miRNA-Based Technologies in Cancer Therapy
Abstract
1. Introduction
2. Transcription and Processing of miRNAs
2.1. miRNA Biogenesis
2.2. miRNA Function
2.2.1. Intracellular miRNAs
2.2.2. Extracellular miRNAs
3. miRNA Dysregulation in Cancer
4. miRNA as a Therapeutic Agent in Cancer
4.1. miRNA Inhibitors
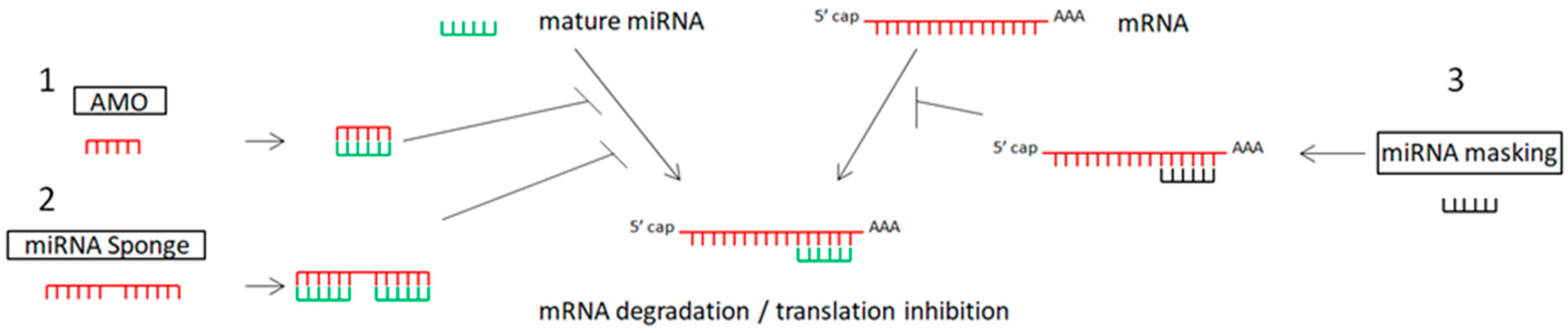
4.1.1. Anti-miRNA Oligonucleotides (AMOs) or Antagomirs
Chemical Modifications of AMOs
4.1.2. miRNA Sponges in Cancer Therapy
Competing Endogenous RNAs (ceRNAs)
4.1.3. miRNA-Masking
4.2. miRNA Mimics
5. Druggable miRNA Systems
5.1. Local Delivery
5.2. Systemic Delivery
6. Categories of Delivery Vehicles for miRNA-Based Therapy
6.1. Viral Vectors
6.2. Non-Viral miRNA Delivery
6.2.1. Lipid-Based Delivery Systems
6.2.2. Polymeric Nanoparticles
6.3. Inorganic Based Delivery
6.4. Exosome-Mediated Delivery of miRNAs
7. miRNA-Based Therapies in Clinical Practice
8. Drug Resistance
Druggable miRNA Metabolic Pathways in Oncology
9. Drug Repurposing Based on Drug–miRNA Associations in Cancer Therapy
9.1. Metformin
9.2. Statins
9.3. Aspirin
9.4. Methotrexate
10. Discussion
11. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.H.; Bernard, P.S.; Perou, C.M. Molecular portraits and the family tree of cancer. Nat. Genet. 2002, 32, 533–540. [Google Scholar] [CrossRef]
- Ramaswamy, M.; Wasan, K.M. Differences in the method by which plasma is separated from whole blood influences amphotericin B plasma recovery and distribution following amphotericin B lipid complex incubation within whole blood. Drug Dev. Ind. Pharm. 2001, 27, 871–875. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Ambros, V.; Horvitz, H.R. Heterochronic mutants of the nematode Caenorhabditis elegans. Science 1984, 226, 409–416. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef] [PubMed]
- Alles, J.; Fehlmann, T.; Fischer, U.; Backes, C.; Galata, V.; Minet, M.; Hart, M.; Abu-Halima, M.; Grasser, F.A.; Lenhof, H.P.; et al. An estimate of the total number of true human miRNAs. Nucleic Acids Res. 2019, 47, 3353–3364. [Google Scholar] [CrossRef]
- Fromm, B.; Zhong, X.; Tarbier, M.; Friedlander, M.R.; Hackenberg, M. The limits of human microRNA annotation have been met. RNA 2022, 28, 781–785. [Google Scholar] [CrossRef] [PubMed]
- da Silveira, W.A.; Renaud, L.; Hazard, E.S.; Hardiman, G. miRNA and lncRNA Expression Networks Modulate Cell Cycle and DNA Repair Inhibition in Senescent Prostate Cells. Genes 2022, 13, 208. [Google Scholar] [CrossRef] [PubMed]
- Brennecke, J.; Stark, A.; Russell, R.B.; Cohen, S.M. Principles of microRNA-target recognition. PLoS Biol. 2005, 3, e85. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; He, Y.; Song, J.; Wang, L.; Guo, P.; Cao, J. MiRNA-181b-5p Modulates Cell Proliferation, Cell Cycle, and Apoptosis by Targeting SSX2IP in Acute Lymphoblastic Leukemia. Turk. J. Haematol. 2022, 39, 160–169. [Google Scholar] [CrossRef]
- Chen, C.Z.; Li, L.; Lodish, H.F.; Bartel, D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science 2004, 303, 83–86. [Google Scholar] [CrossRef]
- Jeong, H.C.; Shukla, S.; Fok, W.C.; Huynh, T.N.; Batista, L.F.Z.; Parker, R. USB1 is a miRNA deadenylase that regulates hematopoietic development. Science 2023, 379, 901–907. [Google Scholar] [CrossRef]
- Esau, C.; Kang, X.; Peralta, E.; Hanson, E.; Marcusson, E.G.; Ravichandran, L.V.; Sun, Y.; Koo, S.; Perera, R.J.; Jain, R.; et al. MicroRNA-143 regulates adipocyte differentiation. J. Biol. Chem. 2004, 279, 52361–52365. [Google Scholar] [CrossRef]
- Tan, X.; Zhu, T.; Zhang, L.; Fu, L.; Hu, Y.; Li, H.; Li, C.; Zhang, J.; Liang, B.; Liu, J. miR-669a-5p promotes adipogenic differentiation and induces browning in preadipocytes. Adipocyte 2022, 11, 120–132. [Google Scholar] [CrossRef]
- Poy, M.N.; Eliasson, L.; Krutzfeldt, J.; Kuwajima, S.; Ma, X.; Macdonald, P.E.; Pfeffer, S.; Tuschl, T.; Rajewsky, N.; Rorsman, P.; et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 2004, 432, 226–230. [Google Scholar] [CrossRef]
- Aghaei, M.; Khodadadian, A.; Elham, K.N.; Nazari, M.; Babakhanzadeh, E. Major miRNA Involved in Insulin Secretion and Production in Beta-Cells. Int. J. Gen. Med. 2020, 13, 89–97. [Google Scholar] [CrossRef]
- Qian, B.; Yang, Y.; Tang, N.; Wang, J.; Sun, P.; Yang, N.; Chen, F.; Wu, T.; Sun, T.; Li, Y.; et al. M1 macrophage-derived exosomes impair beta cell insulin secretion via miR-212-5p by targeting SIRT2 and inhibiting Akt/GSK-3beta/beta-catenin pathway in mice. Diabetologia 2021, 64, 2037–2051. [Google Scholar] [CrossRef]
- Gregory, R.I.; Shiekhattar, R. MicroRNA biogenesis and cancer. Cancer Res. 2005, 65, 3509–3512. [Google Scholar] [CrossRef] [PubMed]
- Dostie, J.; Mourelatos, Z.; Yang, M.; Sharma, A.; Dreyfuss, G. Numerous microRNPs in neuronal cells containing novel microRNAs. RNA 2003, 9, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Rastegar-Moghaddam, S.H.; Ebrahimzadeh-Bideskan, A.; Shahba, S.; Malvandi, A.M.; Mohammadipour, A. Roles of the miR-155 in Neuroinflammation and Neurological Disorders: A Potent Biological and Therapeutic Target. Cell Mol. Neurobiol. 2023, 43, 455–467. [Google Scholar] [CrossRef]
- Kumar, M.; Li, G. Emerging Role of MicroRNA-30c in Neurological Disorders. Int. J. Mol. Sci. 2022, 24, 37. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.; Zavolan, M.; Grasser, F.A.; Chien, M.; Russo, J.J.; Ju, J.; John, B.; Enright, A.J.; Marks, D.; Sander, C.; et al. Identification of virus-encoded microRNAs. Science 2004, 304, 734–736. [Google Scholar] [CrossRef]
- Fernandez-Pato, A.; Virseda-Berdices, A.; Resino, S.; Ryan, P.; Martinez-Gonzalez, O.; Perez-Garcia, F.; Martin-Vicente, M.; Valle-Millares, D.; Brochado-Kith, O.; Blancas, R.; et al. Plasma miRNA profile at COVID-19 onset predicts severity status and mortality. Emerg. Microbes Infect. 2022, 11, 676–688. [Google Scholar] [CrossRef] [PubMed]
- Lee, W. MicroRNA, Insulin Resistance, and Metabolic Disorders. Int. J. Mol. Sci. 2022, 23, 16215. [Google Scholar] [CrossRef]
- Macvanin, M.; Obradovic, M.; Zafirovic, S.; Stanimirovic, J.; Isenovic, E.R. The Role of miRNAs in Metabolic Diseases. Curr. Med. Chem. 2023, 30, 1922–1944. [Google Scholar] [CrossRef]
- Ye, Z.; Wang, S.; Huang, X.; Chen, P.; Deng, L.; Li, S.; Lin, S.; Wang, Z.; Liu, B. Plasma Exosomal miRNAs Associated With Metabolism as Early Predictor of Gestational Diabetes Mellitus. Diabetes 2022, 71, 2272–2283. [Google Scholar] [CrossRef]
- Chen, Y.; Shen, Y.; Lin, P.; Tong, D.; Zhao, Y.; Allesina, S.; Shen, X.; Wu, C.I. Gene regulatory network stabilized by pervasive weak repressions: MicroRNA functions revealed by the May-Wigner theory. Natl. Sci. Rev. 2019, 6, 1176–1188. [Google Scholar] [CrossRef] [PubMed]
- Creemers, E.E.; Tijsen, A.J.; Pinto, Y.M. Circulating microRNAs: Novel biomarkers and extracellular communicators in cardiovascular disease? Circ. Res. 2012, 110, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Xin, L.; Liu, Y.H.; Martin, T.A.; Jiang, W.G. The Era of Multigene Panels Comes? The Clinical Utility of Oncotype DX and MammaPrint. World J. Oncol. 2017, 8, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Avgeris, M.; Mavridis, K.; Scorilas, A. Kallikrein-related peptidases in prostate, breast, and ovarian cancers: From pathobiology to clinical relevance. Biol. Chem. 2012, 393, 301–317. [Google Scholar] [CrossRef]
- Bertoli, G.; Cava, C.; Castiglioni, I. MicroRNAs as Biomarkers for Diagnosis, Prognosis and Theranostics in Prostate Cancer. Int. J. Mol. Sci. 2016, 17, 421. [Google Scholar] [CrossRef]
- Cortez, M.A.; Bueso-Ramos, C.; Ferdin, J.; Lopez-Berestein, G.; Sood, A.K.; Calin, G.A. MicroRNAs in body fluids--the mix of hormones and biomarkers. Nat. Rev. Clin. Oncol. 2011, 8, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef]
- Cruz-Burgos, M.; Cortes-Ramirez, S.A.; Losada-Garcia, A.; Morales-Pacheco, M.; Martinez-Martinez, E.; Morales-Montor, J.G.; Servin-Haddad, A.; Izquierdo-Luna, J.S.; Rodriguez-Martinez, G.; Ramos-Godinez, M.D.P.; et al. Unraveling the Role of EV-Derived miR-150-5p in Prostate Cancer Metastasis and Its Association with High-Grade Gleason Scores: Implications for Diagnosis. Cancers 2023, 15, 4148. [Google Scholar] [CrossRef]
- Melo, S.A.; Sugimoto, H.; O’Connell, J.T.; Kato, N.; Villanueva, A.; Vidal, A.; Qiu, L.; Vitkin, E.; Perelman, L.T.; Melo, C.A.; et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 2014, 26, 707–721. [Google Scholar] [CrossRef]
- Tan, S.; Xia, L.; Yi, P.; Han, Y.; Tang, L.; Pan, Q.; Tian, Y.; Rao, S.; Oyang, L.; Liang, J.; et al. Exosomal miRNAs in tumor microenvironment. J. Exp. Clin. Cancer Res. CR 2020, 39, 67. [Google Scholar] [CrossRef]
