FFPE-Based NGS Approaches into Clinical Practice: The Limits of Glory from a Pathologist Viewpoint
Abstract
:1. Introduction
2. Sequencing Platforms
2.1. Ion Torrent
2.2. Illumina
2.3. GeneReader
2.4. Pacific Biosciences
2.5. Oxford Nanopore Technology
3. NGS in Clinical Practice
4. Preanalytical Issue in NGS
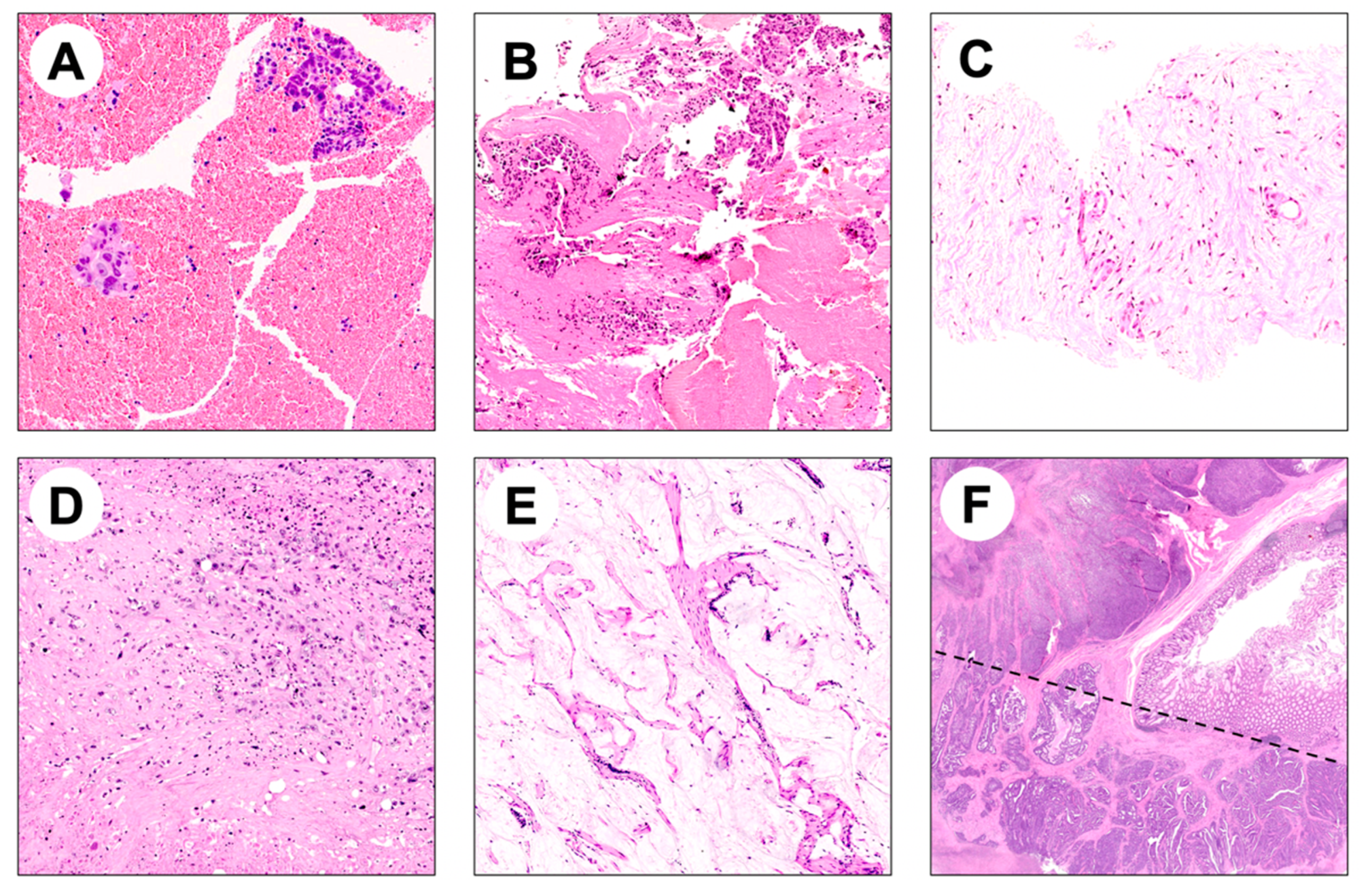
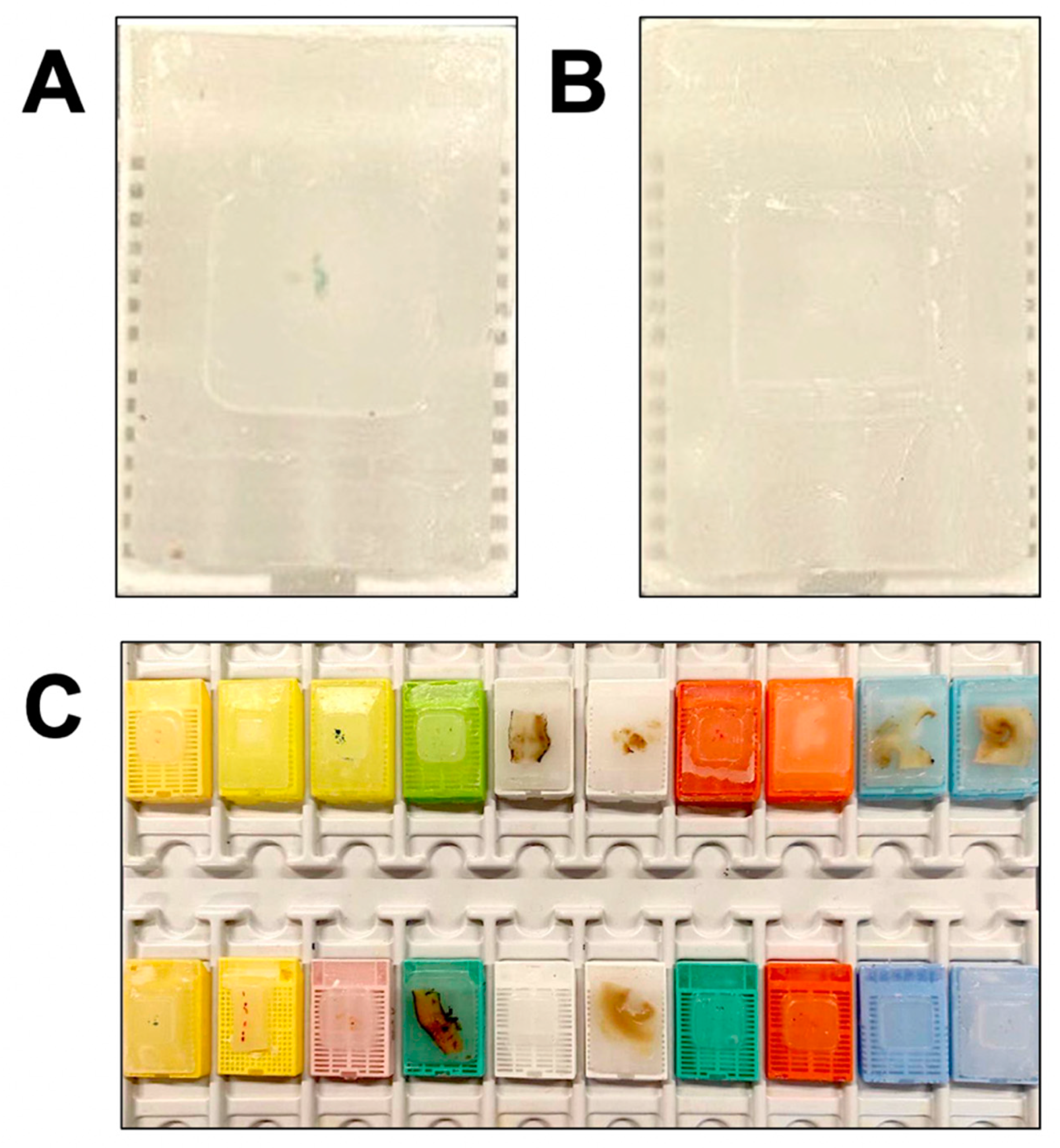
4.1. Histologic Specimens
4.2. Cytologic Specimens
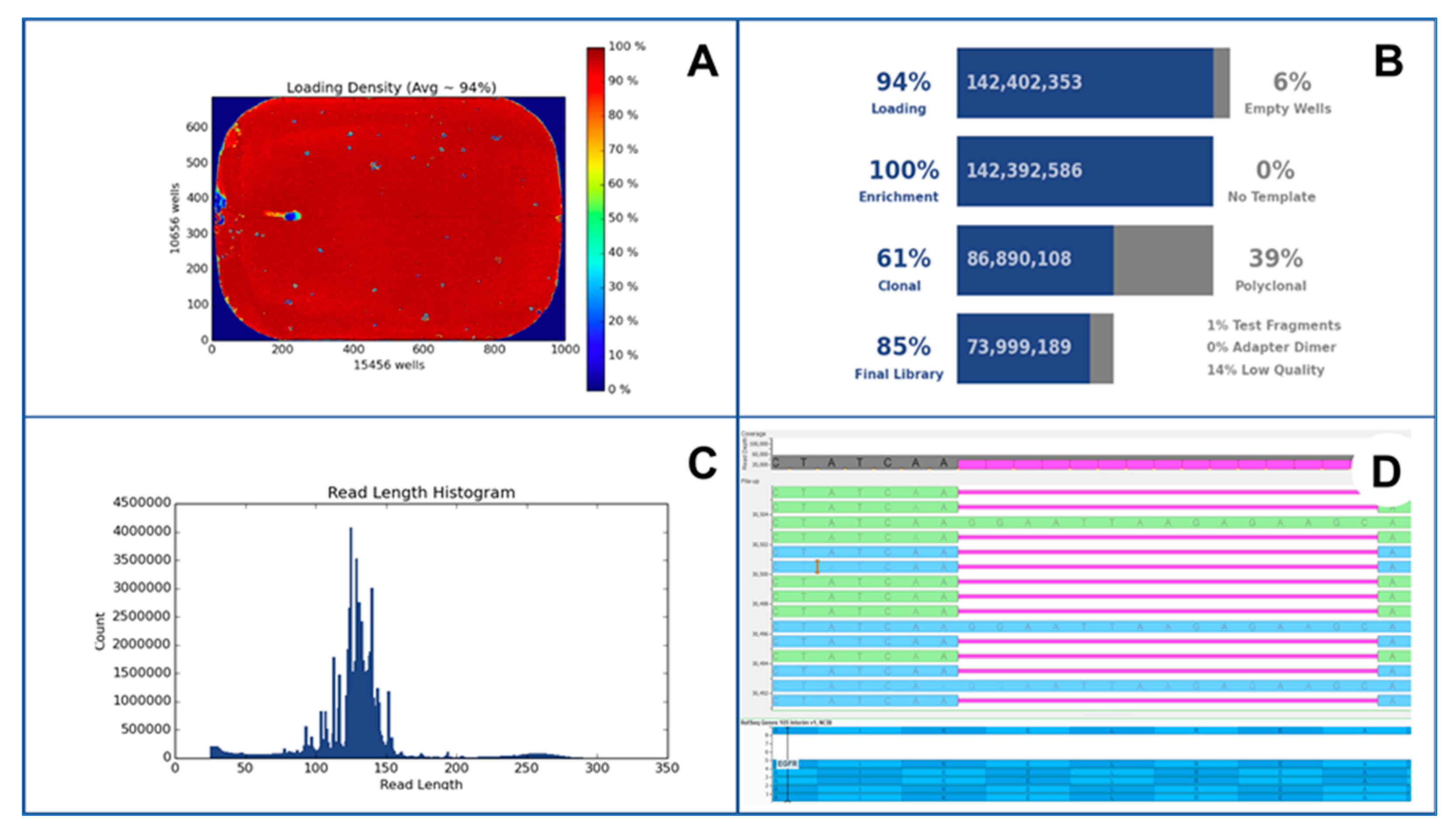
5. Analytical Issue in NGS
6. RNA-Based NGS
7. NGS in Epigenetics
8. Interpretation of DNA Sequencing Data
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Angerilli, V.; Galuppini, F.; Pagni, F.; Fusco, N.; Malapelle, U.; Fassan, M. The Role of the Pathologist in the Next-Generation Era of Tumor Molecular Characterization. Diagnostics 2021, 11, 339. [Google Scholar] [CrossRef] [PubMed]
- Kerick, M.; Isau, M.; Timmermann, B.; Sültmann, H.; Herwig, R.; Krobitsch, S.; Schaefer, G.; Verdorfer, I.; Bartsch, G.; Klocker, H.; et al. Targeted High Throughput Sequencing in Clinical Cancer Settings: Formaldehyde Fixed-Paraffin Embedded (FFPE) Tumor Tissues, Input Amount and Tumor Heterogeneity. BMC Med. Genom. 2011, 4, 68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deans, Z.C.; Costa, J.L.; Cree, I.; Dequeker, E.; Edsjö, A.; Henderson, S.; Hummel, M.; Ligtenberg, M.J.; Loddo, M.; Machado, J.C.; et al. Integration of next-Generation Sequencing in Clinical Diagnostic Molecular Pathology Laboratories for Analysis of Solid Tumours; an Expert Opinion on Behalf of IQN Path ASBL. Virchows Arch. 2017, 470, 5–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA Sequencing with Chain-Terminating Inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pillai, S.; Gopalan, V.; Lam, A.K.-Y. Review of Sequencing Platforms and Their Applications in Phaeochromocytoma and Paragangliomas. Crit. Rev. Oncol. Hematol. 2017, 116, 58–67. [Google Scholar] [CrossRef]
- Mardis, E.R. Next-Generation Sequencing Platforms. Annu. Rev. Anal. Chem. 2013, 6, 287–303. [Google Scholar] [CrossRef] [Green Version]
- Hu, T.; Chitnis, N.; Monos, D.; Dinh, A. Next-Generation Sequencing Technologies: An Overview. Hum. Immunol. 2021, 82, 801–811. [Google Scholar] [CrossRef]
- Schadt, E.E.; Turner, S.; Kasarskis, A. A Window into Third-Generation Sequencing. Hum. Mol. Genet. 2010, 19, R227–R240. [Google Scholar] [CrossRef]
- Sakamoto, Y.; Sereewattanawoot, S.; Suzuki, A. A New Era of Long-Read Sequencing for Cancer Genomics. J. Hum. Genet. 2020, 65, 3–10. [Google Scholar] [CrossRef] [Green Version]
