Protein Succinylation and Malonylation as Potential Biomarkers in Schizophrenia
Abstract
1. Introduction
1.1. Schizophrenia and Antipsychotics
1.2. Succinylation and Malonylation of Lysine
1.3. Models for Studying Schizophrenia and Antipsychotics
2. Materials and Methods
2.1. Postmortem Tissue Preparation
2.2. Cell Line Growth and Preparation
2.3. Liquid Chromatography-Mass Spectrometry
2.4. Protein Identification
2.5. Data Analysis
3. Results
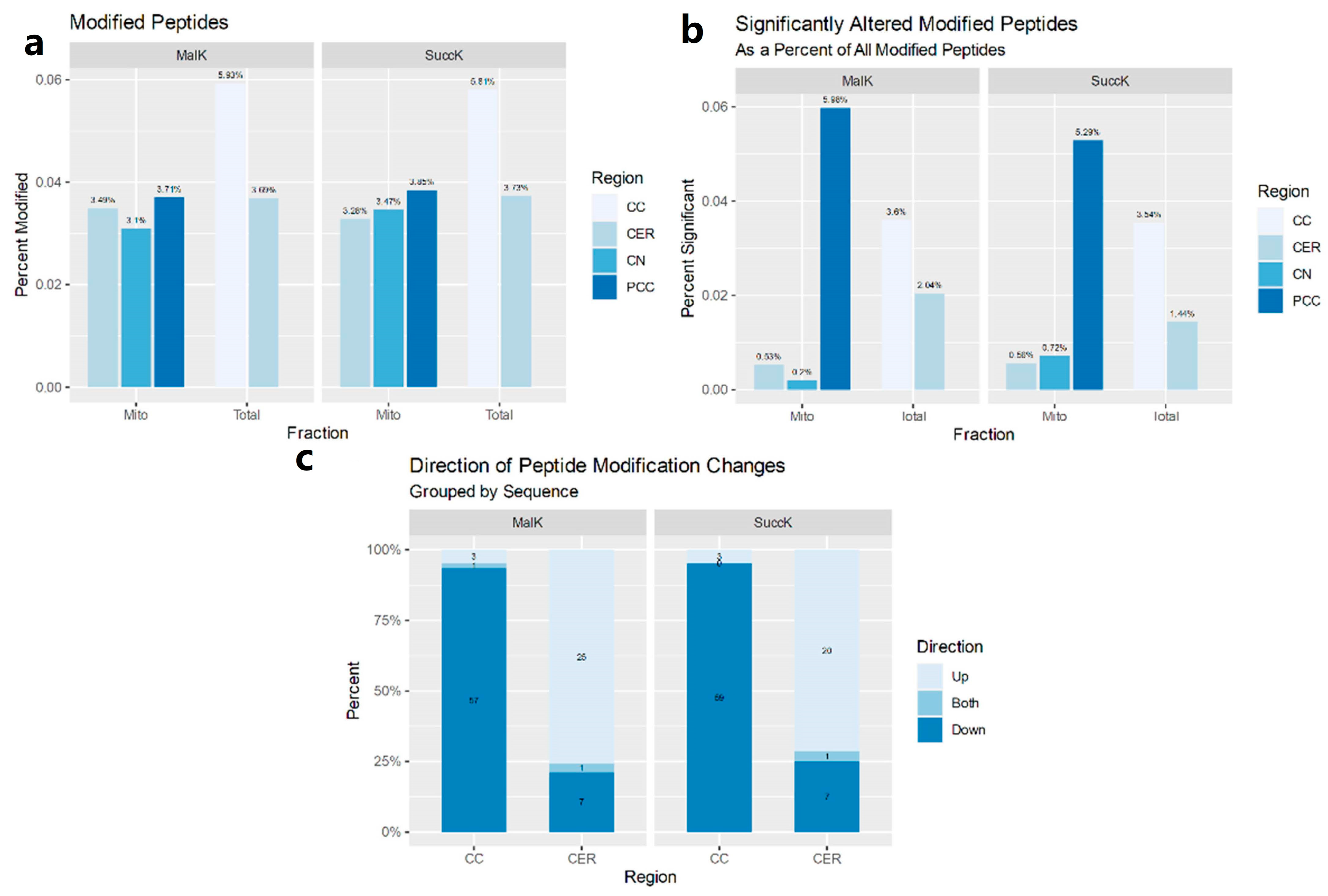
3.1. Postmortem Samples
3.1.1. Mitochondria-Enriched Protein Lysates
3.1.2. Total Cell Lysates
3.2. Expression Profiles in MO3.13
3.2.1. MK-801
3.2.2. Antipsychotics
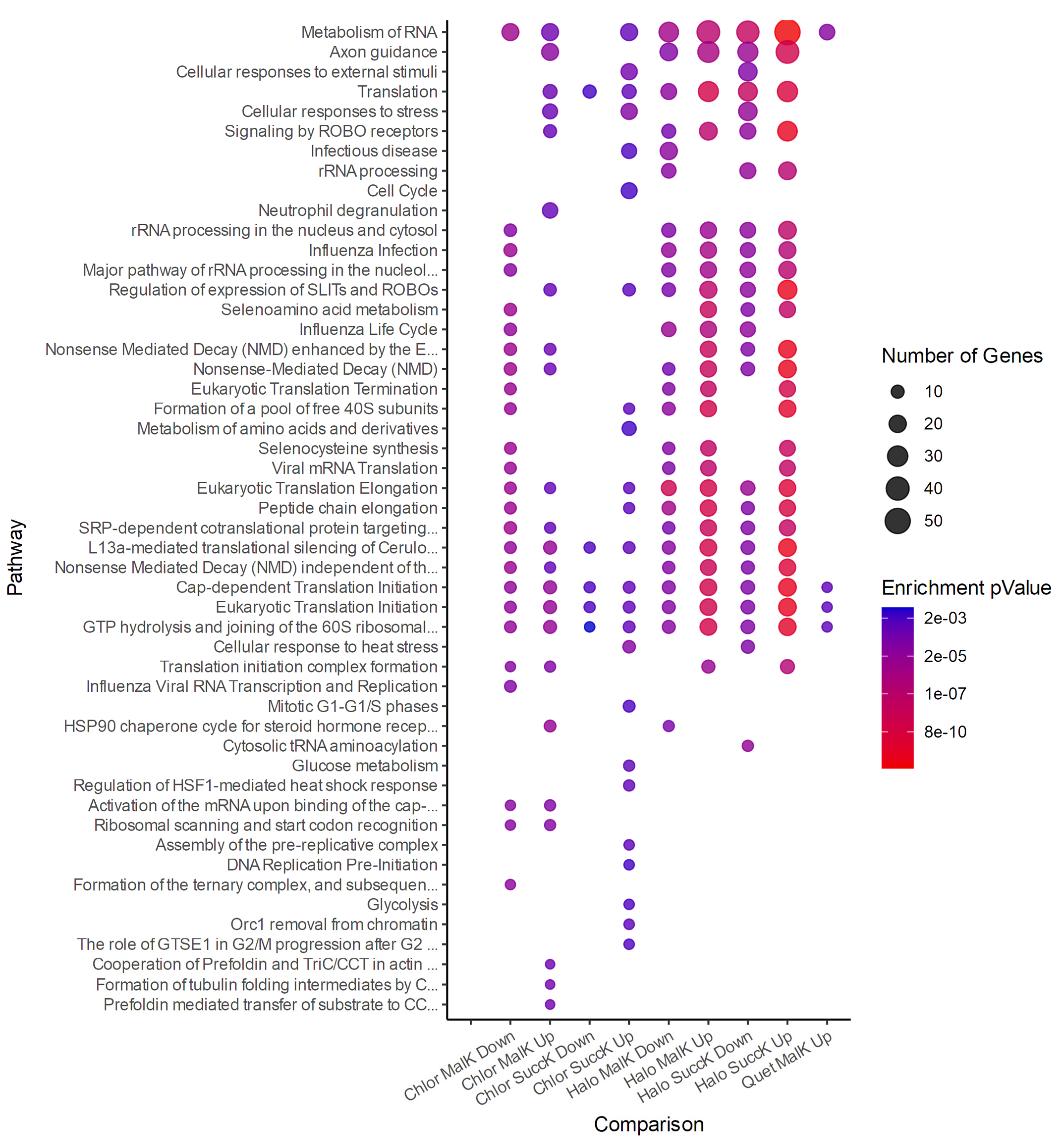
3.2.3. Antipsychotics and Succinylation
3.2.4. Antipsychotics and Malonylation
3.2.5. Attenuation Profiles of Succinylation and Malonylation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Saha, S.; Chant, D.; Welham, J.; McGrath, J. A Systematic Review of the Prevalence of Schizophrenia. PLoS Med. 2005, 2, e141. [Google Scholar] [CrossRef] [PubMed]
- Tsuang, M.T.; Stone, W.S.; Faraone, S.V. Genes, Environment and Schizophrenia. Br. J. Psychiatry 2001, 178, s18–s24. [Google Scholar] [CrossRef] [PubMed]
- Andreasen, N.C.; Arndt, S.; Alliger, R.; Miller, D.; Flaum, M. Symptoms of Schizophrenia: Methods, Meanings, and Mechanisms. Arch. Gen. Psychiatry 1995, 52, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.W. A History of Antipsychotic Drug Development. Compr. Psychiatry 1999, 40, 407–414. [Google Scholar] [CrossRef]
- Correll, C.U.; Schenk, E.M. Tardive Dyskinesia and New Antipsychotics. Curr. Opin. Psychiatry 2008, 21, 151–156. [Google Scholar] [CrossRef]
- Woods, S.W.; Morgenstern, H.; Saksa, J.R.; Walsh, B.C.; Sullivan, M.C.; Money, R.; Hawkins, K.A.; Gueorguieva, R.V.; Glazer, W.M. Incidence of Tardive Dyskinesia with Atypical and Conventional Antipsychotic Medications: Prospective Cohort Study. J. Clin. Psychiatry 2010, 71, 463–474. [Google Scholar] [CrossRef]
- Kane, J.M. Extrapyramidal Side Effects Are Unacceptable. Eur. Neuropsychopharmacol. 2001, 11, S397–S403. [Google Scholar] [CrossRef]
- Kapur, S.; Zipursky, R.; Jones, C.; Shammi, C.S.; Remington, G.; Seeman, P. A Positron Emission Tomography Study of Quetiapine in Schizophrenia: A Preliminary Finding of an Antipsychotic Effect with Only Transiently High Dopamine D2 Receptor Occupancy. Arch. Gen. Psychiatry 2000, 57, 553–559. [Google Scholar] [CrossRef]
- Kapur, S.; Zipursky, R.; Jones, C.; Remington, G.; Houle, S. Relationship between Dopamine D(2) Occupancy, Clinical Response, and Side Effects: A Double-Blind PET Study of First-Episode Schizophrenia. Am. J. Psychiatry 2000, 157, 514–520. [Google Scholar] [CrossRef]
- Guenette, M.D.; Giacca, A.; Hahn, M.; Teo, C.; Lam, L.; Chintoh, A.; Arenovich, T.; Remington, G. Atypical Antipsychotics and Effects of Adrenergic and Serotonergic Receptor Binding on Insulin Secretion In-Vivo: An Animal Model. Schizophr. Res. 2013, 146, 162–169. [Google Scholar] [CrossRef]
- Gault, J.M.; Nussbaum, A.M. Review of Serum Prolactin Levels as an Antipsychotic-Response Biomarker. Open Access. J. Transl. Med. Res. 2018, 2, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Sumiyoshi, T.; Higuchi, Y.; Uehara, T. Neural Basis for the Ability of Atypical Antipsychotic Drugs to Improve Cognition in Schizophrenia. Front. Behav. Neurosci. 2013, 7, 140. [Google Scholar] [CrossRef] [PubMed]
