phowthongkum, P.; Teerapakpinyo, C. Germline Mutations in Thai Patients with Nonmucinous Epithelial Ovarian Cancer. World J. Clin. Oncol. 2019, 10, 358–368. [Google Scholar] [CrossRef] [PubMed]
- Mersch, J.; Brown, N.; Pirzadeh-Miller, S.; Mundt, E.; Cox, H.C.; Brown, K.; Aston, M.; Esterling, L.; Manley, S.; Ross, T. Prevalence of Variant Reclassification Following Hereditary Cancer Genetic Testing. JAMA 2018, 320, 1266–1274. [Google Scholar] [CrossRef] [PubMed]
- Makhnoon, S.; Levin, B.; Ensinger, M.; Mattie, K.; Volk, R.J.; Zhao, Z.; Mendoza, T.; Shete, S.; Samiian, L.; Grana, G.; et al. A Multicenter Study of Clinical Impact of Variant of Uncertain Significance Reclassification in Breast, Ovarian and Colorectal Cancer Susceptibility Genes. Cancer Med. 2023, 12, 2875–2884. [Google Scholar] [CrossRef]
- Win, A.K.; Cleary, S.P.; Dowty, J.G.; Baron, J.A.; Young, J.P.; Buchanan, D.D.; Southey, M.C.; Burnett, T.; Parfrey, P.S.; Green, R.C.; et al. Cancer Risks for Monoallelic Mutyh Mutation Carriers with a Family History of Colorectal Cancer. Int. J. Cancer 2011, 129, 2256–2262. [Google Scholar] [CrossRef]
- Hutchcraft, M.L.; Gallion, H.H.; Kolesar, J.M. Mutyh as an Emerging Predictive Biomarker in Ovarian Cancer. Diagnostics 2021, 11, 413. [Google Scholar] [CrossRef] [PubMed]
- Rennert, G.; Lejbkowicz, F.; Cohen, I.; Pinchev, M.; Rennert, H.S.; Barnett-Griness, O. Mutyh Mutation Carriers Have Increased Breast Cancer Risk. Cancer 2012, 118, 1989–1993. [Google Scholar] [CrossRef] [PubMed]
- Win, A.K.; Reece, J.C.; Dowty, J.G.; Buchanan, D.D.; Clendenning, M.; Rosty, C.; Southey, M.C.; Young, J.P.; Cleary, S.P.; Kim, H.; et al. Risk of Extracolonic Cancers for People with Biallelic and Monoallelic Mutations in Mutyh. Int. J. Cancer 2016, 139, 1557–1563. [Google Scholar] [CrossRef]
- Rizzolo, P.; Silvestri, V.; Bucalo, A.; Zelli, V.; Valentini, V.; Catucci, I.; Zanna, I.; Masala, G.; Bianchi, S.; Spinelli, A.M.; et al. Contribution of Mutyh Variants to Male Breast Cancer Risk: Results from a Multicenter Study in Italy. Front. Oncol. 2018, 8, 583. [Google Scholar] [CrossRef] [PubMed]