- Gupta, P.K. Single-Molecule DNA Sequencing Technologies for Future Genomics Research. Trends Biotechnol. 2008, 26, 602–611. [Google Scholar] [CrossRef]
- Dressman, D.; Yan, H.; Traverso, G.; Kinzler, K.W.; Vogelstein, B. Transforming Single DNA Molecules into Fluorescent Magnetic Particles for Detection and Enumeration of Genetic Variations. Proc. Natl. Acad. Sci. USA 2003, 100, 8817–8822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of Age: Ten Years of next-Generation Sequencing Technologies. Nat. Rev. Genet. 2016, 17, 333–351. [Google Scholar] [CrossRef] [PubMed]
- Rothberg, J.M.; Hinz, W.; Rearick, T.M.; Schultz, J.; Mileski, W.; Davey, M.; Leamon, J.H.; Johnson, K.; Milgrew, M.J.; Edwards, M.; et al. An Integrated Semiconductor Device Enabling Non-Optical Genome Sequencing. Nature 2011, 475, 348–352. [Google Scholar] [CrossRef] [PubMed]
- Merriman, B.; Ion Torrent R&D Team; Rothberg, J.M. Progress in Ion Torrent Semiconductor Chip Based Sequencing. Electrophoresis 2012, 33, 3397–3417. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Xu, F.; Wu, J.; Schubert, J.; Li, M.M. Application of Next Generation Sequencing in Laboratory Medicine. Ann. Lab. Med. 2021, 41, 25–43. [Google Scholar] [CrossRef]
- Koitzsch, U.; Heydt, C.; Attig, H.; Immerschitt, I.; Merkelbach-Bruse, S.; Fammartino, A.; Büttner, R.H.; Kong, Y.; Odenthal, M. Use of the GeneReader NGS System in a Clinical Pathology Laboratory: A Comparative Study. J. Clin. Pathol. 2017, 70, 725–728. [Google Scholar] [CrossRef] [Green Version]
- Heeke, S.; Hofman, V.; Long-Mira, E.; Lespinet, V.; Lalvée, S.; Bordone, O.; Ribeyre, C.; Tanga, V.; Benzaquen, J.; Leroy, S.; et al. Use of the Ion PGM and the GeneReader NGS Systems in Daily Routine Practice for Advanced Lung Adenocarcinoma Patients: A Practical Point of View Reporting a Comparative Study and Assessment of 90 Patients. Cancers 2018, 10, 88. [Google Scholar] [CrossRef] [Green Version]
- Rhoads, A.; Au, K.F. PacBio Sequencing and Its Applications. Genom. Proteom. Bioinform. 2015, 13, 278–289. [Google Scholar] [CrossRef] [Green Version]
- van Dijk, E.L.; Jaszczyszyn, Y.; Naquin, D.; Thermes, C. The Third Revolution in Sequencing Technology. Trends Genet. 2018, 34, 666–681. [Google Scholar] [CrossRef]
- Wenger, A.M.; Peluso, P.; Rowell, W.J.; Chang, P.-C.; Hall, R.J.; Concepcion, G.T.; Ebler, J.; Fungtammasan, A.; Kolesnikov, A.; Olson, N.D.; et al. Accurate Circular Consensus Long-Read Sequencing Improves Variant Detection and Assembly of a Human Genome. Nat. Biotechnol. 2019, 37, 1155–1162. [Google Scholar] [CrossRef]