- Stephens, P. A Review of Clozapine: An Antipsychotic for Treatment-Resistant Schizophrenia. Compr. Psychiatry 1990, 31, 315–326. [Google Scholar] [CrossRef]
- Ojong, M.; Allen, S.N. Management and Prevention of Agranulocytosis in Patients Receiving Clozapine. Ment. Health Clin. 2013, 3, 139–143. [Google Scholar] [CrossRef]
- Sahlholm, K.; Marcellino, D.; Nilsson, J.; Ögren, S.O.; Fuxe, K.; Århem, P. Typical and Atypical Antipsychotics Do Not Differ Markedly in Their Reversibility of Antagonism of the Dopamine D2 Receptor. Int. J. Neuropsychopharmacol. 2014, 17, 149–155. [Google Scholar] [CrossRef]
- Crossley, N.A.; Constante, M.; McGuire, P.; Power, P. Efficacy of Atypical v. Typical Antipsychotics in the Treatment of Early Psychosis: Meta-Analysis. Br. J. Psychiatry 2010, 196, 434–439. [Google Scholar] [CrossRef]
- Weinert, B.T.; Schölz, C.; Wagner, S.A.; Iesmantavicius, V.; Su, D.; Daniel, J.A.; Choudhary, C. Lysine Succinylation Is a Frequently Occurring Modification in Prokaryotes and Eukaryotes and Extensively Overlaps with Acetylation. Cell Rep. 2013, 4, 842–851. [Google Scholar] [CrossRef]
- Zhang, Z.; Tan, M.; Xie, Z.; Dai, L.; Chen, Y.; Zhao, Y. Identification of Lysine Succinylation as a New Post-Translational Modification. Nat. Chem. Biol. 2011, 7, 58–63. [Google Scholar] [CrossRef]
- Chen, H.; Xu, H.; Potash, S.; Starkov, A.; Belousov, V.V.; Bilan, D.S.; Denton, T.T.; Gibson, G.E. Mild Metabolic Perturbations Alter Succinylation of Mitochondrial Proteins. J. Neurosci. Res. 2017, 95, 2244–2252. [Google Scholar] [CrossRef]
- Kurmi, K.; Hitosugi, S.; Wiese, E.K.; Boakye-Agyeman, F.; Gonsalves, W.I.; Lou, Z.; Karnitz, L.M.; Goetz, M.P.; Hitosugi, T. Carnitine Palmitoyltransferase 1A Has a Lysine Succinyltransferase Activity. Cell Rep. 2018, 22, 1365–1373. [Google Scholar] [CrossRef]
- Wang, Y.; Guo, Y.R.; Liu, K.; Yin, Z.; Liu, R.; Xia, Y.; Tan, L.; Yang, P.; Lee, J.-H.; Li, X.; et al. KAT2A Coupled with the α-KGDH Complex Acts as a Histone H3 Succinyltransferase. Nature 2017, 552, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Zhou, Y.; Su, X.; Yu, J.J.; Khan, S.; Jiang, H.; Kim, J.; Woo, J.; Kim, J.H.; Choi, B.H.; et al. Sirt5 Is a NAD-Dependent Protein Lysine Demalonylase and Desuccinylase. Science 2011, 334, 806–809. [Google Scholar] [CrossRef]
- Rardin, M.J.; He, W.; Nishida, Y.; Newman, J.C.; Carrico, C.; Danielson, S.R.; Guo, A.; Gut, P.; Sahu, A.K.; Li, B.; et al. SIRT5 Regulates the Mitochondrial Lysine Succinylome and Metabolic Networks. Cell Metab. 2013, 18, 920–933. [Google Scholar] [CrossRef] [PubMed]
- Nishida, Y.; Rardin, M.J.; Carrico, C.; He, W.; Sahu, A.K.; Gut, P.; Najjar, R.; Fitch, M.; Hellerstein, M.; Gibson, B.W.; et al. SIRT5 Regulates Both Cytosolic and Mitochondrial Protein Malonylation with Glycolysis as a Major Target. Mol. Cell 2015, 59, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Sreedhar, A.; Wiese, E.K.; Hitosugi, T. Enzymatic and Metabolic Regulation of Lysine Succinylation. Genes Dis. 2020, 7, 166–171. [Google Scholar] [CrossRef]
- Bowman, C.E.; Rodriguez, S.; Selen Alpergin, E.S.; Acoba, M.G.; Zhao, L.; Hartung, T.; Claypool, S.M.; Watkins, P.A.; Wolfgang, M.J. The Mammalian Malonyl-CoA Synthetase ACSF3 Is Required for Mitochondrial Protein Malonylation and Metabolic Efficiency. Cell Chem. Biol. 2017, 24, 673–684.e4. [Google Scholar] [CrossRef]
- Dehzangi, A.; López, Y.; Lal, S.P.; Taherzadeh, G.; Sattar, A.; Tsunoda, T.; Sharma, A. Improving Succinylation Prediction Accuracy by Incorporating the Secondary Structure via Helix, Strand and Coil, and Evolutionary Information from Profile Bigrams. PLoS ONE 2018, 13, e0191900. [Google Scholar] [CrossRef]