- Zhou, J.; Nagarkatti, P.; Zhong, Y.; Ginsberg, J.P.; Singh, N.P.; Zhang, J.; Nagarkatti, M. Dysregulation in microRNA expression is associated with alterations in immune functions in combat veterans with post-traumatic stress disorder. PLoS ONE 2014, 9, e94075. [Google Scholar] [CrossRef] [PubMed]
- Kannampuzha, S.; Murali, R.; Gopalakrishnan, A.V.; Mukherjee, A.G.; Wanjari, U.R.; Namachivayam, A.; George, A.; Dey, A.; Vellingiri, B. Novel biomolecules in targeted cancer therapy: A new approach towards precision medicine. Med. Oncol. 2023, 40, 323. [Google Scholar] [CrossRef] [PubMed]
- Katoh, M.; Igarashi, M.; Fukuda, H.; Nakagama, H.; Katoh, M. Cancer genetics and genomics of human FOX family genes. Cancer Lett. 2013, 328, 198–206. [Google Scholar] [CrossRef]
- Antonino, M.; Nicolo, M.; Jerome Renee, L.; Federico, M.; Chiara, V.; Stefano, S.; Maria, S.; Salvatore, C.; Antonio, B.; Calvo-Henriquez, C.; et al. Single-nucleotide polymorphism in chronic rhinosinusitis: A systematic review. Clin. Otolaryngol. 2022, 47, 14–23. [Google Scholar] [CrossRef]
- Franczyk, B.; Rysz, J.; Gluba-Brzozka, A. Pharmacogenetics of Drugs Used in the Treatment of Cancers. Genes 2022, 13, 311. [Google Scholar] [CrossRef] [PubMed]
- Imyanitov, E.N.; Iyevleva, A.G. Molecular tests for prediction of tumor sensitivity to cytotoxic drugs. Cancer Lett. 2022, 526, 41–52. [Google Scholar] [CrossRef]
- Madkour, M.M.; Ramadan, W.S.; Saleh, E.; El-Awady, R. Epigenetic modulations in cancer: Predictive biomarkers and potential targets for overcoming the resistance to topoisomerase I inhibitors. Ann. Med. 2023, 55, 2203946. [Google Scholar] [CrossRef]
- Meng, C.L.; Zhao, W.; Zhong, D.N. Epigenetics and microRNAs in UGT1As. Hum. Genom. 2021, 15, 30. [Google Scholar] [CrossRef]
- Zappe, K.; Cichna-Markl, M. Aberrant DNA Methylation of ABC Transporters in Cancer. Cells 2020, 9, 2281. [Google Scholar] [CrossRef]
- Aboul-Soud, M.A.M.; Alzahrani, A.J.; Mahmoud, A. Decoding variants in drug-metabolizing enzymes and transporters in solid tumor patients by whole-exome sequencing. Saudi J. Biol. Sci. 2021, 28, 628–634. [Google Scholar] [CrossRef]
- Hu, X.; Qin, W.; Li, S.; He, M.; Wang, Y.; Guan, S.; Zhao, H.; Yao, W.; Wei, M.; Liu, M.; et al. Polymorphisms in DNA repair pathway genes and ABCG2 gene in advanced colorectal cancer: Correlation with tumor characteristics and clinical outcome in oxaliplatin-based chemotherapy. Cancer Manag. Res. 2019, 11, 285–297. [Google Scholar] [CrossRef]
- Saini, H.K.; Griffiths-Jones, S.; Enright, A.J. Genomic analysis of human microRNA transcripts. Proc. Natl. Acad. Sci. USA 2007, 104, 17719–17724. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef]
- Bartel, D.P.; Chen, C.Z. Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 2004, 5, 396–400. [Google Scholar] [CrossRef]
- Li, Z.; Rana, T.M. Therapeutic targeting of microRNAs: Current status and future challenges. Nat. Rev. Drug Discov. 2014, 13, 622–638. [Google Scholar] [CrossRef]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes. Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Cullen, B.R. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 2004, 32, 4776–4785. [Google Scholar] [CrossRef] [PubMed]
- Chendrimada, T.P.; Gregory, R.I.; Kumaraswamy, E.; Norman, J.; Cooch, N.; Nishikura, K.; Shiekhattar, R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 2005, 436, 740–744. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Zhang, X.; Graves, P.; Zeng, Y. A comprehensive analysis of precursor microRNA cleavage by human Dicer. Rna 2012, 18, 2083–2092. [Google Scholar] [CrossRef]
- Nakanishi, K. Anatomy of four human Argonaute proteins. Nucleic Acids Res. 2022, 50, 6618–6638. [Google Scholar] [CrossRef]
- Medley, J.C.; Panzade, G.; Zinovyeva, A.Y. microRNA strand selection: Unwinding the rules. Wiley Interdiscip. Rev. RNA 2021, 12, e1627. [Google Scholar] [CrossRef] [PubMed]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009, 10, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, S.J.; White, R.J. Regulation of RNA polymerase III transcription during mammalian cell growth. Cell Cycle 2007, 6, 2323–2326. [Google Scholar] [CrossRef]
- Borchert, G.M.; Lanier, W.; Davidson, B.L. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006, 13, 1097–1101. [Google Scholar] [CrossRef]
- Czech, B.; Hannon, G.J. Small RNA sorting: Matchmaking for Argonautes. Nat. Rev. Genet. 2011, 12, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Perron, M.P.; Provost, P. Protein interactions and complexes in human microRNA biogenesis and function. Front. Biosci. A J. Virtual Libr. 2008, 13, 2537–2547. [Google Scholar] [CrossRef]
- Eamens, A.L.; Wang, M.B. Alternate approaches to repress endogenous microRNA activity in Arabidopsis thaliana. Plant Signal. Behav. 2011, 6, 349–359. [Google Scholar] [CrossRef]
- Zhu, H.; Hu, F.; Wang, R.; Zhou, X.; Sze, S.H.; Liou, L.W.; Barefoot, A.; Dickman, M.; Zhang, X. Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell 2011, 145, 242–256. [Google Scholar] [CrossRef]
- Doxakis, E. Principles of miRNA-target regulation in metazoan models. Int. J. Mol. Sci. 2013, 14, 16280–16302. [Google Scholar] [CrossRef]
- Orom, U.A.; Nielsen, F.C.; Lund, A.H. MicroRNA-10a binds the 5’UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell 2008, 30, 460–471. [Google Scholar] [CrossRef]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Chirshev, E.; Oberg, K.C.; Ioffe, Y.J.; Unternaehrer, J.J. Let-7 as biomarker, prognostic indicator, and therapy for precision medicine in cancer. Clin. Transl. Med. 2019, 8, 24. [Google Scholar] [CrossRef] [PubMed]
- Ameres, S.L.; Zamore, P.D. Diversifying microRNA sequence and function. Nat. Rev. Mol. Cell Biol. 2013, 14, 475–488. [Google Scholar] [CrossRef] [PubMed]
- Chim, S.S.; Shing, T.K.; Hung, E.C.; Leung, T.Y.; Lau, T.K.; Chiu, R.W.; Lo, Y.M. Detection and characterization of placental microRNAs in maternal plasma. Clin. Chem. 2008, 54, 482–490. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef]
- Taylor, D.D.; Gercel-Taylor, C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008, 110, 13–21. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Avgeris, M.; Panoutsopoulou, K.; Papadimitriou, M.A.; Scorilas, A. Circulating exosomal miRNAs: Clinical significance in human cancers. Expert. Rev. Mol. Diagn. 2019, 19, 979–995. [Google Scholar] [CrossRef]
- Alotaibi, F. Exosomal microRNAs in cancer: Potential biomarkers and immunotherapeutic targets for immune checkpoint molecules. Front. Genet. 2023, 14, 1052731. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- Mattick, J.S. The hidden genetic program of complex organisms. Sci. Am. 2004, 291, 60–67. [Google Scholar] [CrossRef]
- Muthamilselvan, S.; Ramasami Sundhar Baabu, P.; Palaniappan, A. Microfluidics for Profiling miRNA Biomarker Panels in AI-Assisted Cancer Diagnosis and Prognosis. Technol. Cancer Res. Treat. 2023, 22, 15330338231185284. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Sevignani, C.; Dumitru, C.D.; Hyslop, T.; Noch, E.; Yendamuri, S.; Shimizu, M.; Rattan, S.; Bullrich, F.; Negrini, M.; et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA 2004, 101, 2999–3004. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Oh, J.E.; Kim, Y.R.; Park, S.W.; Kang, M.R.; Kim, S.S.; Ahn, C.H.; Yoo, N.J.; Lee, S.H. Somatic mutations and losses of expression of microRNA regulation-related genes AGO2 and TNRC6A in gastric and colorectal cancers. J. Pathol. 2010, 221, 139–146. [Google Scholar] [CrossRef]
- Zhang, L.; Huang, J.; Yang, N.; Greshock, J.; Megraw, M.S.; Giannakakis, A.; Liang, S.; Naylor, T.L.; Barchetti, A.; Ward, M.R.; et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc. Natl. Acad. Sci. USA 2006, 103, 9136–9141. [Google Scholar] [CrossRef]
- Kumar, M.S.; Lu, J.; Mercer, K.L.; Golub, T.R.; Jacks, T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 2007, 39, 673–677. [Google Scholar] [CrossRef]
- McManus, M.T. MicroRNAs and cancer. Semin. Cancer Biol. 2003, 13, 253–258. [Google Scholar] [CrossRef]
- Gong, H.; Liu, C.M.; Liu, D.P.; Liang, C.C. The role of small RNAs in human diseases: Potential troublemaker and therapeutic tools. Med. Res. Rev. 2005, 25, 361–381. [Google Scholar] [CrossRef] [PubMed]
- Melo, S.A.; Ropero, S.; Moutinho, C.; Aaltonen, L.A.; Yamamoto, H.; Calin, G.A.; Rossi, S.; Fernandez, A.F.; Carneiro, F.; Oliveira, C.; et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat. Genet. 2009, 41, 365–370. [Google Scholar] [CrossRef]
- Hill, D.A.; Ivanovich, J.; Priest, J.R.; Gurnett, C.A.; Dehner, L.P.; Desruisseau, D.; Jarzembowski, J.A.; Wikenheiser-Brokamp, K.A.; Suarez, B.K.; Whelan, A.J.; et al. DICER1 mutations in familial pleuropulmonary blastoma. Science 2009, 325, 965. [Google Scholar] [CrossRef]