- Baker, E.A.G.; Goodwin, S.; McCombie, W.R.; Mendivil Ramos, O. SiLiCO: A Simulator of Long Read Sequencing in PacBio and Oxford Nanopore. bioRxiv 2016. [Google Scholar] [CrossRef] [Green Version]
- Miga, K.H.; Koren, S.; Rhie, A.; Vollger, M.R.; Gershman, A.; Bzikadze, A.; Brooks, S.; Howe, E.; Porubsky, D.; Logsdon, G.A.; et al. Telomere-to-Telomere Assembly of a Complete Human X Chromosome. Nature 2020, 585, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Logsdon, G.A.; Vollger, M.R.; Eichler, E.E. Long-Read Human Genome Sequencing and Its Applications. Nat. Rev. Genet. 2020, 21, 597–614. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Olsen, H.E.; Paten, B.; Akeson, M. The Oxford Nanopore MinION: Delivery of Nanopore Sequencing to the Genomics Community. Genome Biol. 2016, 17, 239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moorcraft, S.Y.; Gonzalez, D.; Walker, B.A. Understanding next Generation Sequencing in Oncology: A Guide for Oncologists. Crit. Rev. Oncol. Hematol. 2015, 96, 463–474. [Google Scholar] [CrossRef] [PubMed]
- Salk, J.J.; Schmitt, M.W.; Loeb, L.A. Enhancing the Accuracy of next-Generation Sequencing for Detecting Rare and Subclonal Mutations. Nat. Rev. Genet. 2018, 19, 269–285. [Google Scholar] [CrossRef] [PubMed]
- Jennings, L.J.; Arcila, M.E.; Corless, C.; Kamel-Reid, S.; Lubin, I.M.; Pfeifer, J.; Temple-Smolkin, R.L.; Voelkerding, K.V.; Nikiforova, M.N. Guidelines for Validation of Next-Generation Sequencing-Based Oncology Panels: A Joint Consensus Recommendation of the Association for Molecular Pathology and College of American Pathologists. J. Mol. Diagn. 2017, 19, 341–365. [Google Scholar] [CrossRef] [Green Version]
- Malone, E.R.; Oliva, M.; Sabatini, P.J.B.; Stockley, T.L.; Siu, L.L. Molecular Profiling for Precision Cancer Therapies. Genome Med. 2020, 12, 8. [Google Scholar] [CrossRef] [Green Version]
- Corchete, L.A.; Rojas, E.A.; Alonso-López, D.; De Las Rivas, J.; Gutiérrez, N.C.; Burguillo, F.J. Systematic Comparison and Assessment of RNA-Seq Procedures for Gene Expression Quantitative Analysis. Sci. Rep. 2020, 10, 19737. [Google Scholar] [CrossRef]
- Mosele, F.; Remon, J.; Mateo, J.; Westphalen, C.B.; Barlesi, F.; Lolkema, M.P.; Normanno, N.; Scarpa, A.; Robson, M.; Meric-Bernstam, F.; et al. Recommendations for the Use of next-Generation Sequencing (NGS) for Patients with Metastatic Cancers: A Report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2020, 31, 1491–1505. [Google Scholar] [CrossRef]
- De Maglio, G.; Pasello, G.; Dono, M.; Fiorentino, M.; Follador, A.; Sciortino, M.; Malapelle, U.; Tiseo, M. The Storm of NGS in NSCLC Diagnostic-Therapeutic Pathway: How to Sun the Real Clinical Practice. Crit. Rev. Oncol. Hematol. 2022, 169, 103561. [Google Scholar] [CrossRef] [PubMed]
