Automatic Segmentation for Favourable Delineation of Ten Wrist Bones on Wrist Radiographs Using Convolutional Neural Network
Abstract
1. Introduction
2. Methods
2.1. Study Design
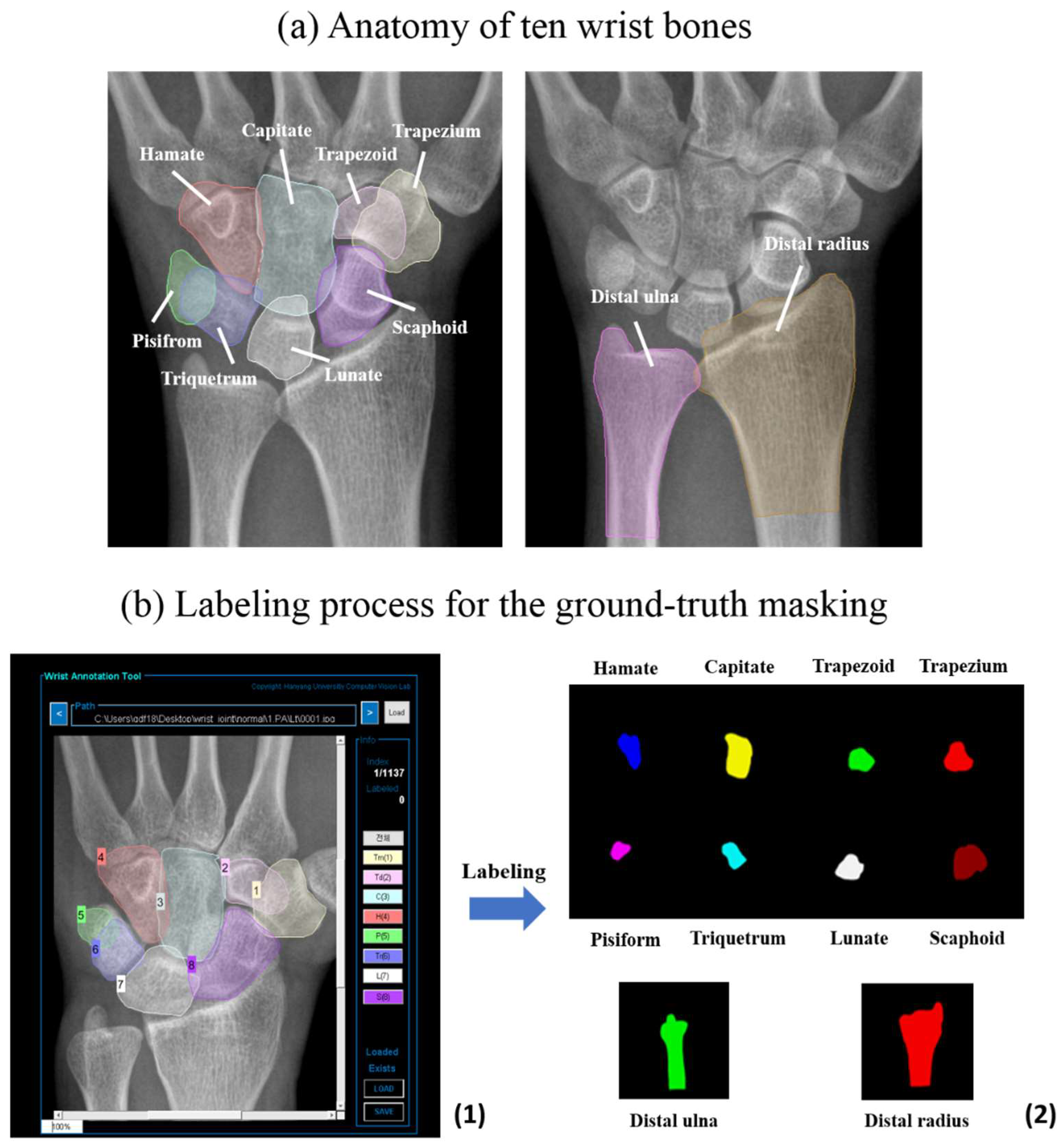
2.2. Dataset of Participants
2.3. Data Pre-Processing
2.4. Network Architecture
2.4.1. Wrist ROI Detection Model Using Single-Shot Multibox Detector (SSD)
2.4.2. Wrist Segmentation Model Using Mask R-CNN with the Extended Mask Head
2.4.3. Training and Validation of Automatic Segmentation Using Fine Mask R-CNN and Mask R-CNN
2.5. Primary Outcomes and Quantitative Evaluation
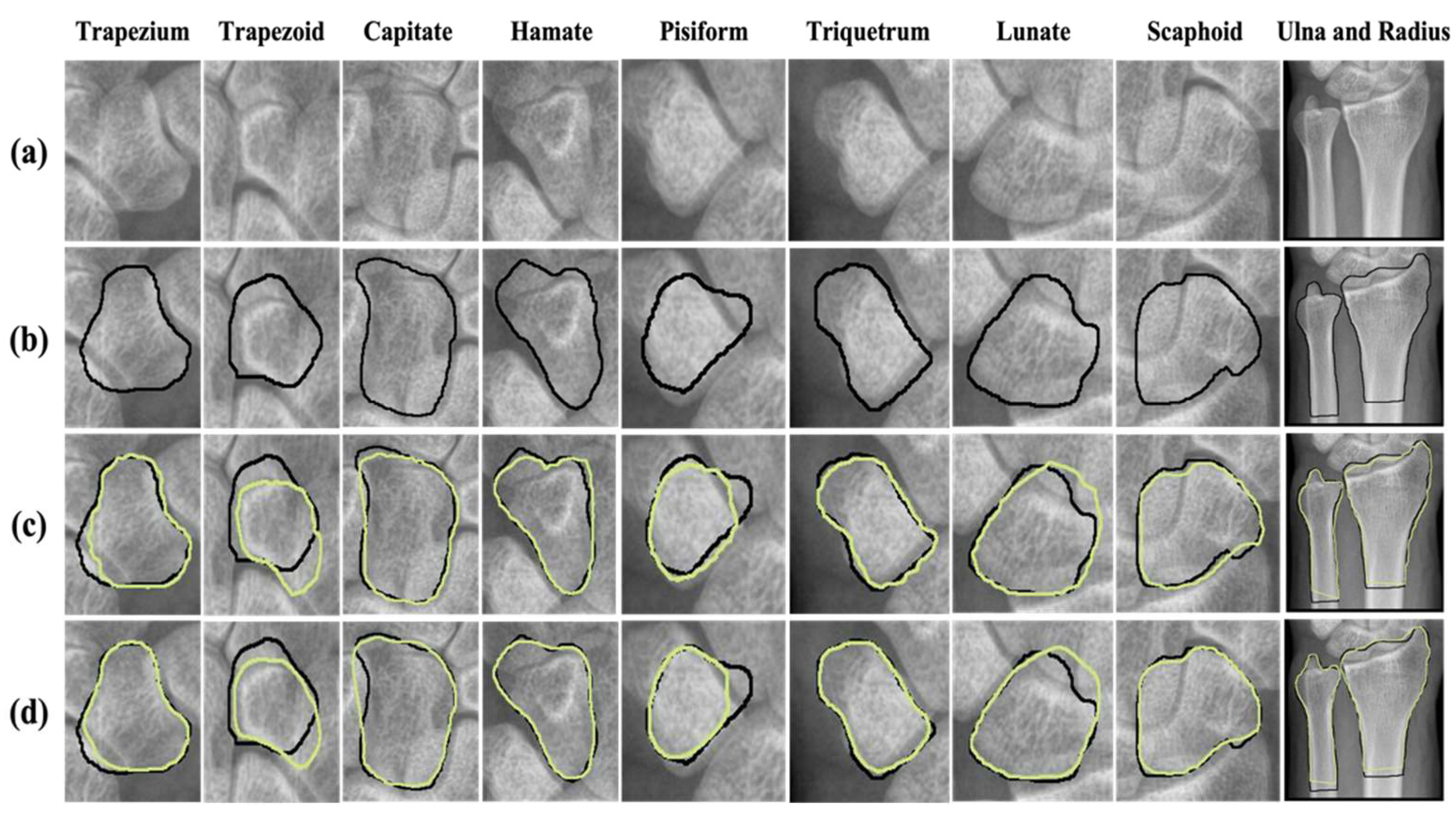
2.6. Visualization of Predicted Masking of Wrist Bones through Automatic Segmentation by Networks
2.7. Statistical Analysis
3. Results
3.1. The Performance Test between the Fine Mask R-CNN and the Mask R-CNN for the Automatic Segmentation of Wrist Bones
3.2. The Turing Test between Ground Truth Masking by Clinicians and Masking Predicted by Fine Mask R-CNN for the Automatic Segmentation of Wrist Bones
3.3. Visualization of Predicted Masking for Wrist Bone Segmentation by Two Networks
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CNN | convolution neural network |
| PA | Posteroanterior |
| AP | Anteroposterior |
| PACS | picture archiving and communication system |
| ROI | region of interest |
| SSD | Single-Shot Multibox Detector |
| SGD | stochastic gradient descent |
| Dice | Dice coefficient |
| IQR | interquartile ranges |
| SD | standard deviation |
| ICC | intraclass correlation coefficient |
References
- Larsen, C.F.; Lauritsen, J. Epidemiology of acute wrist trauma. Int. J. Epidemiol. 1993, 22, 911–916. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, R.; Riley, N.D.; Wijendra, A.; Thurley, N.; Carr, A.J.; Bjf, D. Wrist pain: A systematic review of prevalence and risk factors-What is the role of occupation and activity? BMC Musculoskelet. Disord. 2019, 20, 542. [Google Scholar] [CrossRef] [PubMed]
- Linn, M.R.; Mann, F.A.; Gilula, L.A. Imaging the symptomatic wrist. Orthop. Clin. N. Am. 1990, 21, 515–543. [Google Scholar] [CrossRef]
- Lee, R.K.; Griffith, J.F.; Ng, A.W.; Wong, C.W. Imaging of radial wrist pain. I. Imaging modalities and anatomy. Skelet. Radiol. 2014, 43, 713–724. [Google Scholar] [CrossRef] [PubMed]