- Esterling, L.; Wijayatunge, R.; Brown, K.; Morris, B.; Hughes, E.; Pruss, D.; Manley, S.; Bowles, K.R.; Ross, T.S. Impact of a Cancer Gene Variant Reclassification Program over a 20-Year Period. JCO Precis. Oncol. 2020, 4, 944–954. [Google Scholar] [CrossRef]
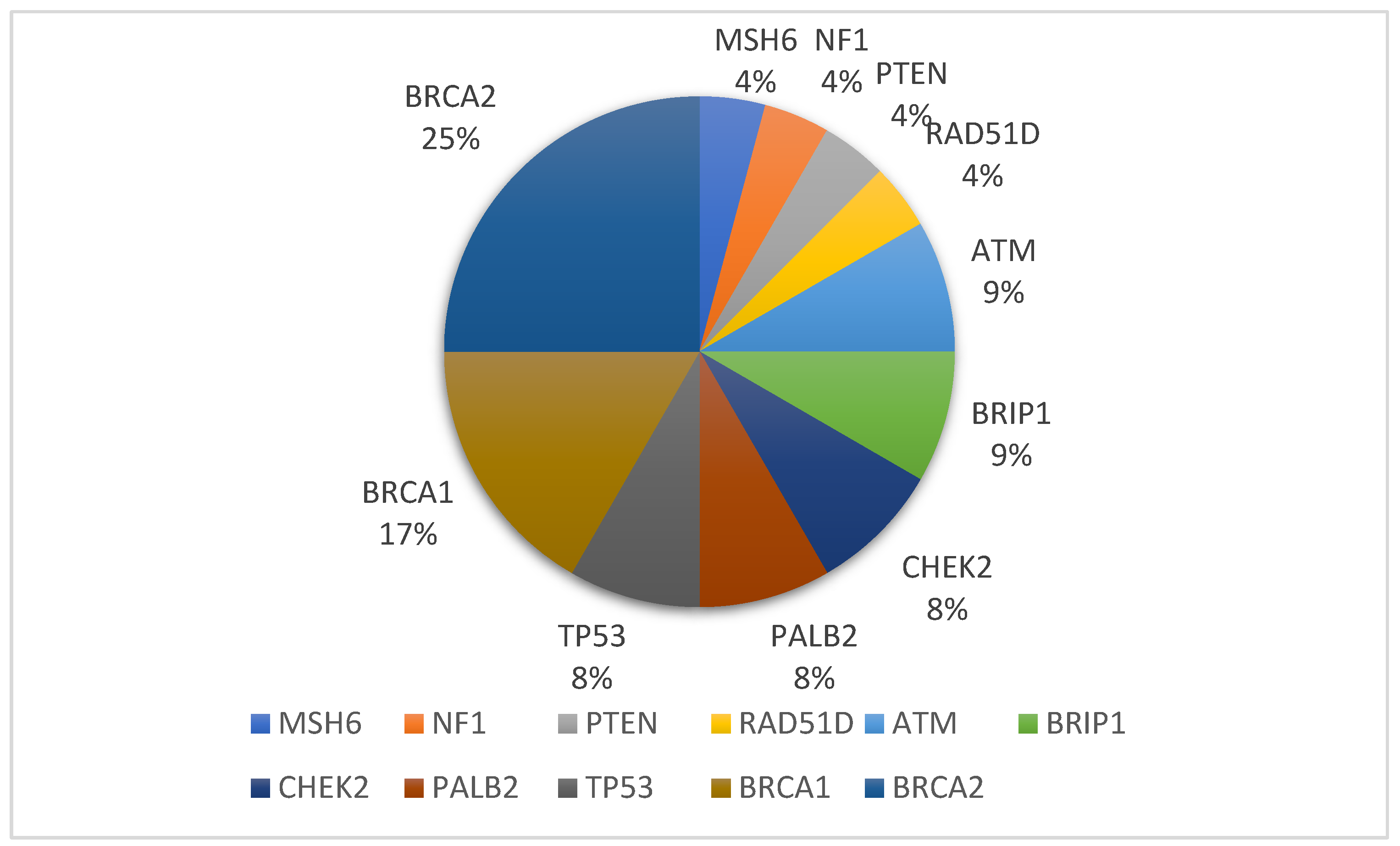
| Study Characteristic (151 Cases) | No (%) | No. among the 23 Cases of PVs/LPs (%) | p-Value |
|---|---|---|---|
| Age at diagnosis (year) | 0.02 | ||
| ≤50 | 91 (60.3) | 19 (20.9) | |
| >50 | 60 (39.7) | 4 (6.7) | |
| Pathological group | 0.13 | ||
| Invasive ductal carcinoma (IDC) | 131 (86.8) | 21 (16.0) | |
| Ductal carcinoma in situ (DCIS) | 6 (4.0) | 2 (33.3) | |
| Others | 14 (9.3) | 0 (0.0) | |
| Molecular subtypes | 0.05 | ||
| Luminal A | 61 (40.4) | 7 (11.5) | |
| Luminal B | 32 (21.2) | 4 (12.5) | |
| HER2-enriched | 30 (19.9) | 3 (10.0) | |
| TNBC | 28 (18.5) | 9 (32.1) | |
| Stage at diagnosis | 0.48 | ||
| DCIS | 6 (4.0) | 2 (33.3) | |
| Stage I | 18 (11.9) | 4 (22.2) | |
| Stage II | 49 (32.5) | 4 (8.2) | |
| Stage III | 53 (35.1) | 8 (15.1) | |
| Stage VI | 25 (16.6) | 5 (20.0) | |
| Religion | 0.16 | ||
| Muslim-Thai | 106 (70.2) | 19 (17.9) | |
| Buddhist-Thai | 45 (29.8) | 4 (8.9) |
| Gene | HGVS | No. of Carriers | Age at Diagnosis (Years) | dbSNP ID | Molecular Subtype |
|---|---|---|---|---|---|
| ATM | c.8373C>A (p.Tyr2791Ter) | 1 | 36 | rs1060504292 | Luminal B |
| c.2377-2A>G (p.Splicing) | 1 | 48 | rs1057516553 | Luminal A | |
| BRCA1 | c.2059C>T (p.Gln689Ter) | 1 | 54 | rs273898674 | TNBC |
| c.1863_1885del (p.His621GlnfsTer7) | 2 | 51, 28 | - | TNBC | |
| c.5068A>C (p.Lys1690Gln) | 1 | 48 | rs397507239 | HER2-enriched | |
| BRCA2 | c.6352_6353del (p.Val2118LysfsTer10) | 1 | 48 | rs80359576 | TNBC |
| c.5164_5165del (p.Ser1722TyrfsTer4) | 1 | 39 | rs80359490 | TNBC | |
| c.7558C>T (p.Arg2520Ter) | 1 | 48 | rs80358981 | Luminal A | |
| c.3680_3681del (p.Leu1227GlnfsTer5) | 1 | 48 | rs80359395 | Luminal A | |
| c.3283C>T (p.Gln1095Ter) | 1 | 48 | rs397507662 | Luminal A | |
| c.262_263del (p.Leu88AlafsTer12) | 1 | 30 | rs276174825 | TNBC | |