- Linardou, H.; Dahabreh, I.J.; Bafaloukos, D.; Kosmidis, P.; Murray, S. Somatic EGFR Mutations and Efficacy of Tyrosine Kinase Inhibitors in NSCLC. Nat. Rev. Clin. Oncol. 2009, 6, 352–366. [Google Scholar] [CrossRef] [PubMed]
- Midha, A.; Dearden, S.; McCormack, R. EGFR Mutation Incidence in Non-Small-Cell Lung Cancer of Adenocarcinoma Histology: A Systematic Review and Global Map by Ethnicity (mutMapII). Am. J. Cancer Res. 2015, 5, 2892–2911. [Google Scholar] [PubMed]
- Shaw, A.T.; Kim, D.-W.; Nakagawa, K.; Seto, T.; Crinó, L.; Ahn, M.-J.; De Pas, T.; Besse, B.; Solomon, B.J.; Blackhall, F.; et al. Crizotinib Versus Chemotherapy in Advanced ALK-Positive Lung Cancer. N. Engl. J. Med. 2013, 368, 2385–2394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solomon, B.J.; Mok, T.; Kim, D.-W.; Wu, Y.-L.; Nakagawa, K.; Mekhail, T.; Felip, E.; Cappuzzo, F.; Paolini, J.; Usari, T.; et al. First-Line Crizotinib Versus Chemotherapy in ALK-Positive Lung Cancer. N. Engl. J. Med. 2014, 371, 2167–2177. [Google Scholar] [CrossRef] [Green Version]
- Tong, J.H.; Yeung, S.F.; Chan, A.W.H.; Chung, L.Y.; Chau, S.L.; Lung, R.W.M.; Tong, C.Y.; Chow, C.; Tin, E.K.Y.; Yu, Y.H.; et al. MET Amplification and Exon 14 Splice Site Mutation Define Unique Molecular Subgroups of Non-Small Cell Lung Carcinoma with Poor Prognosis. Clin. Cancer Res. 2016, 22, 3048–3056. [Google Scholar] [CrossRef] [Green Version]
- Drilon, A.; Clark, J.W.; Weiss, J.; Ou, S.-H.I.; Camidge, D.R.; Solomon, B.J.; Otterson, G.A.; Villaruz, L.C.; Riely, G.J.; Heist, R.S.; et al. Antitumor Activity of Crizotinib in Lung Cancers Harboring a MET Exon 14 Alteration. Nat. Med. 2020, 26, 47–51. [Google Scholar] [CrossRef]
- Planchard, D.; Besse, B.; Groen, H.J.M.; Hashemi, S.M.S.; Mazieres, J.; Kim, T.M.; Quoix, E.; Souquet, P.-J.; Barlesi, F.; Baik, C.; et al. Phase 2 Study of Dabrafenib Plus Trametinib in Patients With BRAF V600E-Mutant Metastatic NSCLC: Updated 5-Year Survival Rates and Genomic Analysis. J. Thorac. Oncol. 2022, 17, 103–115. [Google Scholar] [CrossRef]
- Shaw, A.T.; Riely, G.J.; Bang, Y.-J.; Kim, D.-W.; Camidge, D.R.; Solomon, B.J.; Varella-Garcia, M.; Iafrate, A.J.; Shapiro, G.I.; Usari, T.; et al. Crizotinib in ROS1-Rearranged Advanced Non-Small-Cell Lung Cancer (NSCLC): Updated Results, Including Overall Survival, from PROFILE 1001. Ann. Oncol. 2019, 30, 1121–1126. [Google Scholar] [CrossRef]
