Patient-Level Exposure to Actionable Pharmacogenomic Medications in a Nationally Representative Insurance Claims Database
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Source and Setting
2.2. Study Design and Population
2.3. Measures
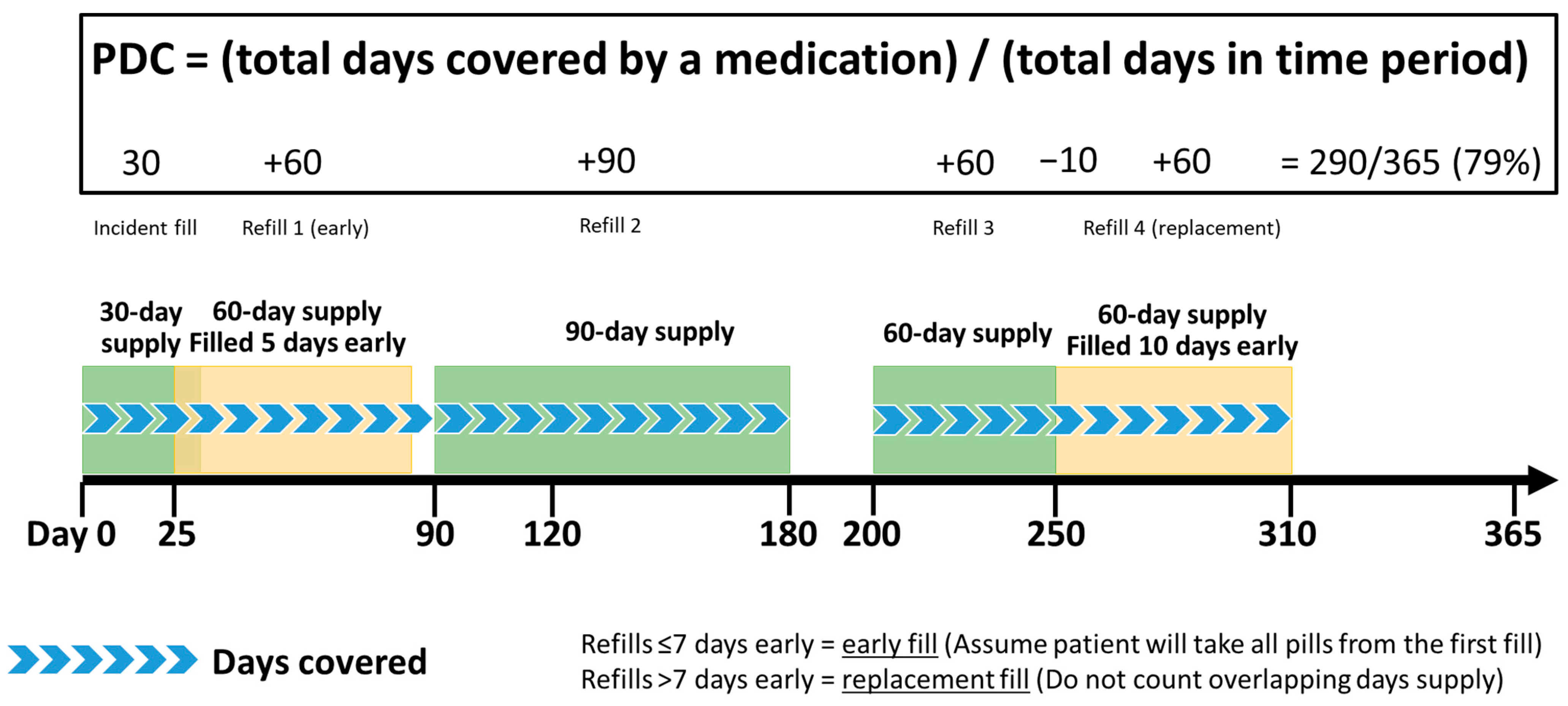
2.3.1. Amount of Exposure to Pharmacogenomic Medications
2.3.2. Clinical and Demographic Measures
2.4. Missing Data
2.5. Statistical Analysis
3. Results
3.1. Cohort Characteristics
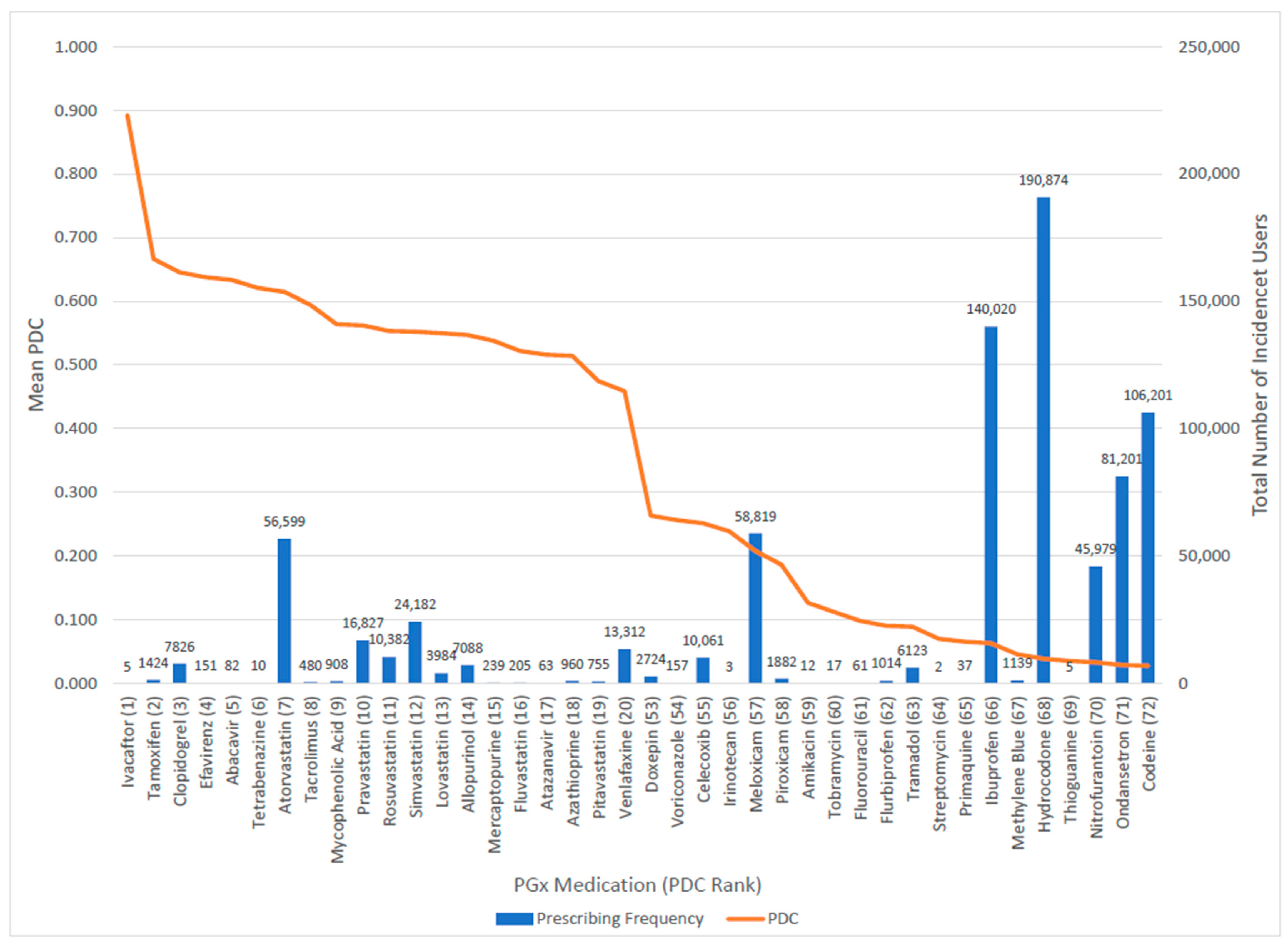
3.2. Amount of Exposure to Pharmacogenomic Medications
3.3. Medication Exposure by Patient Characteristics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roden, D.M.; McLeod, H.L.; Relling, M.V.; Williams, M.S.; Mensah, G.A.; Peterson, J.F.; Van Driest, S.L. Pharmacogenomics. Lancet 2019, 394, 521–532. [Google Scholar] [CrossRef] [PubMed]
- Relling, M.; Klein, T. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin. Pharmacol. Ther. 2011, 89, 464–467. [Google Scholar] [CrossRef] [PubMed]
- Relling, M.V.; Klein, T.E.; Gammal, R.S.; Whirl-Carrillo, M.; Hoffman, J.M.; Caudle, K.E. The Clinical Pharmacogenetics Implementation Consortium: 10 Years Later. Clin. Pharmacol. Ther. 2020, 107, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Caudle, K.E.; Klein, T.E.; Hoffman, J.M.; Muller, D.J.; Whirl-Carrillo, M.; Gong, L.; McDonagh, E.M.; Sangkuhl, K.; Thorn, C.F.; Schwab, M.; et al. Incorporation of Pharmacogenomics into Routine Clinical Practice: The Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline Development Process. Curr. Drug. Metab. 2014, 15, 9–17. [Google Scholar] [CrossRef] [PubMed]
- CPIC. Prioritization. 31 May 2022. Available online: https://cpicpgx.org/prioritization/#:~:text=Level%20Definitions%20for%20CPIC%20Genes%2FDrugs,-CPIC%20Level&text=At%20least%20one%20moderate%20or,but%20prescribing%20actionability%20is%20likely (accessed on 13 April 2023).