- Ning, Q.; Zhao, X.; Bao, L.; Ma, Z.; Zhao, X. Detecting Succinylation Sites from Protein Sequences Using Ensemble Support Vector Machine. BMC Bioinform. 2018, 19, 237. [Google Scholar] [CrossRef]
- Taherzadeh, G.; Yang, Y.; Xu, H.; Xue, Y.; Liew, A.W.-C.; Zhou, Y. Predicting Lysine-Malonylation Sites of Proteins Using Sequence and Predicted Structural Features. J. Comput. Chem. 2018, 39, 1757–1763. [Google Scholar] [CrossRef]
- Gonçalves, V.F.; Zai, C.C.; Tiwari, A.K.; Brandl, E.J.; Derkach, A.; Meltzer, H.Y.; Lieberman, J.A.; Müller, D.J.; Sun, L.; Kennedy, J.L. A Hypothesis-Driven Association Study of 28 Nuclear-Encoded Mitochondrial Genes with Antipsychotic-Induced Weight Gain in Schizophrenia. Neuropsychopharmacology 2014, 39, 1347–1354. [Google Scholar] [CrossRef]
- Martins-de-Souza, D.; Gattaz, W.F.; Schmitt, A.; Novello, J.C.; Marangoni, S.; Turck, C.W.; Dias-Neto, E. Proteome Analysis of Schizophrenia Patients Wernicke’s Area Reveals an Energy Metabolism Dysregulation. BMC Psychiatry 2009, 9, 17. [Google Scholar] [CrossRef]
- Hasnain, M.; Fredrickson, S.K.; Vieweg, W.V.R.; Pandurangi, A.K. Metabolic Syndrome Associated with Schizophrenia and Atypical Antipsychotics. Curr. Diab. Rep. 2010, 10, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Mahadik, S.P.; Evans, D.R. Is Schizophrenia a Metabolic Brain Disorder? Membrane Phospholipid Dysregulation and Its Therapeutic Implications. Psychiatr. Clin. 2003, 26, 85–102. [Google Scholar] [CrossRef]
- Misiak, B.; Łaczmański, Ł.; Słoka, N.K.; Szmida, E.; Piotrowski, P.; Loska, O.; Ślęzak, R.; Kiejna, A.; Frydecka, D. Metabolic Dysregulation in First-Episode Schizophrenia Patients with Respect to Genetic Variation in One-Carbon Metabolism. Psychiatry Res. 2016, 238, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Martins-de-Souza, D.; Harris, L.W.; Guest, P.C.; Bahn, S. The Role of Energy Metabolism Dysfunction and Oxidative Stress in Schizophrenia Revealed by Proteomics. Antioxid. Redox Signal. 2010, 15, 2067–2079. [Google Scholar] [CrossRef] [PubMed]
- Papanastasiou, E. The Prevalence and Mechanisms of Metabolic Syndrome in Schizophrenia: A Review. Ther. Adv. Psychopharmacol. 2013, 3, 33–51. [Google Scholar] [CrossRef]
- Hirschey, M.D.; Zhao, Y. Metabolic Regulation by Lysine Malonylation, Succinylation, and Glutarylation. Mol. Cell Proteom. 2015, 14, 2308–2315. [Google Scholar] [CrossRef]
- Qian, L.; Nie, L.; Chen, M.; Liu, P.; Zhu, J.; Zhai, L.; Tao, S.; Cheng, Z.; Zhao, Y.; Tan, M. Global Profiling of Protein Lysine Malonylation in Escherichia Coli Reveals Its Role in Energy Metabolism. J. Proteome Res. 2016, 15, 2060–2071. [Google Scholar] [CrossRef]
- Hof, P.R.; Haroutunian, V.; Copland, C.; Davis, K.L.; Buxbaum, J.D. Molecular and Cellular Evidence for an Oligodendrocyte Abnormality in Schizophrenia. Neurochem. Res. 2002, 27, 1193–1200. [Google Scholar] [CrossRef]
- Martins-de-Souza, D. Proteome and Transcriptome Analysis Suggests Oligodendrocyte Dysfunction in Schizophrenia. J. Psychiatr. Res. 2010, 44, 149–156. [Google Scholar] [CrossRef]
- Takahashi, N.; Sakurai, T.; Davis, K.L.; Buxbaum, J.D. Linking Oligodendrocyte and Myelin Dysfunction to Neurocircuitry Abnormalities in Schizophrenia. Prog. Neurobiol. 2011, 93, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Tkachev, D.; Mimmack, M.L.; Ryan, M.M.; Wayland, M.; Freeman, T.; Jones, P.B.; Starkey, M.; Webster, M.J.; Yolken, R.H.; Bahn, S. Oligodendrocyte Dysfunction in Schizophrenia and Bipolar Disorder. Lancet 2003, 362, 798–805. [Google Scholar] [CrossRef]