- Porteous, R.; Harish, S.; Parasu, N. Imaging of ulnar-sided wrist pain. Can. Assoc. Radiol. J. 2012, 63, 18–29. [Google Scholar] [CrossRef]
- Bhat, A.K.; Kumar, B.; Acharya, A. Radiographic imaging of the wrist. Indian J. Plast. Surg. 2011, 44, 186–196. [Google Scholar] [CrossRef]
- Hemke, R.; Buckless, C.G.; Tsao, A.; Wang, B.; Torriani, M. Deep learning for automated segmentation of pelvic muscles, fat, and bone from CT studies for body composition assessment. Skelet. Radiol. 2020, 49, 387–395. [Google Scholar] [CrossRef]
- Rehman, F.; Shah, S.I.A.; Riaz, M.N.; Gilani, S.O. A region-based deep level set formulation for vertebral bone segmentation of osteoporotic fractures. J. Digit. Imaging 2020, 33, 191–203. [Google Scholar] [CrossRef]
- Peng, L.-Q.; Guo, Y.-C.; Wan, L.; Liu, T.-A.; Wang, P.; Zhao, H.; Wang, Y.-H. Forensic bone age estimation of adolescent pelvis X-rays based on two-stage convolutional neural network. Int. J. Leg. Med. 2022, 136, 797–810. [Google Scholar] [CrossRef]
- Foster, B.; Joshi, A.; Borgese, M.; Abdelhafez, Y.; Boutin, R.D.; Chaudhari, A.J. WRIST: A WRist Image Segmentation Toolkit for carpal bone delineation from MRI. Comput. Med. Imaging Graph. 2018, 63, 31–40. [Google Scholar] [CrossRef]
- Hendrix, N.; Scholten, E.; Vernhout, B.; Bruijnen, S.; Maresch, B.; de Jong, M.; Diepstraten, S.; Bollen, S.; Schalekamp, S.; de Rooij, M.; et al. Development and Validation of a Convolutional Neural Network for Automated Detection of Scaphoid Fractures on Conventional Radiographs. Radiol. Artif. Intell. 2021, 3, e200260. [Google Scholar] [CrossRef] [PubMed]
- Manos, G.; Cairns, A.; Ricketts, I.; Sinclair, D. Automatic segmentation of hand-wrist radiographs. Image Vis. Comput. 1993, 11, 100–111. [Google Scholar] [CrossRef]
- Zhang, A.; Gertych, A.; Liu, B.J. Automatic bone age assessment for young children from newborn to 7-year-old using carpal bones. Comput. Med. Imaging Graph. 2007, 31, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Włodarczyk, J.; Czaplicka, K.; Tabor, Z.; Wojciechowski, W.; Urbanik, A. Segmentation of bones in magnetic resonance images of the wrist. Int. J. Comput. Assist. Radiol. Surg. 2015, 10, 419–431. [Google Scholar] [CrossRef][Green Version]
- Włodarczyk, J.; Wojciechowski, W.; Czaplicka, K.; Urbanik, A.; Tabor, Z. Fast automated segmentation of wrist bones in magnetic resonance images. Comput. Biol. Med. 2015, 65, 44–53. [Google Scholar] [CrossRef]