| BRIP1 | c.2990_2993del (p.Thr997ArgfsTer61) | 1 | 39 | rs771028677 | Luminal B |
| c.195_196insC (p.Ser66fs) | 1 | 40 | Novel | HER2-enriched | |
| CHEK2 | c.1178C>G (p.Pro393Arg) | 1 | 57 | Novel | Luminal B |
| c.246_260del (p.Asp82_Glu86del) | 1 | 47 | rs587780181 | HER2-enriched | |
| MSH6 | c.4003del (p.Glu1335LysfsTer11) | 1 | 44 | Novel | TNBC |
| NF1 | c.6592_6593del (p.Asp2198fs) | 1 | 40 | rs765010702 | Luminal B |
| PALB2 | c.2257C>T (p.Arg753Ter) | 1 | 45 | rs180177110 | Luminal A |
| c.2101delT (p.Ser701fs) | 1 | 43 | Novel | Luminal A | |
| PTEN | c.154_154+1insGG (p.His53GlyfsTer10) | 1 | 41 | Novel | Luminal A |
| RAD51D | c.680C>T (p.Ser227Leu) | 1 | 51 | rs370228071 | Luminal A |
| TP53 | c.818G>A (p.Arg273His) | 1 | 48 | rs28934576 | TNBC |
| c.1024C>T (p.Arg342Ter) | 1 | 25 | rs730882029 | TNBC |
| Gene | HGVS | No. of Carriers | SNP ID | Molecular Subtype |
|---|---|---|---|---|
| ATM | c.8797A>G (p.Lys2933Glu) | 1 | rs587779875 | Luminal A |
| c.2944C>T (p.Arg982Cys) | 1 | rs587779830 | Luminal A | |
| c.9124C>G (p.Pro3042Ala) | 1 | Novel | TNBC | |
| BARD1 | c.1174_1194delTTGCCTGAATGTTCTTCACCA (p.Leu392_Pro398del) | 1 | Novel | Luminal A |
| c.538T>C (p.Tyr180His) | 1 | rs1060501311 | TNBC | |
| c.127C>T (p.Arg43Cys) | 1 | rs752871324 | Luminal A | |
| BRCA1 | c.3625T>G (p.Leu1209Val) | 1 | rs273900711 | TNBC |
| BRCA2 | c.4089C>G (p.Asn1363Lys) | 1 | Novel | TNBC |
| c.5299_5307del (p.Lys1767_Asp1769del) | 1 | rs80359504 | HER2-enriched | |
| c.5218_5223del (p.Leu1740_Ser1741del) | 1 | rs397507775 | Luminal B | |
| BRIP1 | c.67C>T (p.Pro23Ser) | 1 | Novel | HER2-enriched |
| c.1912A>G (p.Asn638Asp) | 1 | Novel | Luminal A | |
| CDH1 | c.2603G>A (p.Arg868His) | 1 | rs369126891 | TNBC |
| CHEK2 | c.1231G>A (p.Asp411Asn) | 1 | rs755127902 | Luminal B |
| c.976C>T (p.Pro326Ser) | 1 | rs1555917036 | TNBC | |
| MLH1 | c.1730C>T (p.Ser577Leu) | 1 | rs56185292 | Luminal A |
| c.290A>G (p.Tyr97Cys) | 1 | rs773647920 | Luminal A | |
| MSH2 | c.2435C>G (p.Thr812Ser) | 1 | rs1553369826 | TNBC |
| c.2439G>T (p.Met813Ile) | 1 | rs587781678 | Luminal A | |