- Uranova, N.A.; Vostrikov, V.M.; Orlovskaya, D.D.; Rachmanova, V.I. Oligodendroglial Density in the Prefrontal Cortex in Schizophrenia and Mood Disorders: A Study from the Stanley Neuropathology Consortium. Schizophr. Res. 2004, 67, 269–275. [Google Scholar] [CrossRef]
- Karoutzou, G.; Emrich, H.M.; Dietrich, D.E. The Myelin-Pathogenesis Puzzle in Schizophrenia: A Literature Review. Mol. Psychiatry 2008, 13, 245–260. [Google Scholar] [CrossRef]
- Buntinx, M.; Vanderlocht, J.; Hellings, N.; Vandenabeele, F.; Lambrichts, I.; Raus, J.; Ameloot, M.; Stinissen, P.; Steels, P. Characterization of Three Human Oligodendroglial Cell Lines as a Model to Study Oligodendrocyte Injury: Morphology and Oligodendrocyte-Specific Gene Expression. J. Neurocytol. 2003, 32, 25–38. [Google Scholar] [CrossRef]
- Rung, J.P.; Carlsson, A.; Rydén Markinhuhta, K.; Carlsson, M.L. (+)-MK-801 Induced Social Withdrawal in Rats; a Model for Negative Symptoms of Schizophrenia. Prog. Neuropsychopharmacol. Biol. Psychiatry 2005, 29, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Svoboda, J.; Stankova, A.; Entlerova, M.; Stuchlik, A. Acute Administration of MK-801 in an Animal Model of Psychosis in Rats Interferes with Cognitively Demanding Forms of Behavioral Flexibility on a Rotating Arena. Front. Behav. Neurosci. 2015, 9. [Google Scholar] [CrossRef]
- Eyjolfsson, E.M.; Brenner, E.; Kondziella, D.; Sonnewald, U. Repeated Injection of MK801: An Animal Model of Schizophrenia? Neurochem. Int. 2006, 48, 541–546. [Google Scholar] [CrossRef]
- Braun, I.; Genius, J.; Grunze, H.; Bender, A.; Möller, H.-J.; Rujescu, D. Alterations of Hippocampal and Prefrontal GABAergic Interneurons in an Animal Model of Psychosis Induced by NMDA Receptor Antagonism. Schizophr. Res. 2007, 97, 254–263. [Google Scholar] [CrossRef]
- Seillier, A.; Giuffrida, A. Evaluation of NMDA Receptor Models of Schizophrenia: Divergences in the Behavioral Effects of Sub-Chronic PCP and MK-801. Behav. Brain Res. 2009, 204, 410–415. [Google Scholar] [CrossRef]
- Rujescu, D.; Bender, A.; Keck, M.; Hartmann, A.M.; Ohl, F.; Raeder, H.; Giegling, I.; Genius, J.; McCarley, R.W.; Möller, H.-J.; et al. A Pharmacological Model for Psychosis Based on N-Methyl-D-Aspartate Receptor Hypofunction: Molecular, Cellular, Functional and Behavioral Abnormalities. Biol. Psychiatry 2006, 59, 721–729. [Google Scholar] [CrossRef] [PubMed]
- Brandão-Teles, C.; Martins-de-Souza, D.; Guest, P.C.; Cassoli, J.S. MK-801-Treated Oligodendrocytes as a Cellular Model to Study Schizophrenia. Adv. Exp. Med. Biol. 2017, 974, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Ahmadvand, A.; Shahidi, S.B.; Talari, H.; Ghoreishi, F.S.; Mousavi, G.A. Morphology of the Corpus Callosum and Schizophrenia: A Case-Control Study in Kashan, Iran. Electron. Physician 2017, 9, 5478–5486. [Google Scholar] [CrossRef] [PubMed]
- Downhill, J.E.; Buchsbaum, M.S.; Wei, T.; Spiegel-Cohen, J.; Hazlett, E.A.; Haznedar, M.M.; Silverman, J.; Siever, L.J. Shape and Size of the Corpus Callosum in Schizophrenia and Schizotypal Personality Disorder. Schizophr. Res. 2000, 42, 193–208. [Google Scholar] [CrossRef]
- Whitford, T.J.; Kubicki, M.; Schneiderman, J.S.; O’Donnell, L.J.; King, R.; Alvarado, J.L.; Khan, U.; Markant, D.; Nestor, P.G.; Niznikiewicz, M.; et al. Corpus Callosum Abnormalities and Their Association with Psychotic Symptoms in Patients with Schizophrenia. Biol. Psychiatry 2010, 68, 70–77. [Google Scholar] [CrossRef]