- Koch, M.; Schwing, A.G.; Comaniciu, D.; Pollefeys, M. Fully automatic segmentation of wrist bones for arthritis patients. In Proceedings of the 2011 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Chicago, IL, USA, 30 March–2 April 2011; pp. 636–640. [Google Scholar]
- Gou, X.; Rao, Y.; Feng, X.; Yun, Z.; Yang, W. Automatic Segmentation of Ulna and Radius in Forearm Radiographs. Comput. Math Methods Med. 2019, 2019, 6490161. [Google Scholar] [CrossRef]
- Manos, G.K.; Cairns, A.Y.; Rickets, I.W.; Sinclair, D. Segmenting radiographs of the hand and wrist. Comput. Methods Programs Biomed. 1994, 43, 227–237. [Google Scholar] [CrossRef]
- Canny, J. A computational approach to edge detection. IEEE Trans. Pattern Anal. Mach. Intell. 1986, 6, 679–698. [Google Scholar] [CrossRef]
- Li, J.; Nebelung, S.; Schock, J.; Rath, B.; Tingart, M.; Liu, Y.; Siroros, N.; Eschweiler, J. A Novel Combined Level Set Model for Carpus Segmentation from Magnetic Resonance Images with Prior Knowledge aligned in Polar Coordinate System. Comput. Methods Programs Biomed. 2021, 208, 106245. [Google Scholar] [CrossRef]
- Sebastian, T.B.; Tek, H.; Crisco, J.J.; Kimia, B.B. Segmentation of carpal bones from CT images using skeletally coupled deformable models. Med. Image Anal. 2003, 7, 21–45. [Google Scholar] [CrossRef]
- Faisal, A.; Khalil, A.; Chai, H.Y.; Lai, K.W. X-ray carpal bone segmentation and area measurement. Multimed. Tools Appl. 2021, 1–12. [Google Scholar] [CrossRef]
- Giordano, D.; Spampinato, C.; Scarciofalo, G.; Leonardi, R. An automatic system for skeletal bone age measurement by robust processing of carpal and epiphysial/metaphysial bones. IEEE Trans. Instrum. Meas. 2010, 59, 2539–2553. [Google Scholar] [CrossRef]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- Kwon, G.; Ryu, J.; Oh, J.; Lim, J.; Kang, B.-K.; Ahn, C.; Bae, J.; Lee, D.K. Deep learning algorithms for detecting and visualising intussusception on plain abdominal radiography in children: A retrospective multicenter study. Sci. Rep. 2020, 10, 17582. [Google Scholar] [CrossRef] [PubMed]
- Guan, Q.; Huang, Y.; Zhong, Z.; Zheng, Z.; Zheng, L.; Yang, Y. Diagnose like a radiologist: Attention guided convolutional neural network for thorax disease classification. arXiv 2018, arXiv:1801.09927. [Google Scholar]