| MUTYH | c.398A>G (p.Asn133Ser) | 1 | rs771641237 | HER2-enriched |
| c.158-2A>T (Splicing) | 1 | Novel | Luminal A | |
| c.892-2A>G (Splicing) | 5 | rs77542170 | HER2-enriched * 4 Luminal B * 1 | |
| NF1 | c.6743G>A (p.Arg2248His) | 1 | rs562367786 | TNBC |
| PMS2 | c.752T>A (p.Val251Glu) | 1 | Novel | Luminal A |
| RAD51C | c.722T>C (p.Val241Ala) | 1 | rs1555599127 | TNBC |
| RAD54L | c.1862T>C (p.Leu621Pro) | 1 | rs768472959 | Luminal A |
| c.1850A>C (p.Tyr617Ser) | 1 | Novel | Luminal A | |
| TP53 | c.7G>A (p.Glu3Lys) | 1 | Novel | TNBC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sukpan, P.; Sangkhathat, S.; Sriplung, H.; Laochareonsuk, W.; Choochuen, P.; Auseng, N.; Khoonjan, W.; Salaeh, R.; Thangnaphadol, K.; Wanawanakorn, K.; et al. Exome Sequencing Reveals Novel Germline Variants in Breast Cancer Patients in the Southernmost Region of Thailand. J. Pers. Med. 2023, 13, 1587. https://doi.org/10.3390/jpm13111587
Sukpan P, Sangkhathat S, Sriplung H, Laochareonsuk W, Choochuen P, Auseng N, Khoonjan W, Salaeh R, Thangnaphadol K, Wanawanakorn K, et al. Exome Sequencing Reveals Novel Germline Variants in Breast Cancer Patients in the Southernmost Region of Thailand. Journal of Personalized Medicine. 2023; 13(11):1587. https://doi.org/10.3390/jpm13111587
Chicago/Turabian StyleSukpan, Panupong, Surasak Sangkhathat, Hutcha Sriplung, Wison Laochareonsuk, Pongsakorn Choochuen, Nasuha Auseng, Weerawan Khoonjan, Rusta Salaeh, Kornchanok Thangnaphadol, Kasemsun Wanawanakorn, and et al. 2023. "Exome Sequencing Reveals Novel Germline Variants in Breast Cancer Patients in the Southernmost Region of Thailand" Journal of Personalized Medicine 13, no. 11: 1587. https://doi.org/10.3390/jpm13111587
APA StyleSukpan, P., Sangkhathat, S., Sriplung, H., Laochareonsuk, W., Choochuen, P., Auseng, N., Khoonjan, W., Salaeh, R., Thangnaphadol, K., Wanawanakorn, K., & Kanokwiroon, K. (2023). Exome Sequencing Reveals Novel Germline Variants in Breast Cancer Patients in the Southernmost Region of Thailand. Journal of Personalized Medicine, 13(11), 1587. https://doi.org/10.3390/jpm13111587