- Melo, S.A.; Moutinho, C.; Ropero, S.; Calin, G.A.; Rossi, S.; Spizzo, R.; Fernandez, A.F.; Davalos, V.; Villanueva, A.; Montoya, G.; et al. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell 2010, 18, 303–315. [Google Scholar] [CrossRef]
- Rosenfeld, N.; Aharonov, R.; Meiri, E.; Rosenwald, S.; Spector, Y.; Zepeniuk, M.; Benjamin, H.; Shabes, N.; Tabak, S.; Levy, A.; et al. MicroRNAs accurately identify cancer tissue origin. Nat. Biotechnol. 2008, 26, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Wegert, J.; Ishaque, N.; Vardapour, R.; Georg, C.; Gu, Z.; Bieg, M.; Ziegler, B.; Bausenwein, S.; Nourkami, N.; Ludwig, N.; et al. Mutations in the SIX1/2 pathway and the DROSHA/DGCR8 miRNA microprocessor complex underlie high-risk blastemal type Wilms tumors. Cancer Cell 2015, 27, 298–311. [Google Scholar] [CrossRef] [PubMed]
- Walz, A.L.; Ooms, A.; Gadd, S.; Gerhard, D.S.; Smith, M.A.; Guidry Auvil, J.M.; Meerzaman, D.; Chen, Q.R.; Hsu, C.H.; Yan, C.; et al. Recurrent DGCR8, DROSHA, and SIX homeodomain mutations in favorable histology Wilms tumors. Cancer Cell 2015, 27, 286–297. [Google Scholar] [CrossRef]
- Sugito, N.; Ishiguro, H.; Kuwabara, Y.; Kimura, M.; Mitsui, A.; Kurehara, H.; Ando, T.; Mori, R.; Takashima, N.; Ogawa, R.; et al. RNASEN regulates cell proliferation and affects survival in esophageal cancer patients. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2006, 12, 7322–7328. [Google Scholar] [CrossRef]
- Diaz-Garcia, C.V.; Agudo-Lopez, A.; Perez, C.; Lopez-Martin, J.A.; Rodriguez-Peralto, J.L.; de Castro, J.; Cortijo, A.; Martinez-Villanueva, M.; Iglesias, L.; Garcia-Carbonero, R.; et al. DICER1, DROSHA and miRNAs in patients with non-small cell lung cancer: Implications for outcomes and histologic classification. Carcinogenesis 2013, 34, 1031–1038. [Google Scholar] [CrossRef]
- Lin, R.J.; Lin, Y.C.; Chen, J.; Kuo, H.H.; Chen, Y.Y.; Diccianni, M.B.; London, W.B.; Chang, C.H.; Yu, A.L. microRNA signature and expression of Dicer and Drosha can predict prognosis and delineate risk groups in neuroblastoma. Cancer Res. 2010, 70, 7841–7850. [Google Scholar] [CrossRef]
- Doros, L.; Schultz, K.A.; Stewart, D.R.; Bauer, A.J.; Williams, G.; Rossi, C.T.; Carr, A.; Yang, J.; Dehner, L.P.; Messinger, Y.; et al. DICER1-Related Disorders. In GeneReviews®; Pagon, R.A., Adam, M.P., Ardinger, H.H., Wallace, S.E., Amemiya, A., Bean, L.J.H., Bird, T.D., Fong, C.T., Mefford, H.C., Smith, R.J.H., et al., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Sand, M.; Gambichler, T.; Skrygan, M.; Sand, D.; Scola, N.; Altmeyer, P.; Bechara, F.G. Expression levels of the microRNA processing enzymes Drosha and dicer in epithelial skin cancer. Cancer Investig. 2010, 28, 649–653. [Google Scholar] [CrossRef]
- Sand, M.; Gambichler, T.; Sand, D.; Altmeyer, P.; Stuecker, M.; Bechara, F.G. Immunohistochemical expression patterns of the microRNA-processing enzyme Dicer in cutaneous malignant melanomas, benign melanocytic nevi and dysplastic melanocytic nevi. Eur. J. Dermatol. 2011, 21, 18–21. [Google Scholar] [CrossRef] [PubMed]
- Sand, M.; Skrygan, M.; Georgas, D.; Arenz, C.; Gambichler, T.; Sand, D.; Altmeyer, P.; Bechara, F.G. Expression levels of the microRNA maturing microprocessor complex component DGCR8 and the RNA-induced silencing complex (RISC) components argonaute-1, argonaute-2, PACT, TARBP1, and TARBP2 in epithelial skin cancer. Mol. Carcinog. 2012, 51, 916–922. [Google Scholar] [CrossRef] [PubMed]
- Iacomino, G. miRNAs: The Road from Bench to Bedside. Genes 2023, 14, 314. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zhu, L.; Wang, X.; Jin, H. RNA-based therapeutics: An overview and prospectus. Cell Death Dis. 2022, 13, 644. [Google Scholar] [CrossRef]
- Jo, S.J.; Chae, S.U.; Lee, C.B.; Bae, S.K. Clinical Pharmacokinetics of Approved RNA Therapeutics. Int. J. Mol. Sci. 2023, 24, 746. [Google Scholar] [CrossRef]
- Das, N.; Tripathi, N.; Khurana, S. Micro RNA Mimics And Antagonists. Int. J. Sci. Technol. Res. 2015, 4, 176–180. [Google Scholar]
- Manikkath, J.; Jishnu, P.V.; Wich, P.R.; Manikkath, A.; Radhakrishnan, R. Nanoparticulate strategies for the delivery of miRNA mimics and inhibitors in anticancer therapy and its potential utility in oral submucous fibrosis. Nanomedicine 2022, 17, 181–195. [Google Scholar] [CrossRef]
- Kang, E.; Kortylewski, M. Lipid Nanoparticle-Mediated Delivery of miRNA Mimics to Myeloid Cells. Methods Mol. Biol. 2023, 2691, 337–350. [Google Scholar] [CrossRef]
- Fu, Y.; Chen, J.; Huang, Z. Recent progress in microRNA-based delivery systems for the treatment of human disease. ExRNA 2019, 1, 24. [Google Scholar] [CrossRef]
- Reda El Sayed, S.; Cristante, J.; Guyon, L.; Denis, J.; Chabre, O.; Cherradi, N. MicroRNA Therapeutics in Cancer: Current Advances and Challenges. Cancers 2021, 13, 2680. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Zhao, Z.; Cai, Q.; Zhang, Y.; Zhang, P.; Shi, S.; Xie, H.; Peng, X.; Yin, W.; Tao, Y.; et al. miRNA-based biomarkers, therapies, and resistance in Cancer. Int. J. Biol. Sci. 2020, 16, 2628–2647. [Google Scholar] [CrossRef]
- Fu, Z.; Wang, L.; Li, S.; Chen, F.; Au-Yeung, K.K.; Shi, C. MicroRNA as an Important Target for Anticancer Drug Development. Front. Pharmacol. 2021, 12, 736323. [Google Scholar] [CrossRef]
- Otoukesh, B.; Abbasi, M.; Gorgani, H.O.; Farahini, H.; Moghtadaei, M.; Boddouhi, B.; Kaghazian, P.; Hosseinzadeh, S.; Alaee, A. MicroRNAs signatures, bioinformatics analysis of miRNAs, miRNA mimics and antagonists, and miRNA therapeutics in osteosarcoma. Cancer Cell Int. 2020, 20, 254. [Google Scholar] [CrossRef] [PubMed]
- Check Hayden, E. Cancer complexity slows quest for cure. Nature 2008, 455, 148. [Google Scholar] [CrossRef]
- Parsons, D.W.; Jones, S.; Zhang, X.; Lin, J.C.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Siu, I.M.; Gallia, G.L.; et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008, 321, 1807–1812. [Google Scholar] [CrossRef] [PubMed]
- Medina, P.P.; Nolde, M.; Slack, F.J. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature 2010, 467, 86–90. [Google Scholar] [CrossRef]
- Menon, A.; Abd-Aziz, N.; Khalid, K.; Poh, C.L.; Naidu, R. miRNA: A Promising Therapeutic Target in Cancer. Int. J. Mol. Sci. 2022, 23, 11502. [Google Scholar] [CrossRef]
- Garzon, R.; Marcucci, G.; Croce, C.M. Targeting microRNAs in cancer: Rationale, strategies and challenges. Nat. Rev. Drug Discov. 2010, 9, 775–789. [Google Scholar] [CrossRef]
- Cannataro, R.; Cione, E. miRNA as Drug: Antagomir and Beyond. Curr. Pharm. Des. 2023, 29, 462–465. [Google Scholar] [CrossRef]
- Ortega, M.M.; Bouamar, H. Guidelines on Designing MicroRNA Sponges: From Construction to Stable Cell Line. Methods Mol. Biol. 2023, 2595, 171–183. [Google Scholar] [CrossRef]
- Takegawa-Araki, T.; Kumagai, S.; Yasukawa, K.; Kuroda, M.; Sasaki, T.; Obika, S. Structure-Activity Relationships of Anti-microRNA Oligonucleotides Containing Cationic Guanidine-Modified Nucleic Acids. J. Med. Chem. 2022, 65, 2139–2148. [Google Scholar] [CrossRef]
- Hutvagner, G.; Simard, M.J.; Mello, C.C.; Zamore, P.D. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004, 2, E98. [Google Scholar] [CrossRef] [PubMed]
- Orom, U.A.; Kauppinen, S.; Lund, A.H. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene 2006, 372, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Sharma, R.; Ryu, K.; Shen, P.; Salaita, K.; Jo, H. Conditional Antisense Oligonucleotides Triggered by miRNA. ACS Chem. Biol. 2021, 16, 2255–2267. [Google Scholar] [CrossRef] [PubMed]
- Quemener, A.M.; Bachelot, L.; Forestier, A.; Donnou-Fournet, E.; Gilot, D.; Galibert, M.D. The powerful world of antisense oligonucleotides: From bench to bedside. Wiley Interdiscip. Rev. RNA 2020, 11, e1594. [Google Scholar] [CrossRef] [PubMed]
- Seth, P.P.; Siwkowski, A.; Allerson, C.R.; Vasquez, G.; Lee, S.; Prakash, T.P.; Wancewicz, E.V.; Witchell, D.; Swayze, E.E. Short antisense oligonucleotides with novel 2′-4′ conformationaly restricted nucleoside analogues show improved potency without increased toxicity in animals. J. Med. Chem. 2009, 52, 10–13. [Google Scholar] [CrossRef]
- Shadid, M.; Badawi, M.; Abulrob, A. Antisense oligonucleotides: Absorption, distribution, metabolism, and excretion. Expert. Opin. Drug Metab. Toxicol. 2021, 17, 1281–1292. [Google Scholar] [CrossRef]
- Monia, B.P.; Lesnik, E.A.; Gonzalez, C.; Lima, W.F.; McGee, D.; Guinosso, C.J.; Kawasaki, A.M.; Cook, P.D.; Freier, S.M. Evaluation of 2′-modified oligonucleotides containing 2′-deoxy gaps as antisense inhibitors of gene expression. J. Biol. Chem. 1993, 268, 14514–14522. [Google Scholar] [CrossRef] [PubMed]
- Lennox, K.A.; Behlke, M.A. Chemical modification and design of anti-miRNA oligonucleotides. Gene Ther. 2011, 18, 1111–1120. [Google Scholar] [CrossRef]
