Genome-Wide Association Study Identifies Multiple Susceptibility Loci for Malignant Neoplasms of the Brain in Taiwan
Abstract
1. Introduction
2. Materials and Methods
2.1. Genotyping and SNP Selection
2.2. Genotype-Phenotype Association Analysis
2.3. Establishment of Brain Neoplasm-Related SNP Genotype Scores
2.4. i-GSEA4GWAS
2.5. Statistical Analysis
3. Results
3.1. To Identify Brain Neoplasms’ SNP-Related Susceptibility Loci in Han Ancestry Populations
3.2. Relationship between Novel SNPs and the Brain Neoplasm Profile
3.3. Pathway Enrichment of Glioma GWAS SNPs
3.4. Survival Analysis of Malignant Neoplasms in Taiwan
4. Discussion
4.1. Possible Mechanisms That Link These SNPs with Malignant Brain Tumors
4.2. Limitations of Our Study
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Krishnatreya, M.; Kataki, A.C.; Sharma, J.D.; Bhattacharyya, M.; Nandy, P.; Hazarika, M. Brief descriptive epidemiology of primary malignant brain tumors from north-east india. Asian Pac. J. Cancer Prev. 2014, 15, 9871–9873. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 who classification of tumors of the central nervous system: A summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Hess, K.R.; Broglio, K.R.; Bondy, M.L. Adult glioma incidence trends in the united states, 1977–2000. Cancer 2004, 101, 2293–2299. [Google Scholar] [CrossRef] [PubMed]
- Eckel-Passow, J.E.; Lachance, D.H.; Jenkins, R.B. Glioma: Interaction of acquired and germline genetics. Aging 2021, 13, 19085–19087. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.P.Y.; Li, B.K.; Pfaff, E.; Gudenas, B.; Vasiljevic, A.; Orr, B.A.; Dufour, C.; Snuderl, M.; Karajannis, M.A.; Rosenblum, M.K.; et al. Clinical and molecular heterogeneity of pineal parenchymal tumors: A consensus study. Acta Neuropathol. 2021, 141, 771–785. [Google Scholar] [CrossRef] [PubMed]
- Tebani, A.; Jotanovic, J.; Hekmati, N.; Sivertsson, A.; Gudjonsson, O.; Engstrom, B.E.; Wikstrom, J.; Uhlen, M.; Casar-Borota, O.; Ponten, F. Annotation of pituitary neuroendocrine tumors with genome-wide expression analysis. Acta Neuropathol. Commun. 2021, 9, 181. [Google Scholar] [CrossRef]
- Kinnersley, B.; Labussiere, M.; Holroyd, A.; Di Stefano, A.L.; Broderick, P.; Vijayakrishnan, J.; Mokhtari, K.; Delattre, J.Y.; Gousias, K.; Schramm, J.; et al. Genome-wide association study identifies multiple susceptibility loci for glioma. Nat. Commun. 2015, 6, 8559. [Google Scholar] [CrossRef]
- Kinnersley, B.; Houlston, R.S.; Bondy, M.L. Genome-wide association studies in glioma. Cancer Epidemiol. Biomark. Prev. 2018, 27, 418–428. [Google Scholar] [CrossRef]
- Fiatal, S.; Ádány, R. Application of single-nucleotide polymorphism-related risk estimates in identification of increased genetic susceptibility to cardiovascular diseases: A literature review. Front. Public Health 2017, 5, 358. [Google Scholar] [CrossRef]