- Samwald, M.; Xu, H.; Blagec, K.; Empey, P.E.; Malone, D.C.; Ahmed, S.M.; Ryan, P.; Hofer, S.; Boyce, R.D. Incidence of Exposure of Patients in the United States to Multiple Drugs for Which Pharmacogenomic Guidelines Are Available. PLoS ONE 2016, 11, e0164972. [Google Scholar] [CrossRef]
- Chanfreau-Coffinier, C.; Hull, L.E.; Lynch, J.A.; DuVall, S.L.; Damrauer, S.M.; Cunningham, F.E.; Voight, B.F.; Matheny, M.E.; Oslin, D.W.; Icardi, M.S.; et al. Projected Prevalence of Actionable Pharmacogenetic Variants and Level A Drugs Prescribed Among US Veterans Health Administration Pharmacy Users. JAMA Netw. Open 2019, 2, e195345. [Google Scholar] [CrossRef]
- Hicks, J.K.; El Rouby, N.; Ong, H.H.; Schildcrout, J.S.; Ramsey, L.B.; Shi, Y.; Anne Tang, L.; Aquilante, C.L.; Beitelshees, A.L.; Blake, K.V.; et al. Opportunity for Genotype-Guided Prescribing Among Adult Patients in 11 US Health Systems. Clin. Pharmacol. Ther. 2021, 110, 179–188. [Google Scholar] [CrossRef]
- Schildcrout, J.S.; Denny, J.C.; Bowton, E.; Gregg, W.; Pulley, J.M.; Basford, M.A.; Cowan, J.D.; Xu, H.; Ramirez, A.H.; Crawford, D.C.; et al. Optimizing drug outcomes through pharmacogenetics: A case for preemptive genotyping. Clin. Pharmacol. Ther. 2012, 92, 235–242. [Google Scholar] [CrossRef]
- Heise, C.W.; Gallo, T.; Curry, S.C.; Woosley, R.L. Identification of populations likely to benefit from pharmacogenomic testing. Pharmacogenet Genom. 2020, 30, 91–95. [Google Scholar] [CrossRef]
- Ramsey, L.B.; Ong, H.H.; Schildcrout, J.S.; Shi, Y.; Tang, L.A.; Hicks, J.K.; El Rouby, N.; Cavallari, L.H.; Tuteja, S.; Aquilante, C.L.; et al. Prescribing Prevalence of Medications With Potential Genotype-Guided Dosing in Pediatric Patients. JAMA Netw. Open 2020, 3, e2029411. [Google Scholar] [CrossRef]
- Gage, B.F.; Bass, A.R.; Lin, H.; Woller, S.C.; Stevens, S.M.; Al-Hammadi, N.; Li, J.; Rodríguez, T., Jr.; Miller, J.P.; McMillin, G.A.; et al. Effect of Genotype-Guided Warfarin Dosing on Clinical Events and Anticoagulation Control Among Patients Undergoing Hip or Knee Arthroplasty: The GIFT Randomized Clinical Trial. JAMA 2017, 318, 1115–1124. [Google Scholar] [CrossRef] [PubMed]
- Syn, N.L.; Wong, A.L.; Lee, S.C.; Teoh, H.L.; Yip, J.W.L.; Seet, R.C.; Yeo, W.T.; Kristanto, W.; Bee, P.C.; Poon, L.M.; et al. Genotype-guided versus traditional clinical dosing of warfarin in patients of Asian ancestry: A randomized controlled trial. BMC Med. 2018, 16, 104. [Google Scholar] [CrossRef] [PubMed]
- Notarangelo, F.M.; Maglietta, G.; Bevilacqua, P.; Cereda, M.; Merlini, P.A.; Villani, G.Q.; Moruzzi, P.; Patrizi, G.; Malagoli Tagliazucchi, G.; Crocamo, A.; et al. Pharmacogenomic Approach to Selecting Antiplatelet Therapy in Patients With Acute Coronary Syndromes: The PHARMCLO Trial. J. Am. Coll. Cardiol. 2018, 71, 1869–1877. [Google Scholar] [CrossRef] [PubMed]