- Saia-Cereda, V.M.; Cassoli, J.S.; Schmitt, A.; Falkai, P.; Nascimento, J.M.; Martins-de-Souza, D. Proteomics of the Corpus Callosum Unravel Pivotal Players in the Dysfunction of Cell Signaling, Structure, and Myelination in Schizophrenia Brains. Eur. Arch. Psychiatry Clin. Neurosci. 2015, 265, 601–612. [Google Scholar] [CrossRef]
- Saia-Cereda, V.M.; Cassoli, J.S.; Schmitt, A.; Falkai, P.; Martins-de-Souza, D. Differential Proteome and Phosphoproteome May Impact Cell Signaling in the Corpus Callosum of Schizophrenia Patients. Schizophr. Res. 2016, 177, 70–77. [Google Scholar] [CrossRef]
- Sivagnanasundaram, S.; Crossett, B.; Dedova, I.; Cordwell, S.; Matsumoto, I. Abnormal Pathways in the Genu of the Corpus Callosum in Schizophrenia Pathogenesis: A Proteome Study. PROTEOMICS–Clin. Appl. 2007, 1, 1291–1305. [Google Scholar] [CrossRef]
- Innocenti, G.M.; Ansermet, F.; Parnas, J. Schizophrenia, Neurodevelopment and Corpus Callosum. Mol. Psychiatry 2003, 8, 261. [Google Scholar] [CrossRef]
- English, J.A.; Pennington, K.; Dunn, M.J.; Cotter, D.R. The Neuroproteomics of Schizophrenia. Biol. Psychiatry 2011, 69, 163–172. [Google Scholar] [CrossRef]
- Martin, P.; Albers, M. Cerebellum and Schizophrenia: A Selective Review. Schizophr. Bull. 1995, 21, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, L.K.; Giedd, J.N.; Berquin, P.C.; Krain, A.L.; Hamburger, S.D.; Kumra, S.; Rapoport, J.L. Quantitative Morphology of the Cerebellum and Fourth Ventricle in Childhood-Onset Schizophrenia. Am. J. Psychiatry 1997. [Google Scholar] [CrossRef] [PubMed]
- Andreasen, N.C.; Pierson, R. The Role of the Cerebellum in Schizophrenia. Biol. Psychiatry 2008, 64, 81–88. [Google Scholar] [CrossRef]
- Picard, H.; Amado, I.; Mouchet-Mages, S.; Olié, J.-P.; Krebs, M.-O. The Role of the Cerebellum in Schizophrenia: An Update of Clinical, Cognitive, and Functional Evidences. Schizophr. Bull. 2008, 34, 155–172. [Google Scholar] [CrossRef] [PubMed]
- Filiou, M.D.; Teplytska, L.; Otte, D.M.; Zimmer, A.; Turck, C.W. Myelination and Oxidative Stress Alterations in the Cerebellum of the G72/G30 Transgenic Schizophrenia Mouse Model. J. Psychiatr. Res. 2012, 46, 1359–1365. [Google Scholar] [CrossRef] [PubMed]
- Eastwood, S.L.; Law, A.J.; Everall, I.P.; Harrison, P.J. The Axonal Chemorepellant Semaphorin 3A Is Increased in the Cerebellum in Schizophrenia and May Contribute to Its Synaptic Pathology. Mol. Psychiatry 2003, 8, 148. [Google Scholar] [CrossRef] [PubMed]
- Smalla, K.-H.; Mikhaylova, M.; Sahin, J.; Bernstein, H.-G.; Bogerts, B.; Schmitt, A.; van der Schors, R.; Smit, A.B.; Li, K.W.; Gundelfinger, E.D.; et al. A Comparison of the Synaptic Proteome in Human Chronic Schizophrenia and Rat Ketamine Psychosis Suggest That Prohibitin Is Involved in the Synaptic Pathology of Schizophrenia. Mol. Psychiatry 2008, 13, 878–896. [Google Scholar] [CrossRef][Green Version]
- Leech, R.; Sharp, D.J. The Role of the Posterior Cingulate Cortex in Cognition and Disease. Brain 2014, 137, 12–32. [Google Scholar] [CrossRef]
- Sowell, E.R.; Levitt, J.; Thompson, P.M.; Holmes, C.J.; Blanton, R.E.; Kornsand, D.S.; Caplan, R.; McCracken, J.; Asarnow, R.; Toga, A.W. Brain Abnormalities in Early-Onset Schizophrenia Spectrum Disorder Observed with Statistical Parametric Mapping of Structural Magnetic Resonance Images. Am. J. Psychiatry 2000, 157, 1475–1484. [Google Scholar] [CrossRef]