- Meister, G.; Landthaler, M.; Dorsett, Y.; Tuschl, T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. Rna 2004, 10, 544–550. [Google Scholar] [CrossRef]
- Esau, C.C. Inhibition of microRNA with antisense oligonucleotides. Methods 2008, 44, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Spizzo, R.; Rushworth, D.; Guerrero, M.; Calin, G.A. RNA inhibition, microRNAs, and new therapeutic agents for cancer treatment. Clin. Lymphoma Myeloma 2009, 9 (Suppl. S3), S313–S318. [Google Scholar] [CrossRef]
- Zhang, S.; Chen, L.; Jung, E.J.; Calin, G.A. Targeting microRNAs with small molecules: From dream to reality. Clin. Pharmacol. Ther. 2010, 87, 754–758. [Google Scholar] [CrossRef]
- Moriyama, T.; Ohuchida, K.; Mizumoto, K.; Yu, J.; Sato, N.; Nabae, T.; Takahata, S.; Toma, H.; Nagai, E.; Tanaka, M. MicroRNA-21 modulates biological functions of pancreatic cancer cells including their proliferation, invasion, and chemoresistance. Mol. Cancer Ther. 2009, 8, 1067–1074. [Google Scholar] [CrossRef] [PubMed]
- Park, J.K.; Lee, E.J.; Esau, C.; Schmittgen, T.D. Antisense inhibition of microRNA-21 or -221 arrests cell cycle, induces apoptosis, and sensitizes the effects of gemcitabine in pancreatic adenocarcinoma. Pancreas 2009, 38, e190–e199. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhao, L.; Ischenko, I.; Bao, Q.; Schwarz, B.; Niess, H.; Wang, Y.; Renner, A.; Mysliwietz, J.; Jauch, K.W.; et al. Antisense inhibition of microRNA-21 and microRNA-221 in tumor-initiating stem-like cells modulates tumorigenesis, metastasis, and chemotherapy resistance in pancreatic cancer. Target. Oncol. 2015, 10, 535–548. [Google Scholar] [CrossRef] [PubMed]
- Krutzfeldt, J.; Rajewsky, N.; Braich, R.; Rajeev, K.G.; Tuschl, T.; Manoharan, M.; Stoffel, M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005, 438, 685–689. [Google Scholar] [CrossRef] [PubMed]
- Lima, J.F.; Cerqueira, L.; Figueiredo, C.; Oliveira, C.; Azevedo, N.F. Anti-miRNA oligonucleotides: A comprehensive guide for design. RNA Biol. 2018, 15, 338–352. [Google Scholar] [CrossRef]
- Fei, J.; Lan, F.; Guo, M.; Li, Y.; Liu, Y. Inhibitory effects of anti-miRNA oligonucleotides (AMOs) on A549 cell growth. J. Drug Target. 2008, 16, 688–693. [Google Scholar] [CrossRef]
- Baker, B.F.; Lot, S.S.; Condon, T.P.; Cheng-Flournoy, S.; Lesnik, E.A.; Sasmor, H.M.; Bennett, C.F. 2′-O-(2-Methoxy)ethyl-modified anti-intercellular adhesion molecule 1 (ICAM-1) oligonucleotides selectively increase the ICAM-1 mRNA level and inhibit formation of the ICAM-1 translation initiation complex in human umbilical vein endothelial cells. J. Biol. Chem. 1997, 272, 11994–12000. [Google Scholar] [CrossRef] [PubMed]
- Friedman, K.J.; Kole, J.; Cohn, J.A.; Knowles, M.R.; Silverman, L.M.; Kole, R. Correction of aberrant splicing of the cystic fibrosis transmembrane conductance regulator (CFTR) gene by antisense oligonucleotides. J. Biol. Chem. 1999, 274, 36193–36199. [Google Scholar] [CrossRef]
- Mani, S.; Goel, S.; Nesterova, M.; Martin, R.M.; Grindel, J.M.; Rothenberg, M.L.; Zhang, R.; Tortora, G.; Cho-Chung, Y.S. Clinical studies in patients with solid tumors using a second-generation antisense oligonucleotide (GEM 231) targeted against protein kinase A type I. Ann. N. Y. Acad. Sci. 2003, 1002, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Trang, P.; Medina, P.P.; Wiggins, J.F.; Ruffino, L.; Kelnar, K.; Omotola, M.; Homer, R.; Brown, D.; Bader, A.G.; Weidhaas, J.B.; et al. Regression of murine lung tumors by the let-7 microRNA. Oncogene 2010, 29, 1580–1587. [Google Scholar] [CrossRef] [PubMed]
- Si, M.L.; Zhu, S.; Wu, H.; Lu, Z.; Wu, F.; Mo, Y.Y. miR-21-mediated tumor growth. Oncogene 2007, 26, 2799–2803. [Google Scholar] [CrossRef]
- Lennox, K.A.; Owczarzy, R.; Thomas, D.M.; Walder, J.A.; Behlke, M.A. Improved Performance of Anti-miRNA Oligonucleotides Using a Novel Non-Nucleotide Modifier. Mol. Ther.-Nucleic Acids 2013, 2, e117. [Google Scholar] [CrossRef]
- Gleave, M.E.; Monia, B.P. Antisense therapy for cancer. Nat. Rev. Cancer 2005, 5, 468–479. [Google Scholar] [CrossRef]
- Sewell, K.L.; Geary, R.S.; Baker, B.F.; Glover, J.M.; Mant, T.G.; Yu, R.Z.; Tami, J.A.; Dorr, F.A. Phase I trial of ISIS 104838, a 2′-methoxyethyl modified antisense oligonucleotide targeting tumor necrosis factor-alpha. J. Pharmacol. Exp. Ther. 2002, 303, 1334–1343. [Google Scholar] [CrossRef] [PubMed]
- Chi, K.N.; Siu, L.L.; Hirte, H.; Hotte, S.J.; Knox, J.; Kollmansberger, C.; Gleave, M.; Guns, E.; Powers, J.; Walsh, W.; et al. A phase I study of OGX-011, a 2′-methoxyethyl phosphorothioate antisense to clusterin, in combination with docetaxel in patients with advanced cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2008, 14, 833–839. [Google Scholar] [CrossRef] [PubMed]
- Raal, F.J.; Santos, R.D.; Blom, D.J.; Marais, A.D.; Charng, M.J.; Cromwell, W.C.; Lachmann, R.H.; Gaudet, D.; Tan, J.L.; Chasan-Taber, S.; et al. Mipomersen, an apolipoprotein B synthesis inhibitor, for lowering of LDL cholesterol concentrations in patients with homozygous familial hypercholesterolaemia: A randomised, double-blind, placebo-controlled trial. Lancet 2010, 375, 998–1006. [Google Scholar] [CrossRef]
- Vester, B.; Wengel, J. LNA (locked nucleic acid): High-affinity targeting of complementary RNA and DNA. Biochemistry 2004, 43, 13233–13241. [Google Scholar] [CrossRef] [PubMed]
- Valoczi, A.; Hornyik, C.; Varga, N.; Burgyan, J.; Kauppinen, S.; Havelda, Z. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 2004, 32, e175. [Google Scholar] [CrossRef] [PubMed]
- Henry, S.P.; Geary, R.S.; Yu, R.; Levin, A.A. Drug properties of second-generation antisense oligonucleotides: How do they measure up to their predecessors? Curr. Opin. Investig. Drugs 2001, 2, 1444–1449. [Google Scholar]
- Weiler, J.; Hunziker, J.; Hall, J. Anti-miRNA oligonucleotides (AMOs): Ammunition to target miRNAs implicated in human disease? Gene Ther. 2006, 13, 496–502. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, T.; Katsuda, T.; Hagiwara, K.; Kosaka, N.; Yoshioka, Y.; Takahashi, R.U.; Takeshita, F.; Kubota, D.; Kondo, T.; Ichikawa, H.; et al. Clinical relevance and therapeutic significance of microRNA-133a expression profiles and functions in malignant osteosarcoma-initiating cells. Stem Cells 2014, 32, 959–973. [Google Scholar] [CrossRef]
- Davis, S.; Lollo, B.; Freier, S.; Esau, C. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res. 2006, 34, 2294–2304. [Google Scholar] [CrossRef]
- Gallo Cantafio, M.E.; Nielsen, B.S.; Mignogna, C.; Arbitrio, M.; Botta, C.; Frandsen, N.M.; Rolfo, C.; Tagliaferri, P.; Tassone, P.; Di Martino, M.T. Pharmacokinetics and Pharmacodynamics of a 13-mer LNA-inhibitor-miR-221 in Mice and Non-human Primates. Mol. Ther. Nucleic Acids 2016, 5, E326. [Google Scholar] [CrossRef]
- Jie, J.; Liu, D.; Wang, Y.; Wu, Q.; Wu, T.; Fang, R. Generation of MiRNA sponge constructs targeting multiple MiRNAs. J. Clin. Lab. Anal. 2022, 36, e24527. [Google Scholar] [CrossRef]
- Ebert, M.S.; Neilson, J.R.; Sharp, P.A. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 2007, 4, 721–726. [Google Scholar] [CrossRef]
- Xiao, J.; Yang, B.; Lin, H.; Lu, Y.; Luo, X.; Wang, Z. Novel approaches for gene-specific interference via manipulating actions of microRNAs: Examination on the pacemaker channel genes HCN2 and HCN4. J. Cell. Physiol. 2007, 212, 285–292. [Google Scholar] [CrossRef]
- Papapetrou, E.P.; Korkola, J.E.; Sadelain, M. A genetic strategy for single and combinatorial analysis of miRNA function in mammalian hematopoietic stem cells. Stem Cells 2010, 28, 287–296. [Google Scholar] [CrossRef]
- Ma, L.; Young, J.; Prabhala, H.; Pan, E.; Mestdagh, P.; Muth, D.; Teruya-Feldstein, J.; Reinhardt, F.; Onder, T.T.; Valastyan, S.; et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat. Cell Biol. 2010, 12, 247–256. [Google Scholar] [CrossRef]
- Yang, Y.; Meng, H.; Peng, Q.; Yang, X.; Gan, R.; Zhao, L.; Chen, Z.; Lu, J.; Meng, Q.H. Downregulation of microRNA-21 expression restrains non-small cell lung cancer cell proliferation and migration through upregulation of programmed cell death 4. Cancer Gene Ther. 2015, 22, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Bak, R.O.; Mikkelsen, J.G. miRNA sponges: Soaking up miRNAs for regulation of gene expression. Wiley Interdiscip. Rev. RNA 2014, 5, 317–333. [Google Scholar] [CrossRef] [PubMed]
- Krell, J.; Frampton, A.E.; Jacob, J.; Castellano, L.; Stebbing, J. miRNAs in breast cancer: Ready for real time? Pharmacogenomics 2012, 13, 709–719. [Google Scholar] [CrossRef]
- Murakami, K.; Miyagishi, M. Tiny masking locked nucleic acids effectively bind to mRNA and inhibit binding of microRNAs in relation to thermodynamic stability. Biomed. Rep. 2014, 2, 509–512. [Google Scholar] [CrossRef]