- Drilon, A.; Oxnard, G.R.; Tan, D.S.W.; Loong, H.H.F.; Johnson, M.; Gainor, J.; McCoach, C.E.; Gautschi, O.; Besse, B.; Cho, B.C.; et al. Efficacy of Selpercatinib in Fusion-Positive Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2020, 383, 813–824. [Google Scholar] [CrossRef]
- Ricciuti, B.; Brambilla, M.; Metro, G.; Baglivo, S.; Matocci, R.; Pirro, M.; Chiari, R. Targeting NTRK Fusion in Non-Small Cell Lung Cancer: Rationale and Clinical Evidence. Med. Oncol. 2017, 34, 105. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.S.; DuBois, S.G.; Kummar, S.; Farago, A.F.; Albert, C.M.; Rohrberg, K.S.; van Tilburg, C.M.; Nagasubramanian, R.; Berlin, J.D.; Federman, N.; et al. Larotrectinib in Patients with TRK Fusion-Positive Solid Tumours: A Pooled Analysis of Three Phase 1/2 Clinical Trials. Lancet Oncol. 2020, 21, 531–540. [Google Scholar] [CrossRef]
- Dziadziuszko, R.; Krebs, M.G.; De Braud, F.; Siena, S.; Drilon, A.; Doebele, R.C.; Patel, M.R.; Cho, B.C.; Liu, S.V.; Ahn, M.-J.; et al. Updated Integrated Analysis of the Efficacy and Safety of Entrectinib in Locally Advanced or Metastatic Fusion-Positive Non-Small-Cell Lung Cancer. J. Clin. Oncol. 2021, 39, 1253–1263. [Google Scholar] [CrossRef]
- de Bono, J.; Mateo, J.; Fizazi, K.; Saad, F.; Shore, N.; Sandhu, S.; Chi, K.N.; Sartor, O.; Agarwal, N.; Olmos, D.; et al. Olaparib for Metastatic Castration-Resistant Prostate Cancer. N. Engl. J. Med. 2020, 382, 2091–2102. [Google Scholar] [CrossRef]
- Sweeney, C.; Bracarda, S.; Sternberg, C.N.; Chi, K.N.; Olmos, D.; Sandhu, S.; Massard, C.; Matsubara, N.; Alekseev, B.; Parnis, F.; et al. Ipatasertib plus Abiraterone and Prednisolone in Metastatic Castration-Resistant Prostate Cancer (IPATential150): A Multicentre, Randomised, Double-Blind, Phase 3 Trial. Lancet 2021, 398, 131–142. [Google Scholar] [CrossRef]
- Moore, K.; Colombo, N.; Scambia, G.; Kim, B.-G.; Oaknin, A.; Friedlander, M.; Lisyanskaya, A.; Floquet, A.; Leary, A.; Sonke, G.S.; et al. Maintenance Olaparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N. Engl. J. Med. 2018, 379, 2495–2505. [Google Scholar] [CrossRef] [PubMed]
- Fumagalli, C.; Tomao, F.; Betella, I.; Rappa, A.; Calvello, M.; Bonanni, B.; Bernard, L.; Peccatori, F.; Colombo, N.; Viale, G.; et al. Tumor BRCA Test for Patients with Epithelial Ovarian Cancer: The Role of Molecular Pathology in the Era of PARP Inhibitor Therapy. Cancers 2019, 11, 1641. [Google Scholar] [CrossRef] [Green Version]
- Abou-Alfa, G.K.; Macarulla, T.; Javle, M.M.; Kelley, R.K.; Lubner, S.J.; Adeva, J.; Cleary, J.M.; Catenacci, D.V.; Borad, M.J.; Bridgewater, J.; et al. Ivosidenib in IDH1-Mutant, Chemotherapy-Refractory Cholangiocarcinoma (ClarIDHy): A Multicentre, Randomised, Double-Blind, Placebo-Controlled, Phase 3 Study. Lancet Oncol. 2020, 21, 796–807. [Google Scholar] [CrossRef]