- Newell, K.A.; Zavitsanou, K.; Jew, S.K.; Huang, X.-F. Alterations of Muscarinic and GABA Receptor Binding in the Posterior Cingulate Cortex in Schizophrenia. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2007, 31, 225–233. [Google Scholar] [CrossRef]
- Newell, K.A.; Deng, C.; Huang, X.-F. Increased Cannabinoid Receptor Density in the Posterior Cingulate Cortex in Schizophrenia. Exp. Brain Res. 2006, 172, 556–560. [Google Scholar] [CrossRef] [PubMed]
- Haznedar, M.M.; Buchsbaum, M.S.; Luu, C.; Hazlett, E.A.; Siegel, B.V.; Lohr, J.; Wu, J.; Haier, R.J.; Bunney, W.E. Decreased Anterior Cingulate Gyrus Metabolic Rate in Schizophrenia. Am. J. Psychiatry 1997, 154, 682–684. [Google Scholar] [CrossRef] [PubMed]
- Mitelman, S.A.; Byne, W.; Kemether, E.M.; Hazlett, E.A.; Buchsbaum, M.S. Metabolic Disconnection Between the Mediodorsal Nucleus of the Thalamus and Cortical Brodmann’s Areas of the Left Hemisphere in Schizophrenia. Am. J. Psychiatry 2005. [Google Scholar] [CrossRef]
- Owen, F.; Crow, T.J.; Poulter, M.; Cross, A.J.; Longden, A.; Riley, G.J. Increased dopamine-receptor sensitivity in schizophrenia. Lancet 1978, 312, 223–226. [Google Scholar] [CrossRef]
- Sumiyoshi, T.; Stockmeier, C.A.; Overholser, J.C.; Thompson, P.A.; Meltzer, H.Y. Dopamine D4 Receptors and Effects of Guanine Nucleotides on [3H]Raclopride Binding in Postmortem Caudate Nucleus of Subjects with Schizophrenia or Major Depression. Brain Res. 1995, 681, 109–116. [Google Scholar] [CrossRef]
- Breier, A.; Buchanan, R.W.; Elkashef, A.; Munson, R.C.; Kirkpatrick, B.; Gellad, F. Brain Morphology and Schizophrenia: A Magnetic Resonance Imaging Study of Limbic, Prefrontal Cortex, and Caudate Structures. Arch. Gen. Psychiatry 1992, 49, 921–926. [Google Scholar] [CrossRef]
- Resnick, S.M.; Gur, R.E.; Alavi, A.; Gur, R.C.; Reivich, M. Positron Emission Tomography and Subcortical Glucose Metabolism in Schizophrenia. Psychiatry Res. 1988, 24, 1–11. [Google Scholar] [CrossRef]
- Jayakumar, P.N.; Venkatasubramanian, G.; Keshavan, M.S.; Srinivas, J.S.; Gangadhar, B.N. MRI Volumetric and 31P MRS Metabolic Correlates of Caudate Nucleus in Antipsychotic-Naïve Schizophrenia. Acta Psychiatr. Scand. 2006, 114, 346–351. [Google Scholar] [CrossRef]
- Shihabuddin, L.; Buchsbaum, M.S.; Hazlett, E.A.; Haznedar, M.M.; Harvey, P.D.; Newman, A.; Schnur, D.B.; Spiegel-Cohen, J.; Wei, T.; Machac, J.; et al. Dorsal Striatal Size, Shape, and Metabolic Rate in Never-Medicated and Previously Medicated Schizophrenics Performing a Verbal Learning Task. Arch. Gen. Psychiatry 1998, 55, 235–243. [Google Scholar] [CrossRef]
- Wiesel, F.A.; Wik, G.; Sjögren, I.; Blomqvist, G.; Greitz, T.; Stone-Elander, S. Regional Brain Glucose Metabolism in Drug Free Schizophrenic Patients and Clinical Correlates. Acta Psychiatr. Scand. 1987, 76, 628–641. [Google Scholar] [CrossRef]
- Reis-de-Oliveira, G.; Fioramonte, M.; Martins-de-Souza, D. A Complete Proteomic Workflow to Study Brain-Related Disorders via Postmortem Tissue. In Pre-Clinical Models: Techniques and Protocols; Guest, P.C., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2019; pp. 319–328. ISBN 978-1-4939-8994-2. [Google Scholar]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE Database and Related Tools and Resources in 2019: Improving Support for Quantification Data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]