- Kinnersley, B.; Mitchell, J.S.; Gousias, K.; Schramm, J.; Idbaih, A.; Labussiere, M.; Marie, Y.; Rahimian, A.; Wichmann, H.E.; Schreiber, S.; et al. Quantifying the heritability of glioma using genome-wide complex trait analysis. Sci. Rep. 2015, 5, 17267. [Google Scholar] [CrossRef]
- Melin, B.S.; Barnholtz-Sloan, J.S.; Wrensch, M.R.; Johansen, C.; Il'yasova, D.; Kinnersley, B.; Ostrom, Q.T.; Labreche, K.; Chen, Y.; Armstrong, G.; et al. Genome-wide association study of glioma subtypes identifies specific differences in genetic susceptibility to glioblastoma and non-glioblastoma tumors. Nat. Genet. 2017, 49, 789–794. [Google Scholar] [CrossRef] [PubMed]
- Rajaraman, P.; Melin, B.S.; Wang, Z.; McKean-Cowdin, R.; Michaud, D.S.; Wang, S.S.; Bondy, M.; Houlston, R.; Jenkins, R.B.; Wrensch, M.; et al. Genome-wide association study of glioma and meta-analysis. Hum. Genet. 2012, 131, 1877–1888. [Google Scholar] [CrossRef] [PubMed]
- Busek, P.; Stremenova, J.; Sedo, A. Dipeptidyl peptidase-iv enzymatic activity bearing molecules in human brain tumors--good or evil? Front. Biosci. 2008, 13, 2319–2326. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Zhang, Z.; Fregoso, O.; Krainer, A.R. Mechanisms of activation and repression by the alternative splicing factors rbfox1/2. RNA 2012, 18, 274–283. [Google Scholar] [CrossRef]
- Hu, J.; Ho, A.L.; Yuan, L.; Hu, B.; Hua, S.; Hwang, S.S.; Zhang, J.; Hu, T.; Zheng, H.; Gan, B.; et al. From the cover: Neutralization of terminal differentiation in gliomagenesis. Proc. Natl. Acad. Sci. USA 2013, 110, 14520–14527. [Google Scholar] [CrossRef]
- Shen, S.; Yang, C.; Liu, X.; Zheng, J.; Liu, Y.; Liu, L.; Ma, J.; Ma, T.; An, P.; Lin, Y.; et al. Rbfox1 regulates the permeability of the blood-tumor barrier via the linc00673/maff pathway. Mol. Ther. Oncolytics 2020, 17, 138–152. [Google Scholar] [CrossRef]
- Poulos, R.C.; Thoms, J.A.; Shah, A.; Beck, D.; Pimanda, J.E.; Wong, J.W. Systematic screening of promoter regions pinpoints functional cis-regulatory mutations in a cutaneous melanoma genome. Mol. Cancer Res. 2015, 13, 1218–1226. [Google Scholar] [CrossRef]
- Choi, S.; Ha, M.; Lee, J.S.; Heo, H.J.; Kim, G.H.; Oh, S.O.; Lee, J.J.; Goh, T.S.; Kim, Y.H. Novel prognostic factor for uveal melanoma: Bioinformatics analysis of three independent cohorts. Anticancer Res. 2020, 40, 3839–3846. [Google Scholar] [CrossRef]
- Leonardi, R.; Subramanian, C.; Jackowski, S.; Rock, C.O. Cancer-associated isocitrate dehydrogenase mutations inactivate nadph-dependent reductive carboxylation. J. Biol. Chem. 2012, 287, 14615–14620. [Google Scholar] [CrossRef]