- Abou-Alfa, G.K.; Sahai, V.; Hollebecque, A.; Vaccaro, G.; Melisi, D.; Al-Rajabi, R.; Paulson, A.S.; Borad, M.J.; Gallinson, D.; Murphy, A.G.; et al. Pemigatinib for Previously Treated, Locally Advanced or Metastatic Cholangiocarcinoma: A Multicentre, Open-Label, Phase 2 Study. Lancet Oncol. 2020, 21, 671–684. [Google Scholar] [CrossRef]
- Jakubowski, C.D.; Azad, N.S. Immune Checkpoint Inhibitor Therapy in Biliary Tract Cancer (Cholangiocarcinoma). Chin. Clin. Oncol. 2020, 9, 2. [Google Scholar] [CrossRef]
- Doebele, R.C.; Drilon, A.; Paz-Ares, L.; Siena, S.; Shaw, A.T.; Farago, A.F.; Blakely, C.M.; Seto, T.; Cho, B.C.; Tosi, D.; et al. Entrectinib in Patients with Advanced or Metastatic NTRK Fusion-Positive Solid Tumours: Integrated Analysis of Three Phase 1-2 Trials. Lancet Oncol. 2020, 21, 271–282. [Google Scholar] [CrossRef]
- Fassan, M.; Scarpa, A.; Remo, A.; De Maglio, G.; Troncone, G.; Marchetti, A.; Doglioni, C.; Ingravallo, G.; Perrone, G.; Parente, P.; et al. Current Prognostic and Predictive Biomarkers for Gastrointestinal Tumors in Clinical Practice. Pathologica 2020, 112, 248–259. [Google Scholar] [CrossRef] [PubMed]
- Hsu, H.-C.; Thiam, T.K.; Lu, Y.-J.; Yeh, C.Y.; Tsai, W.-S.; You, J.F.; Hung, H.Y.; Tsai, C.-N.; Hsu, A.; Chen, H.-C.; et al. Mutations of KRAS/NRAS/BRAF Predict Cetuximab Resistance in Metastatic Colorectal Cancer Patients. Oncotarget 2016, 7, 22257–22270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kopetz, S.; Grothey, A.; Yaeger, R.; Van Cutsem, E.; Desai, J.; Yoshino, T.; Wasan, H.; Ciardiello, F.; Loupakis, F.; Hong, Y.S.; et al. Encorafenib, Binimetinib, and Cetuximab in V600E-Mutated Colorectal Cancer. N. Engl. J. Med. 2019, 381, 1632–1643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Overman, M.J.; McDermott, R.; Leach, J.L.; Lonardi, S.; Lenz, H.-J.; Morse, M.A.; Desai, J.; Hill, A.; Axelson, M.; Moss, R.A.; et al. Nivolumab in Patients with Metastatic DNA Mismatch Repair-Deficient or Microsatellite Instability-High Colorectal Cancer (CheckMate 142): An Open-Label, Multicentre, Phase 2 Study. Lancet Oncol. 2017, 18, 1182–1191. [Google Scholar] [CrossRef]
- Le, D.T.; Kim, T.W.; Van Cutsem, E.; Geva, R.; Jäger, D.; Hara, H.; Burge, M.; O’Neil, B.; Kavan, P.; Yoshino, T.; et al. Phase II Open-Label Study of Pembrolizumab in Treatment-Refractory, Microsatellite Instability-High/Mismatch Repair-Deficient Metastatic Colorectal Cancer: KEYNOTE-164. J. Clin. Oncol. 2020, 38, 11–19. [Google Scholar] [CrossRef]