- Liu, W.; Anguelov, D.; Erhan, D.; Szegedy, C.; Reed, S.; Fu, C.Y.; Berg, A.C. SSD: Single-Shot Multibox Detector. In European Conference on Computer Vision; Springer: Cham, Switzerland, 2016. [Google Scholar] [CrossRef]
- Alsinan, A.Z.; Patel, V.M.; Hacihaliloglu, I. Automatic segmentation of bone surfaces from ultrasound using a filter-layer-guided CNN. Int. J. Comput. Assist. Radiol. Surg. 2019, 14, 775–783. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhou, X.; Wang, M.; Qiu, J.; Cao, M.; Mao, K. ARU-Net: Research and Application for Wrist Reference Bone Segmentation. In Proceedings of the 2019 International Conference on Wireless and Mobile Computing, Networking and Communications (WiMob), Barcelona, Spain, 21–23 October 2019; pp. 1–5. [Google Scholar]
- Nakatsu, K.; Rahman, R.; Morita, K.; Fujita, D.; Kobashi, S. Automatic Carpal Site Detection Method for Evaluation of Rheumatoid Arthritis Using Deep Learning. J. Adv. Comput. Intell. Intell. Inform. 2022, 26, 42–50. [Google Scholar] [CrossRef]
- Ronneberger, O.; Philipp, F.; Thomas, B. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Kirillov, A.; Wu, Y.; He, K.; Girshick, R. Pointrend: Image segmentation as rendering. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Online, 14–19 June 2020; pp. 9799–9808. [Google Scholar]
- Frid-Adar, M.; Ben-Cohen, A.; Amer, R.; Greenspan, H. Improving the Segmentation of Anatomical Structures in Chest Radiographs Using U-Net with an ImageNet Pre-Trained Encoder. In Image Analysis for Moving Organ, Breast, and Thoracic Images; Springer: Cham, Switzerland, 2018; Volume 11040, pp. 159–168. [Google Scholar] [CrossRef]
- Punn, N.S.; Agarwal, S. Automated Diagnosis of COVID-19 with Limited Posteroanterior Chest X-Ray Images Using Fine-Tuned Deep Neural Networks. Appl. Intell. 2021, 51, 2689–2702. [Google Scholar] [CrossRef]
- Koo, T.K.; Li, M.Y. A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J. Chiropr. Med. 2016, 15, 155–163. [Google Scholar] [CrossRef]
- Meng, L.K.; Khalil, A.; Nizar, M.H.A.; Nisham, M.K.; Pingguan-Murphy, B.; Hum, Y.C.; Salim, M.I.M.; Lai, K.W. Carpal bone segmentation using fully convolutional neural network. Curr. Med. Imaging Rev. 2019, 15, 983–989. [Google Scholar] [CrossRef]
- Su, L.; Fu, X.; Zhang, X.; Cheng, X.; Ma, Y.; Gan, Y.; Hu, Q. Delineation of Carpal Bones From Hand X-Ray Images Through Prior Model, and Integration of Region-Based and Boundary-Based Segmentations. IEEE Access 2018, 6, 19993–20008. [Google Scholar] [CrossRef]