- Ghosh, D.; Funk, C.C.; Caballero, J.; Shah, N.; Rouleau, K.; Earls, J.C.; Soroceanu, L.; Foltz, G.; Cobbs, C.S.; Price, N.D.; et al. A cell-surface membrane protein signature for glioblastoma. Cell Syst 2017, 4, 516–529. [Google Scholar] [CrossRef]
- Mao, H.Z.; Ehrhardt, N.; Bedoya, C.; Gomez, J.A.; DeZwaan-McCabe, D.; Mungrue, I.N.; Kaufman, R.J.; Rutkowski, D.T.; Peterfy, M. Lipase maturation factor 1 (lmf1) is induced by endoplasmic reticulum stress through activating transcription factor 6alpha (atf6alpha) signaling. J. Biol. Chem. 2014, 289, 24417–24427. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.Y.; Pan, P.H.; Wu, C.C.; Liao, S.L.; Chen, W.Y.; Kuan, Y.H.; Wang, W.Y.; Chen, C.J. Endoplasmic reticulum stress contributes to gefitinib-induced apoptosis in glioma. Int. J. Mol. Sci. 2021, 22, 3934. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.C.; Tsai, J.T.; Chao, T.Y.; Ma, H.I.; Chien, C.S.; Liu, W.H. Msi1 associates glioblastoma radioresistance via homologous recombination repair, tumor invasion and cancer stem-like cell properties. Radiother. Oncol. 2018, 129, 352–363. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.C.; Tsai, J.T.; Chao, T.Y.; Ma, H.I.; Liu, W.H. Musashi-1 enhances glioblastoma migration by promoting icam1 translation. Neoplasia 2019, 21, 459–468. [Google Scholar] [CrossRef]
- Lin, J.C.; Kuo, C.Y.; Tsai, J.T.; Liu, W.H. Mir-671-5p inhibition by msi1 promotes glioblastoma tumorigenesis via radioresistance, tumor motility and cancer stem-like cell properties. Biomedicines 2021, 10, 21. [Google Scholar] [CrossRef]
- Oakes, S.A. Endoplasmic reticulum stress signaling in cancer cells. Am. J. Pathol. 2020, 190, 934–946. [Google Scholar] [CrossRef]
- Lee, H.K.; Xiang, C.; Cazacu, S.; Finniss, S.; Kazimirsky, G.; Lemke, N.; Lehman, N.L.; Rempel, S.A.; Mikkelsen, T.; Brodie, C. Grp78 is overexpressed in glioblastomas and regulates glioma cell growth and apoptosis. Neuro Oncol. 2008, 10, 236–243. [Google Scholar] [CrossRef]
- Wei, B.; Wang, R.; Wang, L.; Du, C. Prognostic factor identification by analysis of the gene expression and DNA methylation data in glioma. Math. Biosci. Eng. 2020, 17, 3909–3924. [Google Scholar] [CrossRef]
- Zhang, Z.; Lin, E.; Zhuang, H.; Xie, L.; Feng, X.; Liu, J.; Yu, Y. Construction of a novel gene-based model for prognosis prediction of clear cell renal cell carcinoma. Cancer Cell Int. 2020, 20, 27. [Google Scholar] [CrossRef]
- Gawron-Jakubek, W.; Spaczynska, J.; Pitynski, K.; Loster, B.W. Coexistence of tooth agenesis and ovarian cancer—A systematic literature review. Ginekol. Pol. 2019, 90, 707–710. [Google Scholar] [CrossRef]