- Cavallari, L.H.; Lee, C.R.; Beitelshees, A.L.; Cooper-DeHoff, R.M.; Duarte, J.D.; Voora, D.; Kimmel, S.E.; McDonough, C.W.; Gong, Y.; Dave, C.V.; et al. Multisite Investigation of Outcomes With Implementation of CYP2C19 Genotype-Guided Antiplatelet Therapy After Percutaneous Coronary Intervention. JACC Cardiovasc. Interv. 2018, 11, 181–191. [Google Scholar] [CrossRef] [PubMed]
- IQVIA. IQVIA PharMetrics Plus for Academics Users Guide & Data Dictionary; IQVIA: Plymouth Meeting, PA, USA, 2017. [Google Scholar]
- Cooper-DeHoff, R.M.; Niemi, M.; Ramsey, L.B.; Luzum, J.A.; Tarkiainen, E.K.; Straka, R.J.; Gong, L.; Tuteja, S.; Wilke, R.A.; Wadelius, M.; et al. The Clinical Pharmacogenetics Implementation Consortium Guideline for SLCO1B1, ABCG2, and CYP2C9 genotypes and Statin-Associated Musculoskeletal Symptoms. Clin. Pharmacol. Ther. 2022, 111, 1007–1021. [Google Scholar] [CrossRef]
- Raebel, M.A.; Schmittdiel, J.; Karter, A.J.; Konieczny, J.L.; Steiner, J.F. Standardizing terminology and definitions of medication adherence and persistence in research employing electronic databases. Med. Care 2013, 51 (Suppl. S3), S11–S21. [Google Scholar] [CrossRef]
- Charlson, M.E.; Pompei, P.; Ales, K.L.; MacKenzie, C.R. A new method of classifying prognostic comorbidity in longitudinal studies: Development and validation. J. Chronic Dis. 1987, 40, 373–383. [Google Scholar] [CrossRef]
- Sullivan, M.D.; Edlund, M.J.; Fan, M.Y.; DeVries, A.; Braden, J.B.; Martin, B.C. Trends in use of opioids for non-cancer pain conditions 2000–2005 in commercial and Medicaid insurance plans: The TROUP study. Pain 2008, 138, 440–449. [Google Scholar] [CrossRef]
- Fan, M.; Yarema, M.C.; Box, A.; Hume, S.; Aitchison, K.J.; Bousman, C.A. Identification of high-impact gene-drug pairs for pharmacogenetic testing in Alberta, Canada. Pharmacogenet Genom. 2021, 31, 29–39. [Google Scholar] [CrossRef]
- Kuch, W.; Rinner, C.; Gall, W.; Samwald, M. How Many Patients Could Benefit From Pre-emptive Pharmacogenomic Testing and Decision Support? A Retrospective Study Based on Nationwide Austrian Claims Data. Stud. Health Technol. Inform. 2016, 223, 253–258. [Google Scholar]
- Kimpton, J.E.; Carey, I.M.; Threapleton, C.J.D.; Robinson, A.; Harris, T.; Cook, D.G.; DeWilde, S.; Baker, E.H. Longitudinal exposure of English primary care patients to pharmacogenomic drugs: An analysis to inform design of pre-emptive pharmacogenomic testing. Br. J. Clin. Pharmacol. 2019, 85, 2734–2746. [Google Scholar] [CrossRef] [PubMed]
- Canfield, S.L.; Zuckerman, A.; Anguiano, R.H.; Jolly, J.A.; DeClercq, J.; Wascher, M.; Choi, L.; Knox, S.; Mitchell, D.G. Navigating the Wild West of Medication Adherence Reporting in Specialty Pharmacy. J. Manag. Care Spec. Pharm. 2019, 25, 1073–1077. [Google Scholar] [CrossRef] [PubMed]