- Fabregat, A.; Jupe, S.; Matthews, L.; Sidiropoulos, K.; Gillespie, M.; Garapati, P.; Haw, R.; Jassal, B.; Korninger, F.; May, B.; et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018, 46, D649–D655. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Kabir, M.A.; Uddin, W.; Narayanan, A.; Reddy, P.K.; Jairajpuri, M.A.; Sherman, F.; Ahmad, Z. Functional Subunits of Eukaryotic Chaperonin CCT/TRiC in Protein Folding. J. Amino Acids 2011, 2011, 843206. [Google Scholar] [CrossRef]
- Kucukgoncu, S.; Kosir, U.; Zhou, E.; Sullivan, E.; Srihari, V.H.; Tek, C. Glucose Metabolism Dysregulation at the Onset of Mental Illness Is Not Limited to First Episode Psychosis: A Systematic Review and Meta-Analysis. Early Interv. Psychiatry 2019, 13, 1021–1031. [Google Scholar] [CrossRef]
- Pillinger, T.; McCutcheon, R.A.; Vano, L.; Mizuno, Y.; Arumuham, A.; Hindley, G.; Beck, K.; Natesan, S.; Efthimiou, O.; Cipriani, A.; et al. Comparative Effects of 18 Antipsychotics on Metabolic Function in Patients with Schizophrenia, Predictors of Metabolic Dysregulation, and Association with Psychopathology: A Systematic Review and Network Meta-Analysis. Lancet Psychiatry 2020, 7, 64–77. [Google Scholar] [CrossRef]
- Misiak, B.; Piotrowski, P.; Beszłej, J.A.; Kalinowska, S.; Chęć, M.; Samochowiec, J. Metabolic Dysregulation and Psychosocial Stress in Patients with Schizophrenia Spectrum Disorders: A Case-Control Study. J. Clin. Med. 2020, 9, 3822. [Google Scholar] [CrossRef] [PubMed]
- Moehle, M.S.; Luduena, R.F.; Haroutunian, V.; Meador-Woodruff, J.H.; McCullumsmith, R.E. Regional Differences in Expression of β-Tubulin Isoforms in Schizophrenia. Schizophr. Res. 2012, 135, 181–186. [Google Scholar] [CrossRef]
- Benitez-King, G.; Ramírez-Rodríguez, G.; Ortíz, L.; Meza, I. The Neuronal Cytoskeleton as a Potential Therapeutical Target in Neurodegenerative Diseases and Schizophrenia. Curr. Drug Targets CNS Neurol. Disord. 2004, 3, 515–533. [Google Scholar] [CrossRef]
- Martins-de-Souza, D.; Gattaz, W.F.; Schmitt, A.; Maccarrone, G.; Hunyadi-Gulyás, E.; Eberlin, M.N.; Souza, G.H.M.F.; Marangoni, S.; Novello, J.C.; Turck, C.W.; et al. Proteomic Analysis of Dorsolateral Prefrontal Cortex Indicates the Involvement of Cytoskeleton, Oligodendrocyte, Energy Metabolism and New Potential Markers in Schizophrenia. J. Psychiatr. Res. 2009, 43, 978–986. [Google Scholar] [CrossRef]
- Zhao, Z.; Xu, J.; Chen, J.; Kim, S.; Reimers, M.; Bacanu, S.-A.; Yu, H.; Liu, C.; Sun, J.; Wang, Q.; et al. Transcriptome Sequencing and Genome-Wide Association Analyses Reveal Lysosomal Function and Actin Cytoskeleton Remodeling in Schizophrenia and Bipolar Disorder. Mol. Psychiatry 2015, 20, 563–572. [Google Scholar] [CrossRef]
- Benítez-King, G.; Valdés-Tovar, M.; Trueta, C.; Galván-Arrieta, T.; Argueta, J.; Alarcón, S.; Lora-Castellanos, A.; Solís-Chagoyán, H. The Microtubular Cytoskeleton of Olfactory Neurons Derived from Patients with Schizophrenia or with Bipolar Disorder: Implications for Biomarker Characterization, Neuronal Physiology and Pharmacological Screening. Mol. Cell. Neurosci. 2016, 73, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Bhambhvani, H.P.; Mueller, T.M.; Simmons, M.S.; Meador-Woodruff, J.H. Actin Polymerization Is Reduced in the Anterior Cingulate Cortex of Elderly Patients with Schizophrenia. Transl. Psychiatry 2017, 7, 1278. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Kim, E.; Datta, D.; Lewis, D.A.; Soderling, S.H. Synaptic Actin Dysregulation, a Convergent Mechanism of Mental Disorders? J. Neurosci. 2016, 36, 11411–11417. [Google Scholar] [CrossRef] [PubMed]