- Lawrence, M.G.; Pidsley, R.; Niranjan, B.; Papargiris, M.; Pereira, B.A.; Richards, M.; Teng, L.; Norden, S.; Ryan, A.; Frydenberg, M.; et al. Alterations in the methylome of the stromal tumour microenvironment signal the presence and severity of prostate cancer. Clin. Epigenetics 2020, 12, 48. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Bai, Y.T.; Han, K.; Li, X.D.; Meng, J. Knockdown of ectodysplasin-a receptor-associated adaptor protein exerts a tumor-suppressive effect in tongue squamous cell carcinoma cells. Exp. Ther. Med. 2020, 19, 3337–3347. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Liu, Y.; Liu, Q.; Cummins, S.; Zhao, M. Integrative proteomic analysis reveals potential high-frequency alternative open reading frame-encoded peptides in human colorectal cancer. Life Sci. 2018, 215, 182–189. [Google Scholar] [CrossRef] [PubMed]
- Thon, N.; Kreth, S.; Kreth, F.W. Personalized treatment strategies in glioblastoma: Mgmt promoter methylation status. OncoTargets Ther. 2013, 6, 1363–1372. [Google Scholar] [CrossRef] [PubMed]
- Muller, M.; Morotti, A.; Ponzetto, C. Activation of nf-kappab is essential for hepatocyte growth factor-mediated proliferation and tubulogenesis. Mol. Cell. Biol. 2002, 22, 1060–1072. [Google Scholar] [CrossRef] [PubMed]
- Tomita, N.; Morishita, R.; Taniyama, Y.; Koike, H.; Aoki, M.; Shimizu, H.; Matsumoto, K.; Nakamura, T.; Kaneda, Y.; Ogihara, T. Angiogenic property of hepatocyte growth factor is dependent on upregulation of essential transcription factor for angiogenesis, ets-1. Circulation 2003, 107, 1411–1417. [Google Scholar] [CrossRef]
- Huang, M.; Liu, T.; Ma, P.; Mitteer, R.A.; Zhang, Z.; Kim, H.J.; Yeo, E.; Zhang, D.; Cai, P.; Li, C.; et al. C-met-mediated endothelial plasticity drives aberrant vascularization and chemoresistance in glioblastoma. J. Clin. Invest. 2016, 126, 1801–1814. [Google Scholar] [CrossRef]
- Vaglini, F.; Pardini, C.; Di Desidero, T.; Orlandi, P.; Pasqualetti, F.; Ottani, A.; Pacini, S.; Giuliani, D.; Guarini, S.; Bocci, G. Melanocortin receptor-4 and glioblastoma cells: Effects of the selective antagonist ml00253764 alone and in combination with temozolomide in vitro and in vivo. Mol. Neurobiol. 2018, 55, 4984–4997. [Google Scholar] [CrossRef]
- Rahane, C.S.; Kutzner, A.; Heese, K. A cancer tissue-specific fam72 expression profile defines a novel glioblastoma multiform (gbm) gene-mutation signature. J. Neurooncol. 2019, 141, 57–70. [Google Scholar] [CrossRef]
- Tabouret, E.; Labussiere, M.; Alentorn, A.; Schmitt, Y.; Marie, Y.; Sanson, M. Lrp1b deletion is associated with poor outcome for glioblastoma patients. J. Neurol. Sci. 2015, 358, 440–443. [Google Scholar] [CrossRef]
- Annabi, B.; Doumit, J.; Plouffe, K.; Laflamme, C.; Lord-Dufour, S.; Beliveau, R. Members of the low-density lipoprotein receptor-related proteins provide a differential molecular signature between parental and cd133+ daoy medulloblastoma cells. Mol. Carcinog. 2010, 49, 710–717. [Google Scholar] [CrossRef] [PubMed]