- Colangelo, T.; Polcaro, G.; Ziccardi, P.; Muccillo, L.; Galgani, M.; Pucci, B.; Milone, M.R.; Budillon, A.; Santopaolo, M.; Mazzoccoli, G.; et al. The miR-27a-calreticulin axis affects drug-induced immunogenic cell death in human colorectal cancer cells. Cell Death Dis. 2016, 7, e2108. [Google Scholar] [CrossRef] [PubMed]
- Saiyed, A.N.; Vasavada, A.R.; Johar, S.R.K. Recent trends in miRNA therapeutics and the application of plant miRNA for prevention and treatment of human diseases. Futur. J. Pharm. Sci. 2022, 8, 24. [Google Scholar] [CrossRef]
- Yang, H.; Liu, Y.; Chen, L.; Zhao, J.; Guo, M.; Zhao, X.; Wen, Z.; He, Z.; Chen, C.; Xu, L. MiRNA-Based Therapies for Lung Cancer: Opportunities and Challenges? Biomolecules 2023, 13, 877. [Google Scholar] [CrossRef]
- Hossain, A.; Kuo, M.T.; Saunders, G.F. Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol. Cell. Biol. 2006, 26, 8191–8201. [Google Scholar] [CrossRef]
- Hutvagner, G.; Zamore, P.D. RNAi: Nature abhors a double-strand. Curr. Opin. Genet. Dev. 2002, 12, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Takeshita, F.; Patrawala, L.; Osaki, M.; Takahashi, R.U.; Yamamoto, Y.; Kosaka, N.; Kawamata, M.; Kelnar, K.; Bader, A.G.; Brown, D.; et al. Systemic delivery of synthetic microRNA-16 inhibits the growth of metastatic prostate tumors via downregulation of multiple cell-cycle genes. Mol. Ther. J. Am. Soc. Gene Ther. 2010, 18, 181–187. [Google Scholar] [CrossRef]
- Wiggins, J.F.; Ruffino, L.; Kelnar, K.; Omotola, M.; Patrawala, L.; Brown, D.; Bader, A.G. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 2010, 70, 5923–5930. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.M.; Phillips, B.L.; Patel, R.S.; Cohen, D.M.; Jakymiw, A.; Kong, W.W.; Cheng, J.Q.; Chan, E.K. Keratinization-associated miR-7 and miR-21 regulate tumor suppressor reversion-inducing cysteine-rich protein with kazal motifs (RECK) in oral cancer. J. Biol. Chem. 2012, 287, 29261–29272. [Google Scholar] [CrossRef] [PubMed]
- Segal, M.; Biscans, A.; Gilles, M.E.; Anastasiadou, E.; De Luca, R.; Lim, J.; Khvorova, A.; Slack, F.J. Hydrophobically Modified let-7b miRNA Enhances Biodistribution to NSCLC and Downregulates HMGA2 In Vivo. Mol. Ther. Nucleic Acids 2020, 19, 267–277. [Google Scholar] [CrossRef]
- Segal, M.; Slack, F.J. Challenges identifying efficacious miRNA therapeutics for cancer. Expert. Opin. Drug Discov. 2020, 15, 987–992. [Google Scholar] [CrossRef]
- Li, X.; Corbett, A.L.; Taatizadeh, E.; Tasnim, N.; Little, J.P.; Garnis, C.; Daugaard, M.; Guns, E.; Hoorfar, M.; Li, I.T.S. Challenges and opportunities in exosome research-Perspectives from biology, engineering, and cancer therapy. APL Bioeng. 2019, 3, 011503. [Google Scholar] [CrossRef]
- Stepanov, G.; Zhuravlev, E.; Shender, V.; Nushtaeva, A.; Balakhonova, E.; Mozhaeva, E.; Kasakin, M.; Koval, V.; Lomzov, A.; Pavlyukov, M.; et al. Nucleotide Modifications Decrease Innate Immune Response Induced by Synthetic Analogs of snRNAs and snoRNAs. Genes 2018, 9, 531. [Google Scholar] [CrossRef]
- Denzler, R.; McGeary, S.E.; Title, A.C.; Agarwal, V.; Bartel, D.P.; Stoffel, M. Impact of MicroRNA Levels, Target-Site Complementarity, and Cooperativity on Competing Endogenous RNA-Regulated Gene Expression. Mol. Cell 2016, 64, 565–579. [Google Scholar] [CrossRef]
- Abd-Aziz, N.; Kamaruzman, N.I.; Poh, C.L. Development of MicroRNAs as Potential Therapeutics against Cancer. J. Oncol. 2020, 2020, 8029721. [Google Scholar] [CrossRef]
- Bost, J.P.; Barriga, H.; Holme, M.N.; Gallud, A.; Maugeri, M.; Gupta, D.; Lehto, T.; Valadi, H.; Esbjorner, E.K.; Stevens, M.M.; et al. Delivery of Oligonucleotide Therapeutics: Chemical Modifications, Lipid Nanoparticles, and Extracellular Vesicles. ACS Nano 2021, 15, 13993–14021. [Google Scholar] [CrossRef]
- Inoue, J.; Fujiwara, K.; Hamamoto, H.; Kobayashi, K.; Inazawa, J. Improving the Efficacy of EGFR Inhibitors by Topical Treatment of Cutaneous Squamous Cell Carcinoma with miR-634 Ointment. Mol. Ther. Oncolytics 2020, 19, 294–307. [Google Scholar] [CrossRef] [PubMed]
- Romano, G.; Acunzo, M.; Nana-Sinkam, P. microRNAs as Novel Therapeutics in Cancer. Cancers 2021, 13, 1526. [Google Scholar] [CrossRef]
- Lennox, K.A.; Behlke, M.A. A direct comparison of anti-microRNA oligonucleotide potency. Pharm. Res. 2010, 27, 1788–1799. [Google Scholar] [CrossRef] [PubMed]
- Moraes, F.C.; Pichon, C.; Letourneur, D.; Chaubet, F. miRNA Delivery by Nanosystems: State of the Art and Perspectives. Pharmaceutics 2021, 13, 1901. [Google Scholar] [CrossRef]
- Ma, S.C.; Zhang, J.Q.; Yan, T.H.; Miao, M.X.; Cao, Y.M.; Cao, Y.B.; Zhang, L.C.; Li, L. Novel strategies to reverse chemoresistance in colorectal cancer. Cancer Med. 2023, 12, 11073–11096. [Google Scholar] [CrossRef] [PubMed]
- Arghiani, N.; Shah, K. Modulating microRNAs in cancer: Next-generation therapies. Cancer Biol. Med. 2021, 19, 289–304. [Google Scholar] [CrossRef]
- Holjencin, C.; Jakymiw, A. MicroRNAs and Their Big Therapeutic Impacts: Delivery Strategies for Cancer Intervention. Cells 2022, 11, 2332. [Google Scholar] [CrossRef]
- Hosseinahli, N.; Aghapour, M.; Duijf, P.H.G.; Baradaran, B. Treating cancer with microRNA replacement therapy: A literature review. J. Cell. Physiol. 2018, 233, 5574–5588. [Google Scholar] [CrossRef]
- Dasgupta, I.; Chatterjee, A. Recent Advances in miRNA Delivery Systems. Methods Protoc. 2021, 4, 10. [Google Scholar] [CrossRef]
- Jayandharan, G. Gene and Cell Therapy: Biology and Applications; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar]
- Kortylewski, M.; Nechaev, S. How to train your dragon: Targeted delivery of microRNA to cancer cells in vivo. Mol. Ther. J. Am. Soc. Gene Ther. 2014, 22, 1070–1071. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bulcha, J.T.; Wang, Y.; Ma, H.; Tai, P.W.L.; Gao, G. Viral vector platforms within the gene therapy landscape. Signal Transduct. Target. Ther. 2021, 6, 53. [Google Scholar] [CrossRef] [PubMed]
- Ang, L.; Guo, L.; Wang, J.; Huang, J.; Lou, X.; Zhao, M. Oncolytic virotherapy armed with an engineered interfering lncRNA exhibits antitumor activity by blocking the epithelial mesenchymal transition in triple-negative breast cancer. Cancer Lett. 2020, 479, 42–53. [Google Scholar] [CrossRef] [PubMed]
- Bhere, D.; Arghiani, N.; Lechtich, E.R.; Yao, Y.; Alsaab, S.; Bei, F.; Matin, M.M.; Shah, K. Simultaneous downregulation of miR-21 and upregulation of miR-7 has anti-tumor efficacy. Sci. Rep. 2020, 10, 1779. [Google Scholar] [CrossRef]
- Kim, M.W.; Kwon, S.H.; Choi, J.H.; Lee, A. A Promising Biocompatible Platform: Lipid-Based and Bio-Inspired Smart Drug Delivery Systems for Cancer Therapy. Int. J. Mol. Sci. 2018, 19, 3859. [Google Scholar] [CrossRef]
- O’Neill, C.P.; Dwyer, R.M. Nanoparticle-Based Delivery of Tumor Suppressor microRNA for Cancer Therapy. Cells 2020, 9, 521. [Google Scholar] [CrossRef]
- Wang, H.; Liu, S.; Jia, L.; Chu, F.; Zhou, Y.; He, Z.; Guo, M.; Chen, C.; Xu, L. Nanostructured lipid carriers for MicroRNA delivery in tumor gene therapy. Cancer Cell Int. 2018, 18, 101. [Google Scholar] [CrossRef]
- Beg, M.S.; Brenner, A.J.; Sachdev, J.; Borad, M.; Kang, Y.K.; Stoudemire, J.; Smith, S.; Bader, A.G.; Kim, S.; Hong, D.S. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Investig. New Drugs 2017, 35, 180–188. [Google Scholar] [CrossRef]
- Hong, D.S.; Kang, Y.K.; Borad, M.; Sachdev, J.; Ejadi, S.; Lim, H.Y.; Brenner, A.J.; Park, K.; Lee, J.L.; Kim, T.Y.; et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br. J. Cancer 2020, 122, 1630–1637. [Google Scholar] [CrossRef]
- de Antonellis, P.; Liguori, L.; Falanga, A.; Carotenuto, M.; Ferrucci, V.; Andolfo, I.; Marinaro, F.; Scognamiglio, I.; Virgilio, A.; De Rosa, G.; et al. MicroRNA 199b-5p delivery through stable nucleic acid lipid particles (SNALPs) in tumorigenic cell lines. Naunyn Schmiedebergs Arch. Pharmacol. 2013, 386, 287–302. [Google Scholar] [CrossRef]
- van den Berg, A.I.S.; Yun, C.O.; Schiffelers, R.M.; Hennink, W.E. Polymeric delivery systems for nucleic acid therapeutics: Approaching the clinic. J. Control Release 2021, 331, 121–141. [Google Scholar] [CrossRef] [PubMed]
- Madkhali, O.; Mekhail, G.; Wettig, S.D. Modified gelatin nanoparticles for gene delivery. Int. J. Pharm. 2019, 554, 224–234. [Google Scholar] [CrossRef] [PubMed]
- Garg, U.; Chauhan, S.; Nagaich, U.; Jain, N. Current Advances in Chitosan Nanoparticles Based Drug Delivery and Targeting. Adv. Pharm. Bull. 2019, 9, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Wei, P.; Cornel, E.J.; Du, J. Ultrasound-responsive polymer-based drug delivery systems. Drug Deliv. Transl. Res. 2021, 11, 1323–1339. [Google Scholar] [CrossRef]
- McClorey, G.; Banerjee, S. Cell-Penetrating Peptides to Enhance Delivery of Oligonucleotide-Based Therapeutics. Biomedicines 2018, 6, 51. [Google Scholar] [CrossRef]