- Kim, M.W.; Jung, J.; Park, S.J.; Park, Y.S.; Yi, J.H.; Yang, W.S.; Kim, J.H.; Cho, B.J.; Ha, S.O. Application of convolutional neural networks for distal radio-ulnar fracture detection on plain radiographs in the emergency room. Clin. Exp. Emerg. Med. 2021, 8, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Maki, S.; Yamazaki, T.; Wakita, H.; Toguchi, Y.; Horii, M.; Yamauchi, T.; Kawamura, K.; Aramomi, M.; Sugiyama, H.; et al. Detecting Distal Radial Fractures from Wrist Radiographs Using a Deep Convolutional Neural Network with an Accuracy Comparable to Hand Orthopedic Surgeons. J. Digit. Imaging 2022, 35, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Somkantha, K.; Theera-Umpon, N.; Auephanwiriyakul, S. Bone age assessment in young children using automatic carpal bone feature extraction and support vector regression. J. Digit. Imaging 2011, 24, 1044–1058. [Google Scholar] [CrossRef]
- Gilula, L.A. Carpal injuries: Analytical approach and case exercises. AJR Am. J. Roentgenol. 1979, 133, 513–517. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Kang, K.; Han, C.; Wang, S.; Chen, Q.; Chen, Y.; Zhang, F.; Liu, Z. A blind randomized validated convolutional neural network for auto-segmentation of clinical target volume in rectal cancer patients receiving neoadjuvant radiotherapy. Cancer Med. 2022, 11, 166–175. [Google Scholar] [CrossRef]

| Tm | Td | C | H | P | Tr | L | S | Carpal | R | U | Forearm | Total | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mask R-CNN Dice, mean [SD] | 0.92 (0.03) | 0.90 (0.05) | 0.93 (0.04) | 0.93 (0.02) | 0.91 (0.05) | 0.93 (0.02) | 0.93 (0.02) | 0.93 (0.02) | 0.92 (0.01) | 0.94 (0.02) | 0.93 (0.02) | 0.94 (0.01) | 0.93 (0.01) |
| Fine Mask R-CNN Dice, mean [SD] | 0.93 (0.03) | 0.91 (0.05) | 0.95 (0.04) | 0.95 (0.02) | 0.93 (0.04) | 0.95 (0.02) | 0.95 (0.02) | 0.96 (0.02) | 0.94 (0.01) | 0.96 (0.01) | 0.96 (0.02) | 0.96 (0.01) | 0.95 (0.01) |
| Comparison between two networks’ p-values | <0.001 * | <0.001 * | <0.001 * | <0.001 * | <0.001 * | <0.001 * | <0.001 * | <0.001 * | <0.001 * | <0.001 * | <0.001 * | <0.001 * | <0.001 * |
| Tm | Td | C | H | P | Tr | L | S | Carpal | R | U | Forearm | Total | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Prediction | Score | Median | 4 | 5 | 4 | 5 | 5 | 5 | 5 | 5 | 37 | 5 | 5 | 10 | 47 |