- Ni, X.; Ji, C.; Cao, G.; Cheng, H.; Guo, L.; Gu, S.; Ying, K.; Zhao, R.C.; Mao, Y. Molecular cloning and characterization of the protein 4.1o gene, a novel member of the protein 4.1 family with focal expression in ovary. J. Hum. Genet. 2003, 48, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Haase, D.; Meister, M.; Muley, T.; Hess, J.; Teurich, S.; Schnabel, P.; Hartenstein, B.; Angel, P. Frmd3, a novel putative tumour suppressor in nsclc. Oncogene 2007, 26, 4464–4468. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Loftus, J.C.; Yang, Z.; Kloss, J.; Dhruv, H.; Tran, N.L.; Riggs, D.L. A novel interaction between pyk2 and map4k4 is integrated with glioma cell migration. J. Signal Transduct. 2013, 2013, 956580. [Google Scholar] [CrossRef]
- Chen, T.J.; Chou, C.L.; Tian, Y.F.; Yeh, C.F.; Chan, T.C.; He, H.L.; Li, W.S.; Tsai, H.H.; Li, C.F.; Lai, H.Y. High frmd3 expression is prognostic for worse survival in rectal cancer patients treated with ccrt. Int. J. Clin. Oncol. 2021, 26, 1689–1697. [Google Scholar] [CrossRef]
- Sweetser, D.A.; Peniket, A.J.; Haaland, C.; Blomberg, A.A.; Zhang, Y.; Zaidi, S.T.; Dayyani, F.; Zhao, Z.; Heerema, N.A.; Boultwood, J.; et al. Delineation of the minimal commonly deleted segment and identification of candidate tumor-suppressor genes in del(9q) acute myeloid leukemia. Genes Chromosomes Cancer 2005, 44, 279–291. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, K.; Yu, Q. Frmd6 inhibits human glioblastoma growth and progression by negatively regulating activity of receptor tyrosine kinases. Oncotarget 2016, 7, 70080–70091. [Google Scholar] [CrossRef][Green Version]
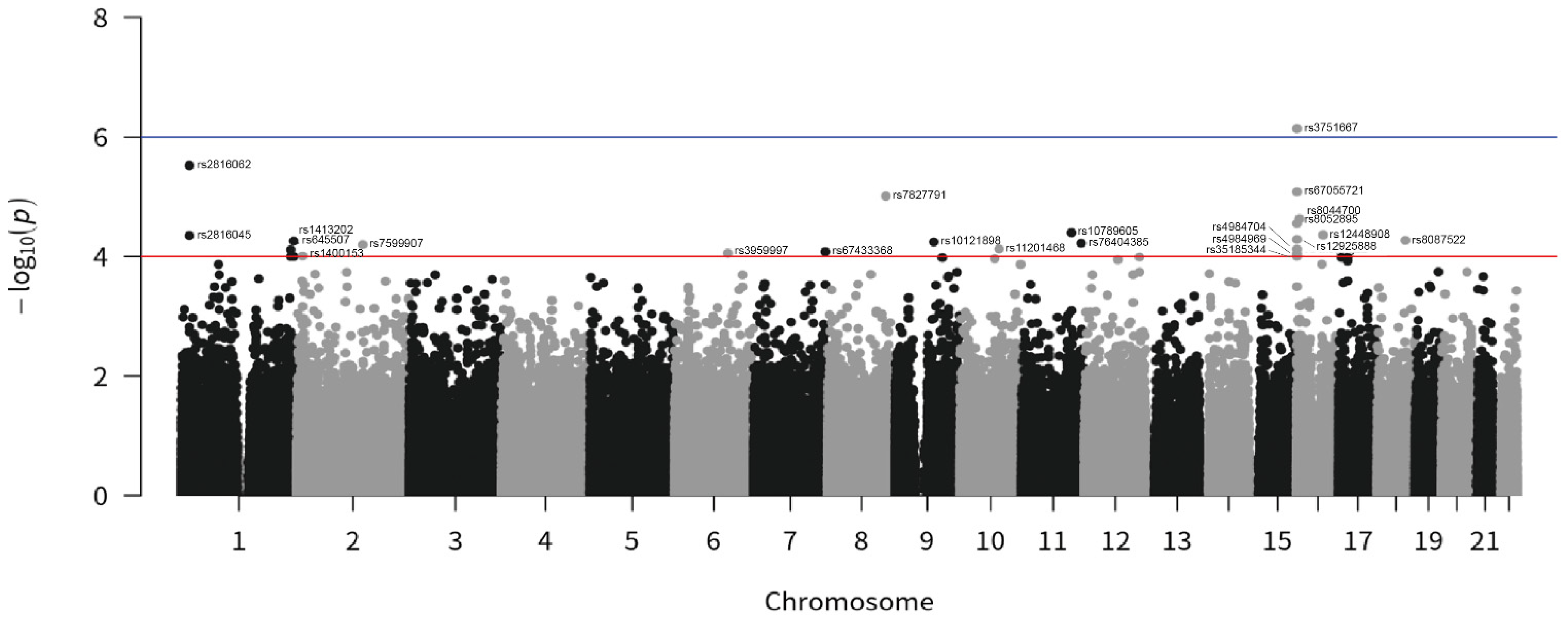



| SNP | Symbol | Nearest Gene | Cytogenetic Band | Loci (GRCh38.p13) | All Cases | East Asia | |
|---|---|---|---|---|---|---|---|
| p | OR | Alternative Allele | |||||
| rs2816045 | - | PAX7 | 1p36.13 | 1:18566505-18566505 | 4.44 × 10−5 | 1.86 | G = 0.3800 |
| rs2816062 | - | PAX7 | 1p36.13 | 1:18577408-18577408 | 2.99 × 10−6 | 2.03 | A = 0.3720 |
| rs645507 | EDARADD | - | 1q42.3 | 1:236409960-236409960 | 7.71 × 10−5 | 1.89 | A = 0.3167 |
| rs653545 | EDARADD | - | 1q42.3 | 1:236422530-236422530 | 1.01 × 10−4 | 1.91 | C = 0.3000 |
| rs1413202 | - | PLD5 | 1q43 | 1:242708730-242708730 | 5.49 × 10−5 | 1.83 | C = 0.4033 |
| rs34992957 | - | PLD5 | 1q43 | 1:242712199-242712199 | 1.01 × 10−4 | 1.83 | C = 0.3400 |
| rs11201468 | - | ENST00000656796 | 10q23.1 | 10:85206976-85206976 | 7.47 × 10−5 | 0.31 | T = 0.0500 |
| rs10789605 | - | ENSG00000261098 | 10q23.1 | 11:107313859-107313859 | 3.97 × 10−5 | 0.51 | G = 0.2867 |