- Mease, P.J.; Young, P.; Gruben, D.; Fallon, L.; Germino, R.; Kavanaugh, A. Early Real-World Experience of Tofacitinib for Psoriatic Arthritis: Data from a United States Healthcare Claims Database. Adv. Ther. 2022, 39, 2932–2945. [Google Scholar] [CrossRef] [PubMed]
- Mannino, D.; Bogart, M.; Wu, B.; Germain, G.; Laliberté, F.; MacKnight, S.D.; Jung, Y.; Stiegler, M.; Duh, M.S. Adherence and persistence to once-daily single-inhaler versus multiple-inhaler triple therapy among patients with chronic obstructive pulmonary disease in the USA: A real-world study. Respir. Med. 2022, 197, 106807. [Google Scholar] [CrossRef]
| Demographics | Total Cohort 1 (N = 605,355) | CPIC Level A (N = 487,800) | CPIC Level A/B (N = 21,059) | CPIC Level B (N = 258,994) |
|---|---|---|---|---|
| Age, mean (SD, min–max) | 45.1 (15.2, 18–83) | 45.7 (15.1, 18–83) | 45.6 (15.0, 18–83) | 43.5 (15.3, 18–83) |
| Female sex, n (%) | 361,569 (59.7) | 291,211 (59.7) | 14,065 (66.8) | 162,038 (62.6) |
| Region 2 | ||||
| 154,575 (25.5) | 123,622 (25.3) | 4700 (22.3) | 67,586 (26.1) |
| 146,698 (24.2) | 118,806 (24.4) | 5024 (23.9) | 66,671 (25.7) | |
| 140,823 (23.3) | 116,162 (23.8) | 4610 (22.0) | 51,676 (20.0) | |
| 139,532 (23.1) | 109,900 (22.5) | 5853 (27.8) | 62,445 (24.1) | |
| Insurance Type | ||||
| 456,052 (75.3) | 360,156 (73.8) | 13,650 (64.8) | 194,944 (75.3) |
| 142,250 (23.5) | 121,436 (24.9) | 7073 (33.6) | 60,544 (23.4) | |
| 7053 (1.2) | 6208 (1.3) | 336 (1.6) | 3506 (1.4) | |
| Charlson Comorbidity Index, mean (SD, min-max) 3 | 0.6 (1.3, 0–21) | 0.6 (1.4, 0–21) | 1.1 (1.9, 0–21) | 0.6 (1.3, 0–21) |
| Total pharmacogenomic medications initiated 4, mean (SD, min-max) | 1.7 (1.1, 1–18) | 1.9 (1.4, 1–18) | 2.9 (1.8, 1–16) | 2.1 (1.3, 1–18) |
| Total Cohort (N = 1,035,918) 1 | CPIC Level A (N = 730,736) 1 | CPIC Level A/B (N = 21,663) 1 | CPIC Level B (N = 283,519) 1 | |
|---|---|---|---|---|
| Mean (SD, min–max) Median (IQR) | Mean (SD, min–max) Median (IQR) | Mean (SD, min–max) Median (IQR) | Mean (SD, min–max) Median (IQR) | |
| Proportion of days covered 2 | 0.21 (0.3, 0.003–1.0) 0.07 (0.02–0.25) | 0.24 (0.3, 0.003–1.0) 0.08 (0.02–0.35) | 0.44 (0.3, 0.003–1.0) 0.33 (0.08–0.79) | 0.10 (0.2, 0.003–1.0) 0.02 (0.01–0.05) |
| Number of fills 2 | 2.8 (3.0, 1–81) 1 (1–3) | 3.0 (3.1, 1–81) 1 (1–4) | 5.1 (4.2, 1–47) 4 (1–8) | 2.1 (2.5, 1–45) 1 (1–2) |
| Average days supplied across all fills 2 | 22.3 (22.3, 1–180) 15 (5–30) | 26.2 (23.5, 1–180) 30 (7–30) | 36.0 (17.1, 1–100) 30 (30–30) | 11.2 (14.3, 1–120) 5 (3–10) |
| Age Group 1 | N (%) | Unique Patients (N = 605,355) 2 | Males (N = 243,764) 1 | Females (N = 361,569) 1 |
|---|---|---|---|---|
| PDC, Mean (SD) | PDC, Mean (SD) | PDC, Mean (SD) | ||
| 18–29 | 120,432 (19.9) | 0.10 (0.19) | 0.10 (0.20) | 0.10 (0.19) |
| 30–39 | 106,302 (17.6) | 0.14 (0.24) | 0.15 (0.25) | 0.13 (0.22) |
| 40–49 | 119,793 (19.8) | 0.19 (0.28) | 0.21 (0.30) | 0.18 (0.27) |
| 50–59 | 146,903 (24.3) | 0.25 (0.32) | 0.27 (0.34) | 0.23 (0.31) |