| IQR | 4, 5 | 5, 5 | 4, 5 | 4, 5 | 4, 5 | 4, 5 | 5, 5 | 5, 5 | 36, 38 | 5, 5 | 5, 5 | 9, 10 | 45, 48 | ||
| ICC | Mean | 0.58 | 0.59 | 0.60 | 0.60 | 0.54 | 0.77 | 0.71 | 0.31 | 0.51 | 0.61 | 0.51 | 0.56 | 0.54 | |
| 95% CI | 0.45, 0.69 | 0.46, 0.70 | 0.47, 0.70 | 0.46, 0.70 | 0.39, 0.65 | 0.70, 0.83 | 0.62, 0.78 | 0.10, 0.48 | 0.35, 0.64 | 0.48, 0.71 | 0.36, 0.63 | 0.42, 0.67 | 0.36, 0.66 | ||
| Ground Truth | Score | Median | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 39 | 5 | 5 | 10 | 48 |
| IQR | 4, 5 | 5, 5 | 4, 5 | 5, 5 | 5, 5 | 5, 5 | 5, 5 | 5, 5 | 37, 39 | 5, 5 | 5, 5 | 10, 10 | 47, 49 | ||
| ICC | Mean | 0.57 | 0.04 | 0.39 | 0.56 | 0.42 | 0.61 | 0.55 | 0.52 | 0.48 | 0.65 | 0.40 | 0.57 | 0.54 | |
| 95% CI | 0.36, 0.70 | 0.25, 0.27 | 0.21, 0.54 | 0.42, 0.67 | 0.24, 0.56 | 0.49, 0.71 | 0.41, 0.66 | 0.36, 0.64 | 0.27, 0.63 | 0.54, 0.74 | 0.22, 0.55 | 0.44, 0.68 | 0.34, 0.67 | ||
| Score between two maskings | p-value | <0.001 * | 0.25 | <0.001 * | <0.001 * | <0.001 * | <0.001 * | <0.001 * | 0.39 | <0.001 * | <0.001 * | <0.001 * | <0.001 * | <0.001 * | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, B.-k.; Han, Y.; Oh, J.; Lim, J.; Ryu, J.; Yoon, M.S.; Lee, J.; Ryu, S. Automatic Segmentation for Favourable Delineation of Ten Wrist Bones on Wrist Radiographs Using Convolutional Neural Network. J. Pers. Med. 2022, 12, 776. https://doi.org/10.3390/jpm12050776
Kang B-k, Han Y, Oh J, Lim J, Ryu J, Yoon MS, Lee J, Ryu S. Automatic Segmentation for Favourable Delineation of Ten Wrist Bones on Wrist Radiographs Using Convolutional Neural Network. Journal of Personalized Medicine. 2022; 12(5):776. https://doi.org/10.3390/jpm12050776
Chicago/Turabian StyleKang, Bo-kyeong, Yelin Han, Jaehoon Oh, Jongwoo Lim, Jongbin Ryu, Myeong Seong Yoon, Juncheol Lee, and Soorack Ryu. 2022. "Automatic Segmentation for Favourable Delineation of Ten Wrist Bones on Wrist Radiographs Using Convolutional Neural Network" Journal of Personalized Medicine 12, no. 5: 776. https://doi.org/10.3390/jpm12050776
APA StyleKang, B.-k., Han, Y., Oh, J., Lim, J., Ryu, J., Yoon, M. S., Lee, J., & Ryu, S. (2022). Automatic Segmentation for Favourable Delineation of Ten Wrist Bones on Wrist Radiographs Using Convolutional Neural Network. Journal of Personalized Medicine, 12(5), 776. https://doi.org/10.3390/jpm12050776