- Silva, S.; Almeida, A.J.; Vale, N. Combination of Cell-Penetrating Peptides with Nanoparticles for Therapeutic Application: A Review. Biomolecules 2019, 9, 22. [Google Scholar] [CrossRef]
- Hobel, S.; Aigner, A. Polyethylenimines for siRNA and miRNA delivery in vivo. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnolgy 2013, 5, 484–501. [Google Scholar] [CrossRef]
- Lim, C.C.; Chia, L.Y.; Kumar, P.V. Dendrimer-based nanocomposites for the production of RNA delivery systems. OpenNano 2023, 13, 100173. [Google Scholar] [CrossRef]
- Berger, A.G.; Chou, J.J.; Hammond, P.T. Approaches to Modulate the Chronic Wound Environment Using Localized Nucleic Acid Delivery. Adv. Wound Care New Rochelle 2021, 10, 503–528. [Google Scholar] [CrossRef]
- Mintzer, M.A.; Grinstaff, M.W. Biomedical applications of dendrimers: A tutorial. Chem. Soc. Rev. 2011, 40, 173–190. [Google Scholar] [CrossRef]
- Dzmitruk, V.; Apartsin, E.; Ihnatsyeu-Kachan, A.; Abashkin, V.; Shcharbin, D.; Bryszewska, M. Dendrimers Show Promise for siRNA and microRNA Therapeutics. Pharmaceutics 2018, 10, 126. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Zhong, W.; Cao, X.; Pan, G.; Xu, M.; Zheng, J.; Chen, H.; Feng, X.; Luo, C.; Lu, C.; et al. Loss of miR-31-5p drives hematopoietic stem cell malignant transformation and restoration eliminates leukemia stem cells in mice. Sci. Transl. Med. 2022, 14, eabh2548. [Google Scholar] [CrossRef]
- Pedziwiatr-Werbicka, E.; Gorzkiewicz, M.; Horodecka, K.; Abashkin, V.; Klajnert-Maculewicz, B.; Pena-Gonzalez, C.E.; Sanchez-Nieves, J.; Gomez, R.; de la Mata, F.J.; Bryszewska, M. Silver Nanoparticles Surface-Modified with Carbosilane Dendrons as Carriers of Anticancer siRNA. Int. J. Mol. Sci. 2020, 21, 4647. [Google Scholar] [CrossRef] [PubMed]
- Zenze, M.; Daniels, A.; Singh, M. Dendrimers as Modifiers of Inorganic Nanoparticles for Therapeutic Delivery in Cancer. Pharmaceutics 2023, 15, 398. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Song, C.; Li, G.; Guo, Y.; Wang, Z.; Guo, H.; Xia, J.; Tao, Y.; Shi, Q.; Shi, X.; et al. Ultrasound-enhanced theranostics of orthotopic breast cancer through a multifunctional core-shell tecto dendrimer-based nanomedicine platform. Biomater. Sci. 2023, 11, 4385–4396. [Google Scholar] [CrossRef]
- Marcinkowska, M.; Sobierajska, E.; Stanczyk, M.; Janaszewska, A.; Chworos, A.; Klajnert-Maculewicz, B. Conjugate of PAMAM Dendrimer, Doxorubicin and Monoclonal Antibody-Trastuzumab: The New Approach of a Well-Known Strategy. Polymers 2018, 10, 187. [Google Scholar] [CrossRef]
- Maghsoudnia, N.; Eftekhari, R.B.; Sohi, A.N.; Dorkoosh, F.A. Chloroquine Assisted Delivery of microRNA Mimic Let-7b to NSCLC Cell Line by PAMAM (G5)—HA Nano-Carrier. Curr. Drug Deliv. 2021, 18, 31–43. [Google Scholar] [CrossRef] [PubMed]
- Mir, M.; Ahmed, N.; Rehman, A.U. Recent applications of PLGA based nanostructures in drug delivery. Colloids Surf. B Biointerfaces 2017, 159, 217–231. [Google Scholar] [CrossRef] [PubMed]
- Sadeghipour, N.; Kumar, S.U.; Massoud, T.F.; Paulmurugan, R. A rationally identified panel of microRNAs targets multiple oncogenic pathways to enhance chemotherapeutic effects in glioblastoma models. Sci. Rep. 2022, 12, 12017. [Google Scholar] [CrossRef]
- Labatut, A.E.; Mattheolabakis, G. Non-viral based miR delivery and recent developments. Eur. J. Pharm. Biopharm. 2018, 128, 82–90. [Google Scholar] [CrossRef]
- Lu, M.; Xing, H.; Xun, Z.; Yang, T.; Ding, P.; Cai, C.; Wang, D.; Zhao, X. Exosome-based small RNA delivery: Progress and prospects. Asian J. Pharm. Sci. 2018, 13, 1–11. [Google Scholar] [CrossRef]
- Shimbo, K.; Miyaki, S.; Ishitobi, H.; Kato, Y.; Kubo, T.; Shimose, S.; Ochi, M. Exosome-formed synthetic microRNA-143 is transferred to osteosarcoma cells and inhibits their migration. Biochem. Biophys. Res. Commun. 2014, 445, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Bonneau, E.; Neveu, B.; Kostantin, E.; Tsongalis, G.J.; De Guire, V. How close are miRNAs from clinical practice? A perspective on the diagnostic and therapeutic market. EJIFCC 2019, 30, 114–127. [Google Scholar]
- Chakraborty, C.; Sharma, A.R.; Sharma, G.; Lee, S.S. Therapeutic advances of miRNAs: A preclinical and clinical update. J. Adv. Res. 2021, 28, 127–138. [Google Scholar] [CrossRef]
- Seto, A.G.; Beatty, X.; Lynch, J.M.; Hermreck, M.; Tetzlaff, M.; Duvic, M.; Jackson, A.L. Cobomarsen, an oligonucleotide inhibitor of miR-155, co-ordinately regulates multiple survival pathways to reduce cellular proliferation and survival in cutaneous T-cell lymphoma. Br. J. Haematol. 2018, 183, 428–444. [Google Scholar] [CrossRef]
- Anastasiadou, E.; Seto, A.G.; Beatty, X.; Hermreck, M.; Gilles, M.E.; Stroopinsky, D.; Pinter-Brown, L.C.; Pestano, L.; Marchese, C.; Avigan, D.; et al. Cobomarsen, an Oligonucleotide Inhibitor of miR-155, Slows DLBCL Tumor Cell Growth In Vitro and In Vivo. Clin. Cancer Res. 2021, 27, 1139–1149. [Google Scholar] [CrossRef] [PubMed]
- Witten, L.; Slack, F.J. miR-155 as a novel clinical target for hematological malignancies. Carcinogenesis 2020, 41, 2–7. [Google Scholar] [CrossRef] [PubMed]
- Querfeld, C.; Pacheco, T.; Foss, F.M.; Halwani, A.S.; Porcu, P.; Seto, A.G.; Ruckman, J.; Landry, M.L.; Jackson, A.L.; Pestano, L.A.; et al. Preliminary Results of a Phase 1 Trial Evaluating MRG-106, a Synthetic microRNA Antagonist (LNA antimiR) of microRNA-155, in Patients with CTCL. Blood 2016, 128, 1829. [Google Scholar] [CrossRef]
- Reid, G.; Kao, S.C.; Pavlakis, N.; Brahmbhatt, H.; MacDiarmid, J.; Clarke, S.; Boyer, M.; van Zandwijk, N. Clinical development of TargomiRs, a miRNA mimic-based treatment for patients with recurrent thoracic cancer. Epigenomics 2016, 8, 1079–1085. [Google Scholar] [CrossRef]
- van Zandwijk, N.; Pavlakis, N.; Kao, S.C.; Linton, A.; Boyer, M.J.; Clarke, S.; Huynh, Y.; Chrzanowska, A.; Fulham, M.J.; Bailey, D.L.; et al. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: A first-in-man, phase 1, open-label, dose-escalation study. Lancet. Oncol. 2017, 18, 1386–1396. [Google Scholar] [CrossRef]
- Reid, G.; Pel, M.E.; Kirschner, M.B.; Cheng, Y.Y.; Mugridge, N.; Weiss, J.; Williams, M.; Wright, C.; Edelman, J.J.; Vallely, M.P.; et al. Restoring expression of miR-16: A novel approach to therapy for malignant pleural mesothelioma. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2013, 24, 3128–3135. [Google Scholar] [CrossRef]
- Calin, G.A.; Cimmino, A.; Fabbri, M.; Ferracin, M.; Wojcik, S.E.; Shimizu, M.; Taccioli, C.; Zanesi, N.; Garzon, R.; Aqeilan, R.I.; et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc. Natl. Acad. Sci. USA. 2008, 105, 5166–5171. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Ashraf, M.U.; Kumar, A.; Bae, Y.S. Therapeutic Potential of microRNA Against Th2-associated Immune Disorders. Curr. Top. Med. Chem. 2021, 21, 753–766. [Google Scholar] [CrossRef] [PubMed]
- Kasina, V.; Wahane, A.; Liu, C.H.; Yang, L.; Nieh, M.P.; Slack, F.J.; Bahal, R. Next-generation poly-L-histidine formulations for miRNA mimic delivery. Mol. Ther. Methods Clin. Dev. 2023, 29, 271–283. [Google Scholar] [CrossRef]
- Fariha, A.; Hami, I.; Tonmoy, M.I.Q.; Akter, S.; Al Reza, H.; Bahadur, N.M.; Rahaman, M.M.; Hossain, M.S. Cell cycle associated miRNAs as target and therapeutics in lung cancer treatment. Heliyon 2022, 8, e11081. [Google Scholar] [CrossRef]
- Bouchie, A. First microRNA mimic enters clinic. Nat. Biotechnol. 2013, 31, 577. [Google Scholar] [CrossRef]
- Lithwick-Yanai, G.; Dromi, N.; Shtabsky, A.; Morgenstern, S.; Strenov, Y.; Feinmesser, M.; Kravtsov, V.; Leon, M.E.; Hajduch, M.; Ali, S.Z.; et al. Multicentre validation of a microRNA-based assay for diagnosing indeterminate thyroid nodules utilising fine needle aspirate smears. J. Clin. Pathol. 2017, 70, 500–507. [Google Scholar] [CrossRef]
- Chakraborty, C.; Sharma, A.R.; Sharma, G.; Doss, C.G.P.; Lee, S.S. Therapeutic miRNA and siRNA: Moving from Bench to Clinic as Next Generation Medicine. Mol. Ther. Nucleic Acids 2017, 8, 132–143. [Google Scholar] [CrossRef]
- Kohler, B.; Dubovik, S.; Horterer, E.; Wilk, U.; Stockl, J.B.; Tekarslan-Sahin, H.; Ljepoja, B.; Paulitschke, P.; Frohlich, T.; Wagner, E.; et al. Combating Drug Resistance by Exploiting miRNA-200c-Controlled Phase II Detoxification. Cancers 2022, 14, 5554. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A. MicroRNAs and cancer: What we know and what we still have to learn. Genome Med. 2009, 1, 78. [Google Scholar] [CrossRef]
- Available online: https://www.prnewswire.com/news-releases/sirnaomics-sirna-therapeutic-candidate-stp705-granted-orphan-drug-designation-by-us-fda-for-treatment-of-hepatocellular-carcinoma-301002347.html (accessed on 24 June 2022).