- Paz-Ares, L.; Barlesi, F.; Siena, S.; Ahn, M.-J.; Drilon, A.; Conley, A.; Rolfo, C.; Wolf, J.; Seto, T.; Doebele, R.; et al. Patient-Reported Outcomes from STARTRK-2: A Global Phase II Basket Study of Entrectinib for ROS1 Fusion-Positive Non-Small-Cell Lung Cancer and NTRK Fusion-Positive Solid Tumours. ESMO Open 2021, 6, 100113. [Google Scholar] [CrossRef]
- Sartore-Bianchi, A.; Trusolino, L.; Martino, C.; Bencardino, K.; Lonardi, S.; Bergamo, F.; Zagonel, V.; Leone, F.; Depetris, I.; Martinelli, E.; et al. Dual-Targeted Therapy with Trastuzumab and Lapatinib in Treatment-Refractory, KRAS Codon 12/13 Wild-Type, HER2-Positive Metastatic Colorectal Cancer (HERACLES): A Proof-of-Concept, Multicentre, Open-Label, Phase 2 Trial. Lancet Oncol. 2016, 17, 738–746. [Google Scholar] [CrossRef]
- Bang, Y.-J.; Ueno, M.; Malka, D.; Chung, H.C.; Nagrial, A.; Kelley, R.K.; Piha-Paul, S.A.; Ros, W.; Italiano, A.; Nakagawa, K.; et al. Pembrolizumab (Pembro) for Advanced Biliary Adenocarcinoma: Results from the KEYNOTE-028 (KN028) and KEYNOTE-158 (KN158) Basket Studies. J. Clin. Oncol. 2019, 37, 4079. [Google Scholar] [CrossRef]
- Cristescu, R.; Mogg, R.; Ayers, M.; Albright, A.; Murphy, E.; Yearley, J.; Sher, X.; Liu, X.Q.; Lu, H.; Nebozhyn, M.; et al. Pan-Tumor Genomic Biomarkers for PD-1 Checkpoint Blockade-Based Immunotherapy. Science 2018, 362, eaar3593. [Google Scholar] [CrossRef] [Green Version]
- Galuppini, F.; Dal Pozzo, C.A.; Deckert, J.; Loupakis, F.; Fassan, M.; Baffa, R. Tumor Mutation Burden: From Comprehensive Mutational Screening to the Clinic. Cancer Cell Int. 2019, 19, 209. [Google Scholar] [CrossRef] [PubMed]
- Susman, S.; Berindan-Neagoe, I.; Petrushev, B.; Pirlog, R.; Florian, I.-S.; Mihu, C.-M.; Berce, C.; Craciun, L.; Grewal, R.; Tomuleasa, C. The Role of the Pathology Department in the Preanalytical Phase of Molecular Analyses. Cancer Manag. Res. 2018, 10, 745–753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freidin, M.B.; Bhudia, N.; Lim, E.; Nicholson, A.G.; Cookson, W.O.; Moffatt, M.F. Impact of Collection and Storage of Lung Tumor Tissue on Whole Genome Expression Profiling. J. Mol. Diagn. 2012, 14, 140–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, D.; Wang, A.; Xie, T.; Zhang, S.; Cao, D.; Sun, J. Effects of Ex Vivo Ischemia Time and Delayed Processing on Quality of Specimens in Tissue Biobank. Mol. Med. Rep. 2020, 22, 4278–4288. [Google Scholar] [CrossRef]
- Guerrera, F.; Tabbò, F.; Bessone, L.; Maletta, F.; Gaudiano, M.; Ercole, E.; Annaratone, L.; Todaro, M.; Boita, M.; Filosso, P.L.; et al. The Influence of Tissue Ischemia Time on RNA Integrity and Patient-Derived Xenografts (PDX) Engraftment Rate in a Non-Small Cell Lung Cancer (NSCLC) Biobank. PLoS ONE 2016, 11, e0145100. [Google Scholar] [CrossRef]