| 60–69 | 86,954 (14.4) | 0.30 (0.35) | 0.32 (0.36) | 0.28 (0.34) |
| 70–79 | 23,101 (3.8) | 0.37 (0.38) | 0.39 (0.39) | 0.35 (0.37) |
| 80+ | 1870 (0.3) | 0.42 (0.40) | 0.46 (0.40) | 0.40 (0.39) |
| Diagnoses from the Charlson Comorbidity Index | N (%) | PDC, Mean (SD, Min–Max) |
|---|---|---|
| No Charlson Comorbidity Index diagnoses | 428,691 (70.8) | 0.17 (0.28, 0.003–1.0) 1 |
| ≥1 Charlson Comorbidity Index diagnoses | 176,664 (29.2) | 0.27 (0.33, 0.003–1.0) 1 |
| Specific Diagnoses | ||
| Myocardial Infarction | 6933 (1.2) | 0.41 (0.3, 0.003–1.0) |
| Diabetes without complications | 69,993 (12.2) | 0.31 (0.3, 0.003–1.0) |
| Diabetes with complications | 15,486 (2.7) | 0.33 (0.3, 0.003–1.0) |
| Paraplegia and Hemiplegia | 2607 (0.5) | 0.33 (0.3, 0.003–1.0) |
| Renal Disease | 11,201 (2.0) | 0.34 (0.3, 0.003–1.0) |
| Cancer | 21,821 (3.8) | 0.26 (0.3, 0.003–1.0) |
| Moderate or Severe Liver Disease | 1029 (0.2) | 0.28 (0.3, 0.003–1.0) |
| Metastatic Carcinoma | 3027 (0.5) | 0.24 (0.2, 0.003–1.0) |
| AIDS/HIV | 1577 (0.3) | 0.25 (0.3, 0.003–1.0) |
| Congestive Heart Failure | 11,601 (2.0) | 0.35 (0.3, 0.003–1.0) |
| Peripheral Vascular Disease | 13,673 (2.4) | 0.32 (0.3, 0.003–1.0) |
| Cerebrovascular Disease | 15,715 (2.7) | 0.33 (0.3, 0.003–1.0) |
| Dementia | 11,029 (0.2) | 0.39 (0.3, 0.003–1.0) |
| Chronic Pulmonary Disease | 65,379 (11.4) | 0.23 (0.3, 0.003–1.0) |
| Connective Tissue Disease-Rheumatic Disease | 10,042 (1.8) | 0.26 (0.3, 0.003–1.0) |
| Peptic Ulcer Disease | 3571 (0.6) | 0.27 (0.2, 0.003–1.0) |
| Mild Liver Disease | 19,219 (3.4) | 0.24 (0.2, 0.003–1.0) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bianchini, M.L.; Aquilante, C.L.; Kao, D.P.; Martin, J.L.; Anderson, H.D. Patient-Level Exposure to Actionable Pharmacogenomic Medications in a Nationally Representative Insurance Claims Database. J. Pers. Med. 2023, 13, 1574. https://doi.org/10.3390/jpm13111574
Bianchini ML, Aquilante CL, Kao DP, Martin JL, Anderson HD. Patient-Level Exposure to Actionable Pharmacogenomic Medications in a Nationally Representative Insurance Claims Database. Journal of Personalized Medicine. 2023; 13(11):1574. https://doi.org/10.3390/jpm13111574
Chicago/Turabian StyleBianchini, Monica L., Christina L. Aquilante, David P. Kao, James L. Martin, and Heather D. Anderson. 2023. "Patient-Level Exposure to Actionable Pharmacogenomic Medications in a Nationally Representative Insurance Claims Database" Journal of Personalized Medicine 13, no. 11: 1574. https://doi.org/10.3390/jpm13111574
APA StyleBianchini, M. L., Aquilante, C. L., Kao, D. P., Martin, J. L., & Anderson, H. D. (2023). Patient-Level Exposure to Actionable Pharmacogenomic Medications in a Nationally Representative Insurance Claims Database. Journal of Personalized Medicine, 13(11), 1574. https://doi.org/10.3390/jpm13111574