- Kobeissi, I.; Eljilany, I.; Achkar, T.; LaFramboise, W.A.; Santana-Santos, L.; Tarhini, A.A. A Tumor and Immune-Related Micro-RNA Signature Predicts Relapse-Free Survival of Melanoma Patients Treated with Ipilimumab. Int. J. Mol. Sci. 2023, 24, 8167. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Li, D.; Leng, S.; Zhu, X. RNA-based pharmacotherapy for tumors: From bench to clinic and back. Biomed. Pharmacother. 2020, 125, 109997. [Google Scholar] [CrossRef]
- Si, W.; Shen, J.; Zheng, H.; Fan, W. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin. Epigenetics 2019, 11, 25. [Google Scholar] [CrossRef] [PubMed]
- Giovannetti, E.; Erozenci, A.; Smit, J.; Danesi, R.; Peters, G.J. Molecular mechanisms underlying the role of microRNAs (miRNAs) in anticancer drug resistance and implications for clinical practice. Crit. Rev. Oncol./Hematol. 2012, 81, 103–122. [Google Scholar] [CrossRef] [PubMed]
- To, K.K. MicroRNA: A prognostic biomarker and a possible druggable target for circumventing multidrug resistance in cancer chemotherapy. J. Biomed. Sci. 2013, 20, 99. [Google Scholar] [CrossRef]
- Meng, F.; Henson, R.; Lang, M.; Wehbe, H.; Maheshwari, S.; Mendell, J.T.; Jiang, J.; Schmittgen, T.D.; Patel, T. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology 2006, 130, 2113–2129. [Google Scholar] [CrossRef]
- Munoz, J.L.; Bliss, S.A.; Greco, S.J.; Ramkissoon, S.H.; Ligon, K.L.; Rameshwar, P. Delivery of Functional Anti-miR-9 by Mesenchymal Stem Cell-derived Exosomes to Glioblastoma Multiforme Cells Conferred Chemosensitivity. Mol. Ther. Nucleic Acids 2013, 2, e126. [Google Scholar] [CrossRef]
- Li, Y.; Li, L.; Guan, Y.; Liu, X.; Meng, Q.; Guo, Q. MiR-92b regulates the cell growth, cisplatin chemosensitivity of A549 non small cell lung cancer cell line and target PTEN. Biochem. Biophys. Res. Commun. 2013, 440, 604–610. [Google Scholar] [CrossRef]
- Pedroza-Torres, A.; Romero-Cordoba, S.L.; Justo-Garrido, M.; Salido-Guadarrama, I.; Rodriguez-Bautista, R.; Montano, S.; Muniz-Mendoza, R.; Arriaga-Canon, C.; Fragoso-Ontiveros, V.; Alvarez-Gomez, R.M.; et al. MicroRNAs in Tumor Cell Metabolism: Roles and Therapeutic Opportunities. Front. Oncol. 2019, 9, 1404. [Google Scholar] [CrossRef]
- Subramaniam, S.; Jeet, V.; Clements, J.A.; Gunter, J.H.; Batra, J. Emergence of MicroRNAs as Key Players in Cancer Cell Metabolism. Clin. Chem. 2019, 65, 1090–1101. [Google Scholar] [CrossRef]
- Jiang, S.; Zhang, L.F.; Zhang, H.W.; Hu, S.; Lu, M.H.; Liang, S.; Li, B.; Li, Y.; Li, D.; Wang, E.D.; et al. A novel miR-155/miR-143 cascade controls glycolysis by regulating hexokinase 2 in breast cancer cells. EMBO J. 2012, 31, 1985–1998. [Google Scholar] [CrossRef] [PubMed]
- Sikand, K.; Singh, J.; Ebron, J.S.; Shukla, G.C. Housekeeping gene selection advisory: Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and beta-actin are targets of miR-644a. PLoS ONE 2012, 7, e47510. [Google Scholar] [CrossRef] [PubMed]
- Khanna, M.; Saini, S.; Shariff, M.; Ronsard, L.; Singh, J.K.; Kumar, H. Data highlighting miR-155 and GAPDH correlation. Data Brief. 2019, 24, 103945. [Google Scholar] [CrossRef]
- Qu, W.; Ding, S.M.; Cao, G.; Wang, S.J.; Zheng, X.H.; Li, G.H. miR-132 mediates a metabolic shift in prostate cancer cells by targeting Glut1. FEBS Open Bio 2016, 6, 735–741. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Gao, J.; Huang, Q.; Jin, Y.; Wei, Z. Downregulating microRNA-144 mediates a metabolic shift in lung cancer cells by regulating GLUT1 expression. Oncol. Lett. 2016, 11, 3772–3776. [Google Scholar] [CrossRef]
- Kinoshita, T.; Nohata, N.; Yoshino, H.; Hanazawa, T.; Kikkawa, N.; Fujimura, L.; Chiyomaru, T.; Kawakami, K.; Enokida, H.; Nakagawa, M.; et al. Tumor suppressive microRNA-375 regulates lactate dehydrogenase B in maxillary sinus squamous cell carcinoma. Int. J. Oncol. 2012, 40, 185–193. [Google Scholar] [CrossRef]
- Xiao, X.; Huang, X.; Ye, F.; Chen, B.; Song, C.; Wen, J.; Zhang, Z.; Zheng, G.; Tang, H.; Xie, X. The miR-34a-LDHA axis regulates glucose metabolism and tumor growth in breast cancer. Sci. Rep. 2016, 6, 21735. [Google Scholar] [CrossRef]
- Chen, H.; Gao, S.; Cheng, C. MiR-323a-3p suppressed the glycolysis of osteosarcoma via targeting LDHA. Hum. Cell 2018, 31, 300–309. [Google Scholar] [CrossRef]
- He, Y.; Chen, X.; Yu, Y.; Li, J.; Hu, Q.; Xue, C.; Chen, J.; Shen, S.; Luo, Y.; Ren, F.; et al. LDHA is a direct target of miR-30d-5p and contributes to aggressive progression of gallbladder carcinoma. Mol. Carcinog. 2018, 57, 772–783. [Google Scholar] [CrossRef]
- Pullen, T.J.; da Silva Xavier, G.; Kelsey, G.; Rutter, G.A. miR-29a and miR-29b contribute to pancreatic beta-cell-specific silencing of monocarboxylate transporter 1 (Mct1). Mol. Cell. Biol. 2011, 31, 3182–3194. [Google Scholar] [CrossRef]
- Romero-Cordoba, S.L.; Rodriguez-Cuevas, S.; Bautista-Pina, V.; Maffuz-Aziz, A.; D’Ippolito, E.; Cosentino, G.; Baroni, S.; Iorio, M.V.; Hidalgo-Miranda, A. Loss of function of miR-342-3p results in MCT1 over-expression and contributes to oncogenic metabolic reprogramming in triple negative breast cancer. Sci. Rep. 2018, 8, 12252. [Google Scholar] [CrossRef]
- Dong, J.; Xiao, D.; Zhao, Z.; Ren, P.; Li, C.; Hu, Y.; Shi, J.; Su, H.; Wang, L.; Liu, H.; et al. Epigenetic silencing of microRNA-137 enhances ASCT2 expression and tumor glutamine metabolism. Oncogenesis 2017, 6, e356. [Google Scholar] [CrossRef] [PubMed]
- Anderton, B.; Camarda, R.; Balakrishnan, S.; Balakrishnan, A.; Kohnz, R.A.; Lim, L.; Evason, K.J.; Momcilovic, O.; Kruttwig, K.; Huang, Q.; et al. MYC-driven inhibition of the glutamate-cysteine ligase promotes glutathione depletion in liver cancer. EMBO Rep. 2017, 18, 569–585. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Wang, H.; Chen, J.; Li, Z.; Li, S.; Hu, Z.; Huang, S.; Zhao, Y.; He, X. MicroRNA-129-5p Regulates Glycolysis and Cell Proliferation by Targeting the Glucose Transporter SLC2A3 in Gastric Cancer Cells. Front. Pharmacol. 2018, 9, 502. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Qiu, Z.; Wang, Z.; Wang, Q.; Tan, N.; Chen, T.; Chen, Z.; Huang, S.; Gu, J.; Li, J.; et al. MiR-199a-5p is negatively associated with malignancies and regulates glycolysis and lactate production by targeting hexokinase 2 in liver cancer. Hepatology 2015, 62, 1132–1144. [Google Scholar] [CrossRef]
- Yang, Y.; Ishak Gabra, M.B.; Hanse, E.A.; Lowman, X.H.; Tran, T.Q.; Li, H.; Milman, N.; Liu, J.; Reid, M.A.; Locasale, J.W.; et al. MiR-135 suppresses glycolysis and promotes pancreatic cancer cell adaptation to metabolic stress by targeting phosphofructokinase-1. Nat. Commun. 2019, 10, 809. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, Z.; Xuan, Z.; Xu, P.; Wang, W.; Chen, Z.; Wang, S.; Sun, G.; Xu, J.; Xu, Z. Novel role of miR-133a-3p in repressing gastric cancer growth and metastasis via blocking autophagy-mediated glutaminolysis. J. Exp. Clin. Cancer Res. CR 2018, 37, 320. [Google Scholar] [CrossRef]
- Esau, C.; Davis, S.; Murray, S.F.; Yu, X.X.; Pandey, S.K.; Pear, M.; Watts, L.; Booten, S.L.; Graham, M.; McKay, R.; et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006, 3, 87–98. [Google Scholar] [CrossRef]
- Schiliro, C.; Firestein, B.L. Mechanisms of Metabolic Reprogramming in Cancer Cells Supporting Enhanced Growth and Proliferation. Cells 2021, 10, 1056. [Google Scholar] [CrossRef]
- Moldogazieva, N.T.; Mokhosoev, I.M.; Terentiev, A.A. Metabolic Heterogeneity of Cancer Cells: An Interplay between HIF-1, GLUTs, and AMPK. Cancers 2020, 12, 862. [Google Scholar] [CrossRef]
- Li, W.; Hao, J.; Zhang, L.; Cheng, Z.; Deng, X.; Shu, G. Astragalin Reduces Hexokinase 2 through Increasing miR-125b to Inhibit the Proliferation of Hepatocellular Carcinoma Cells in Vitro and in Vivo. J. Agric. Food Chem. 2017, 65, 5961–5972. [Google Scholar] [CrossRef] [PubMed]
- Nosengo, N. Can you teach old drugs new tricks? Nature 2016, 534, 314–316. [Google Scholar] [CrossRef]
- Pantziarka, P.; Bouche, G.; Meheus, L.; Sukhatme, V.; Sukhatme, V.P.; Vikas, P. The Repurposing Drugs in Oncology (ReDO) Project. Ecancermedicalscience 2014, 8, 442. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Raese, R.A.; Luo, D.; Feng, J.; Xin, W.; Dong, C.; Qian, Y.; Guo, N.L. MicroRNA-Based Discovery of Biomarkers, Therapeutic Targets, and Repositioning Drugs for Breast Cancer. Cells 2023, 12, 1917. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.M.; Donnelly, L.A.; Emslie-Smith, A.M.; Alessi, D.R.; Morris, A.D. Metformin and reduced risk of cancer in diabetic patients. Bmj 2005, 330, 1304–1305. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, T.I.; Jeon, S.M.; Hong, S.P.; Cheon, J.H.; Kim, W.H. The effects of metformin on the survival of colorectal cancer patients with diabetes mellitus. Int. J. Cancer 2012, 131, 752–759. [Google Scholar] [CrossRef]
- Noto, H.; Goto, A.; Tsujimoto, T.; Noda, M. Cancer risk in diabetic patients treated with metformin: A systematic review and meta-analysis. PLoS ONE 2012, 7, e33411. [Google Scholar] [CrossRef]
- Gandini, S.; Puntoni, M.; Heckman-Stoddard, B.M.; Dunn, B.K.; Ford, L.; DeCensi, A.; Szabo, E. Metformin and cancer risk and mortality: A systematic review and meta-analysis taking into account biases and confounders. Cancer Prev. Res. 2014, 7, 867–885. [Google Scholar] [CrossRef]
- Shaw, R.J.; Kosmatka, M.; Bardeesy, N.; Hurley, R.L.; Witters, L.A.; DePinho, R.A.; Cantley, L.C. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc. Natl. Acad. Sci. USA 2004, 101, 3329–3335. [Google Scholar] [CrossRef]
- Oliveras-Ferraros, C.; Cufi, S.; Vazquez-Martin, A.; Torres-Garcia, V.Z.; Del Barco, S.; Martin-Castillo, B.; Menendez, J.A. Micro(mi)RNA expression profile of breast cancer epithelial cells treated with the anti-diabetic drug metformin: Induction of the tumor suppressor miRNA let-7a and suppression of the TGFbeta-induced oncomiR miRNA-181a. Cell Cycle 2011, 10, 1144–1151. [Google Scholar] [CrossRef]