- Rosenmund, C.; Westbrook, G.L. Calcium-Induced Actin Depolymerization Reduces NMDA Channel Activity. Neuron 1993, 10, 805–814. [Google Scholar] [CrossRef]
- Allison, D.W.; Gelfand, V.I.; Spector, I.; Craig, A.M. Role of Actin in Anchoring Postsynaptic Receptors in Cultured Hippocampal Neurons: Differential Attachment of NMDA versus AMPA Receptors. J. Neurosci. 1998, 18, 2423–2436. [Google Scholar] [CrossRef]
- Morishita, W.; Marie, H.; Malenka, R.C. Distinct Triggering and Expression Mechanisms Underlie LTD of AMPA and NMDA Synaptic Responses. Nat. Neurosci. 2005, 8, 1043–1050. [Google Scholar] [CrossRef]
- Olney, J.W.; Newcomer, J.W.; Farber, N.B. NMDA Receptor Hypofunction Model of Schizophrenia. J. Psychiatr. Res. 1999, 33, 523–533. [Google Scholar] [CrossRef]
- Balu, D.T. Chapter Twelve-The NMDA Receptor and Schizophrenia: From Pathophysiology to Treatment. In Advances in Pharmacology; Schwarcz, R., Ed.; Neuropsychopharmacology: A Tribute to Joseph T. Coyle; Academic Press: Cambridge, MA, USA, 2016; Volume 76, pp. 351–382. [Google Scholar]
| Haloperidol | Chlorpromazine | Quetiapine | |||||||
|---|---|---|---|---|---|---|---|---|---|
| <10% | 10–25% | 25–50% | <10% | 10–25% | 25–50% | <10% | 10–25% | 25–50% | |
| SuccK | SNRNP70 ** | HSP90AA1 * | RPF2 | MATR3 ** | NPM1 * | ACLY | CNP ** | HSPD1 * | ACTBL2 |
| DHX9 * | RPS3A * | ACLY * | RDX * | CPT2 | RPS15 ** | PARP1 * | BCLAF1 | ||
| FRMPD2 | SEPT11 * | MAPRE1 | BTF3 | ITGB1 | ILF3 * | RAB5C * | DDX49 | ||
| TOP2A | LRPPRC # | PTBP3 | DDX5 | ||||||
| NACA | DYNC1H1 | ||||||||
| PCNA | FIP1L1 | ||||||||
| PKM | HSPA7 | ||||||||
| SLIRP | NEFM | ||||||||
| TOP1 | NUP62 | ||||||||
| RPS10 | |||||||||
| TUBB | |||||||||
| MalK | CCT2 * | ALDOB * | ABCE1 | ACLY ** | API5 * | DSTN | CLTCL1 ** | POLDIP3 * | ARF1 |
| KATNAL2 * | PRKCSH * | DLG3 | ESD ** | HNRNPLL * | HSPA4 | GNL3 ** | BUD31 | ||
| RPLP0 * | PPP2R1A | HIST1H1E ** | RPL27A * | HSPA7 | COPA | ||||
| BANF1 | RAB7A | DDX21 | RUVBL2 * | LDHC | EMG1 | ||||
| SEC61A1 | HSP90B1 | ACO2 | MYH13 | HNRNPU | |||||
| TBC1D4 | HNRNPM | PHGDH | MYL6 | ||||||
| HNRNPU | POTEKP | SLC3A2 | |||||||
| HSP90B1 | SPIN3 | ||||||||
| RPS14 | |||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smith, B.J.; Brandão-Teles, C.; Zuccoli, G.S.; Reis-de-Oliveira, G.; Fioramonte, M.; Saia-Cereda, V.M.; Martins-de-Souza, D. Protein Succinylation and Malonylation as Potential Biomarkers in Schizophrenia. J. Pers. Med. 2022, 12, 1408. https://doi.org/10.3390/jpm12091408
Smith BJ, Brandão-Teles C, Zuccoli GS, Reis-de-Oliveira G, Fioramonte M, Saia-Cereda VM, Martins-de-Souza D. Protein Succinylation and Malonylation as Potential Biomarkers in Schizophrenia. Journal of Personalized Medicine. 2022; 12(9):1408. https://doi.org/10.3390/jpm12091408
Chicago/Turabian StyleSmith, Bradley Joseph, Caroline Brandão-Teles, Giuliana S. Zuccoli, Guilherme Reis-de-Oliveira, Mariana Fioramonte, Verônica M. Saia-Cereda, and Daniel Martins-de-Souza. 2022. "Protein Succinylation and Malonylation as Potential Biomarkers in Schizophrenia" Journal of Personalized Medicine 12, no. 9: 1408. https://doi.org/10.3390/jpm12091408
APA StyleSmith, B. J., Brandão-Teles, C., Zuccoli, G. S., Reis-de-Oliveira, G., Fioramonte, M., Saia-Cereda, V. M., & Martins-de-Souza, D. (2022). Protein Succinylation and Malonylation as Potential Biomarkers in Schizophrenia. Journal of Personalized Medicine, 12(9), 1408. https://doi.org/10.3390/jpm12091408