| rs76404385 | ETS1 | - | 11q24.3 | 11:128463160-128463160 | 6.00 × 10−5 | 0.44 | T = 0.152 |
| rs4984704 | LMF1 | - | 16p13.3 | 16:890713-890713 | 7.57 × 10−5 | 1.86 | G = 0.354 |
| rs8052895 | LMF1 | - | 16p13.3 | 16:897817-897817 | 2.82 × 10−5 | 1.91 | T = 0.382 |
| rs4984969 | LMF1 | - | 16p13.3 | 16:907885-907885 | 9.11 × 10−5 | 1.95 | G = 0.293 |
| rs12925888 | LMF1 | - | 16p13.3 | 16:927944-927944 | 5.16 × 10−5 | 1.94 | G = 0.34 |
| rs3751667 | LMF1 | - | 16p13.3 | 16:954554-954554 | 7.24 × 10−7 | 2.17 | T = 0.412 |
| rs35185344 | LMF1 | - | 16p13.3 | 16:966608-966612 | 9.92 × 10−5 | 1.79 | ATATA = 0.44 |
| rs67055721 | LMF1 | - | 16p13.3 | 16:972244-972249 | 8.28 × 10−6 | 2.09 | C = 0.34 |
| rs8044700 | RBFOX1 | - | 16p13.3 | 16:6093700-6093700 | 2.35 × 10−5 | 2.36 | A = 0.84 |
| rs12448908 | - | FTO | 16q12.2 | 16:54205958-54205958 | 4.64 × 10−5 | 0.53 | C = 0.69 |
| rs8087522 | MC4R | - | 18q21.32 | 18:60373245-60373245 | 5.35 × 10−5 | 0.37 | A = 0.123 |
| rs1400153 | - | ENSG00000285876 | 2p24.3 | 2:13029448-13029448 | 9.79 × 10−5 | 0.57 | G = 0.472 |
| rs7599907 | LRP1B | - | 2q22.2 | 2:141433506-141433506 | 6.29 × 10−5 | 0.52 | C = 0.70 |
| rs3959997 | - | FRK | 6q22.1 | 6:115838393-115838393 | 8.82 × 10−5 | 0.48 | T = 0.19 |
| rs67433368 | DPP6 | - | 7q36.2 | 7:154353490-154353490 | 8.32 × 10−5 | 3.13 | A = 0.01 |
| rs7827791 | NDUFB9 | - | 8q24.13 | 8:124549270-124549270 | 9.73 × 10−6 | 1.91 | G = 0.52 |
| rs10121898 | FRMD3 | - | 9q21.32 | 9:83324595-83324595 | 5.70 × 10−5 | 0.46 | A = 0.07 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, J.-C.; Wu, Y.-C.; Yang, F.-C.; Tsai, J.-T.; Huang, D.Y.; Liu, W.-H. Genome-Wide Association Study Identifies Multiple Susceptibility Loci for Malignant Neoplasms of the Brain in Taiwan. J. Pers. Med. 2022, 12, 1161. https://doi.org/10.3390/jpm12071161
Lin J-C, Wu Y-C, Yang F-C, Tsai J-T, Huang DY, Liu W-H. Genome-Wide Association Study Identifies Multiple Susceptibility Loci for Malignant Neoplasms of the Brain in Taiwan. Journal of Personalized Medicine. 2022; 12(7):1161. https://doi.org/10.3390/jpm12071161
Chicago/Turabian StyleLin, Jang-Chun, Yi-Chieh Wu, Fu-Chi Yang, Jo-Ting Tsai, David YC Huang, and Wei-Hsiu Liu. 2022. "Genome-Wide Association Study Identifies Multiple Susceptibility Loci for Malignant Neoplasms of the Brain in Taiwan" Journal of Personalized Medicine 12, no. 7: 1161. https://doi.org/10.3390/jpm12071161
APA StyleLin, J.-C., Wu, Y.-C., Yang, F.-C., Tsai, J.-T., Huang, D. Y., & Liu, W.-H. (2022). Genome-Wide Association Study Identifies Multiple Susceptibility Loci for Malignant Neoplasms of the Brain in Taiwan. Journal of Personalized Medicine, 12(7), 1161. https://doi.org/10.3390/jpm12071161