- Sharma, P.; Singh, S. Combinatorial Effect of DCA and Let-7a on Triple-Negative MDA-MB-231 Cells: A Metabolic Approach of Treatment. Integr. Cancer Ther. 2020, 19, 1534735420911437. [Google Scholar] [CrossRef]
- Istvan, E.S.; Deisenhofer, J. Structural mechanism for statin inhibition of HMG-CoA reductase. Science 2001, 292, 1160–1164. [Google Scholar] [CrossRef]
- Wiley, J.J.; Baxter, M.P. Tibial spine fractures in children. Clin. Orthop. Relat. Res. 1990, 255, 54–60. [Google Scholar] [CrossRef]
- Lin, J.J.; Ezer, N.; Sigel, K.; Mhango, G.; Wisnivesky, J.P. The effect of statins on survival in patients with stage IV lung cancer. Lung Cancer 2016, 99, 137–142. [Google Scholar] [CrossRef]
- Hung, M.S.; Chen, I.C.; Lee, C.P.; Huang, R.J.; Chen, P.C.; Tsai, Y.H.; Yang, Y.H. Statin improves survival in patients with EGFR-TKI lung cancer: A nationwide population-based study. PLoS ONE 2017, 12, e0171137. [Google Scholar] [CrossRef]
- Tilija Pun, N.; Jeong, C.H. Statin as a Potential Chemotherapeutic Agent: Current Updates as a Monotherapy, Combination Therapy, and Treatment for Anti-Cancer Drug Resistance. Pharmaceuticals 2021, 14, 470. [Google Scholar] [CrossRef] [PubMed]
- Beckwitt, C.H.; Brufsky, A.; Oltvai, Z.N.; Wells, A. Statin drugs to reduce breast cancer recurrence and mortality. Breast Cancer Res. BCR 2018, 20, 144. [Google Scholar] [CrossRef] [PubMed]
- Karlic, H.; Thaler, R.; Gerner, C.; Grunt, T.; Proestling, K.; Haider, F.; Varga, F. Inhibition of the mevalonate pathway affects epigenetic regulation in cancer cells. Cancer Genet. 2015, 208, 241–252. [Google Scholar] [CrossRef]
- Hundal, R.S.; Petersen, K.F.; Mayerson, A.B.; Randhawa, P.S.; Inzucchi, S.; Shoelson, S.E.; Shulman, G.I. Mechanism by which high-dose aspirin improves glucose metabolism in type 2 diabetes. J. Clin. Investig. 2002, 109, 1321–1326. [Google Scholar] [CrossRef]
- Paez Espinosa, E.V.; Murad, J.P.; Khasawneh, F.T. Aspirin: Pharmacology and clinical applications. Thrombosis 2012, 2012, 173124. [Google Scholar] [CrossRef]
- Barnard, M.E.; Poole, E.M.; Curhan, G.C.; Eliassen, A.H.; Rosner, B.A.; Terry, K.L.; Tworoger, S.S. Association of Analgesic Use With Risk of Ovarian Cancer in the Nurses’ Health Studies. JAMA Oncol. 2018, 4, 1675–1682. [Google Scholar] [CrossRef]
- Simon, T.G.; Ma, Y.; Ludvigsson, J.F.; Chong, D.Q.; Giovannucci, E.L.; Fuchs, C.S.; Meyerhardt, J.A.; Corey, K.E.; Chung, R.T.; Zhang, X.; et al. Association Between Aspirin Use and Risk of Hepatocellular Carcinoma. JAMA Oncol. 2018, 4, 1683–1690. [Google Scholar] [CrossRef]
- Jiang, M.J.; Chen, Y.Y.; Dai, J.J.; Gu, D.N.; Mei, Z.; Liu, F.R.; Huang, Q.; Tian, L. Dying tumor cell-derived exosomal miR-194-5p potentiates survival and repopulation of tumor repopulating cells upon radiotherapy in pancreatic cancer. Mol. Cancer 2020, 19, 68. [Google Scholar] [CrossRef]
- Gan, H.; Lin, L.; Hu, N.; Yang, Y.; Gao, Y.; Pei, Y.; Chen, K.; Sun, B. Aspirin ameliorates lung cancer by targeting the miR-98/WNT1 axis. Thorac. Cancer 2019, 10, 744–750. [Google Scholar] [CrossRef]
- Watari, J.; Ito, C.; Shimoda, T.; Tomita, T.; Oshima, T.; Fukui, H.; Das, K.M.; Miwa, H. DNA methylation silencing of microRNA gene methylator in the precancerous background mucosa with and without gastric cancer: Analysis of the effects of H. pylori eradication and long-term aspirin use. Sci. Rep. 2019, 9, 12559. [Google Scholar] [CrossRef]
- Yiannakopoulou, E. Targeting epigenetic mechanisms and microRNAs by aspirin and other non steroidal anti-inflammatory agents--implications for cancer treatment and chemoprevention. Cell. Oncol. 2014, 37, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Bazavar, M.; Fazli, J.; Valizadeh, A.; Ma, B.; Mohammadi, E.; Asemi, Z.; Alemi, F.; Maleki, M.; Xing, S.; Yousefi, B. miR-192 enhances sensitivity of methotrexate drug to MG-63 osteosarcoma cancer cells. Pathol. Res. Pract. 2020, 216, 153176. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Danesh, F.R. Promises and challenges of miRNA therapeutics. Am. J. Physiol. Ren. Physiol. 2022, 323, F673–F674. [Google Scholar] [CrossRef] [PubMed]
- Lam, J.K.; Chow, M.Y.; Zhang, Y.; Leung, S.W. siRNA Versus miRNA as Therapeutics for Gene Silencing. Mol. Ther. Nucleic Acids 2015, 4, e252. [Google Scholar] [CrossRef]
- Bumcrot, D.; Manoharan, M.; Koteliansky, V.; Sah, D.W. RNAi therapeutics: A potential new class of pharmaceutical drugs. Nat. Chem. Biol. 2006, 2, 711–719. [Google Scholar] [CrossRef]
- Chakraborty, C.; Wen, Z.H.; Agoramoorthy, G.; Lin, C.S. Therapeutic microRNA Delivery Strategies with Special Emphasis on Cancer Therapy and Tumorigenesis: Current Trends and Future Challenges. Curr. Drug Metab. 2016, 17, 469–477. [Google Scholar] [CrossRef] [PubMed]
- Saraniti, C.; Speciale, R.; Santangelo, M.; Massaro, N.; Maniaci, A.; Gallina, S.; Serra, A.; Cocuzza, S. Functional outcomes after supracricoid modified partial laryngectomy. J. Biol. Regul. Homeost. Agents 2019, 33, 1903–1907. [Google Scholar] [CrossRef]
- Orellana, E.A.; Abdelaal, A.M.; Rangasamy, L.; Tenneti, S.; Myoung, S.; Low, P.S.; Kasinski, A.L. Enhancing MicroRNA Activity through Increased Endosomal Release Mediated by Nigericin. Mol. Ther. Nucleic Acids 2019, 16, 505–518. [Google Scholar] [CrossRef] [PubMed]
- Milone, M.C.; O’Doherty, U. Clinical use of lentiviral vectors. Leukemia 2018, 32, 1529–1541. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tan, M.; Wang, Y.; Liu, X.; Lin, A. Advances in modification and delivery of nucleic acid drugs. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2023, 52, 417–428. [Google Scholar] [CrossRef]
- Gareev, I.; de Jesus Encarnacion Ramirez, M.; Goncharov, E.; Ivliev, D.; Shumadalova, A.; Ilyasova, T.; Wang, C. MiRNAs and lncRNAs in the regulation of innate immune signaling. Noncoding RNA Res. 2023, 8, 534–541. [Google Scholar] [CrossRef]
| Method of Delivery | Characteristics |
|---|---|
| anti-miRNA ASOs (AMOs) | complementary sequence to endogenous miRNA |
| Modified AMOs with 2′-O-methyl-group (OMe) | Increase binding affinity and nuclease resistance |
| Modified AMOs with 2′ methoxyethyl (MOE) | More stable and specific |
| Modified AMOs with locked nucleic acid (LNA) | High binding affinity and low toxicity |
| miRNA sponges | Inhibit a whole family of associated miRNAs |
| miRNA masking | Gene-specific |
| Biotech Company | Experimental Product | Target miRNA | Pathologic Condition | Clinical Phase |
|---|---|---|---|---|
| MiRagen Therapeutics | MRG-106 | miR-155 | Lymphoma and Leukemia | I, II |
| ENGeneIC | Mesomir | miR-16 | Mesothelioma | III |
| Synlogic | PLGA-poly-L-His NPs | miR-34a | Advanced solid tumors | Pre-Clinical |
| Asbestos Diseases ResearchFoundation | TargomiRs | miR-16 | Malignant Pleural Mesothelioma, NS-CLC | I |
| Alnylam Pharmaceuticals | Screening | |||
| Interna Technologies | Screening | |||
| Mello Biotech | Screening | |||
| Opko | Screening |
| Small Molecules | Proteins and Antibodies | miRNA Therapeutics | |
|---|---|---|---|
| Nature of action | Activation or Inhibition of target | Activation or Inhibition of targets | Inhibition of targets |
| Site of target proteins | Extracellular and intracellular targets | Extracellular targets | All targets including non-druggable targets |
| Selectivity and potency | Variable, depending on binding site and ligand specificity their affinity efficacy | Highly selective and potent | Highly selective and potent |
| Lead optimization | Lead ID and optimization slow | Lead ID and optimization slow | Rapid lead ID and optimization |
| Manufacture | Easy to synthesize | Difficult to produce | Easy to synthesize |
| Stability | Stable | Unstable | Stable |
| Delivery | Easy | Difficult | Difficult |
| Safety/toxicity | Risk of off-target effects | Risk of immunogenicity | Risk of immunogenicity |
| Viral-Based Delivery Vectors | Advantages | Disadvantages |
|---|---|---|
| Retroviral vectors | Stability of transgene expression | Propensity for carcinogenesis due to insertional mutagenesis Unable to transduce not dividing cells. |
| Lentiviral vectors (LVs) | Stability of transgene expression Can transduce both dividing and nondividing cells | Lower risk of insertional mutagenesis and oncogenesis |
| Adenoviral (AdVs) and Adeno-associated vectors (AAVs) | Low immunogenicity High transduction efficiency in a variety of cells | Packaging capacity low Expensive manufacturing |
| Lipid-based delivery | Non-Immunogenic Biocompatible Easy production | Cytotoxicity |
| Polymeric deliver | High packaging capacity | Cytotoxicity Nonspecific delivery |
| Inorganic | Non-Immunogenic Biocompatible Easy production Nontoxic Control of physical features | Low efficacy |
| Exosome based delivery | Biocompatible Non immunogenic Tissue organ-specific delivery | Lack of strong experimental evidence/data |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pagoni, M.; Cava, C.; Sideris, D.C.; Avgeris, M.; Zoumpourlis, V.; Michalopoulos, I.; Drakoulis, N. miRNA-Based Technologies in Cancer Therapy. J. Pers. Med. 2023, 13, 1586. https://doi.org/10.3390/jpm13111586
Pagoni M, Cava C, Sideris DC, Avgeris M, Zoumpourlis V, Michalopoulos I, Drakoulis N. miRNA-Based Technologies in Cancer Therapy. Journal of Personalized Medicine. 2023; 13(11):1586. https://doi.org/10.3390/jpm13111586
Chicago/Turabian StylePagoni, Maria, Claudia Cava, Diamantis C. Sideris, Margaritis Avgeris, Vassilios Zoumpourlis, Ioannis Michalopoulos, and Nikolaos Drakoulis. 2023. "miRNA-Based Technologies in Cancer Therapy" Journal of Personalized Medicine 13, no. 11: 1586. https://doi.org/10.3390/jpm13111586
APA StylePagoni, M., Cava, C., Sideris, D. C., Avgeris, M., Zoumpourlis, V., Michalopoulos, I., & Drakoulis, N. (2023). miRNA-Based Technologies in Cancer Therapy. Journal of Personalized Medicine, 13(11), 1586. https://doi.org/10.3390/jpm13111586