- van Maldegem, F.; de Wit, M.; Morsink, F.; Musler, A.; Weegenaar, J.; van Noesel, C.J.M. Effects of Processing Delay, Formalin Fixation, and Immunohistochemistry on RNA Recovery From Formalin-Fixed Paraffin-Embedded Tissue Sections. Diagn. Mol. Pathol. 2008, 17, 51–58. [Google Scholar] [CrossRef]
- Do, H.; Dobrovic, A. Sequence Artifacts in DNA from Formalin-Fixed Tissues: Causes and Strategies for Minimization. Clin. Chem. 2015, 61, 64–71. [Google Scholar] [CrossRef] [Green Version]
- Arreaza, G.; Qiu, P.; Pang, L.; Albright, A.; Hong, L.Z.; Marton, M.J.; Levitan, D. Pre-Analytical Considerations for Successful Next-Generation Sequencing (NGS): Challenges and Opportunities for Formalin-Fixed and Paraffin-Embedded Tumor Tissue (FFPE) Samples. Int. J. Mol. Sci. 2016, 17, 1579. [Google Scholar] [CrossRef]
- Dotti, I.; Bonin, S.; Basili, G.; Nardon, E.; Balani, A.; Siracusano, S.; Zanconati, F.; Palmisano, S.; De Manzini, N.; Stanta, G. Effects of Formalin, Methacarn, and fineFIX Fixatives on RNA Preservation. Diagn. Mol. Pathol. 2010, 19, 112–122. [Google Scholar] [CrossRef]
- Prentice, L.M.; Miller, R.R.; Knaggs, J.; Mazloomian, A.; Aguirre Hernandez, R.; Franchini, P.; Parsa, K.; Tessier-Cloutier, B.; Lapuk, A.; Huntsman, D.; et al. Formalin Fixation Increases Deamination Mutation Signature but Should Not Lead to False Positive Mutations in Clinical Practice. PLoS ONE 2018, 13, e0196434. [Google Scholar] [CrossRef] [Green Version]
- Miquelestorena-Standley, E.; Jourdan, M.-L.; Collin, C.; Bouvier, C.; Larousserie, F.; Aubert, S.; Gomez-Brouchet, A.; Guinebretière, J.-M.; Tallegas, M.; Brulin, B.; et al. Effect of Decalcification Protocols on Immunohistochemistry and Molecular Analyses of Bone Samples. Mod. Pathol. 2020, 33, 1505–1517. [Google Scholar] [CrossRef] [PubMed]
- Groelz, D.; Viertler, C.; Pabst, D.; Dettmann, N.; Zatloukal, K. Impact of Storage Conditions on the Quality of Nucleic Acids in Paraffin Embedded Tissues. PLoS ONE 2018, 13, e0203608. [Google Scholar] [CrossRef] [PubMed]
- Guyard, A.; Boyez, A.; Pujals, A.; Robe, C.; Tran Van Nhieu, J.; Allory, Y.; Moroch, J.; Georges, O.; Fournet, J.-C.; Zafrani, E.-S.; et al. DNA Degrades during Storage in Formalin-Fixed and Paraffin-Embedded Tissue Blocks. Virchows Arch. 2017, 471, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Kuwata, T.; Wakabayashi, M.; Hatanaka, Y.; Morii, E.; Oda, Y.; Taguchi, K.; Noguchi, M.; Ishikawa, Y.; Nakajima, T.; Sekine, S.; et al. Impact of DNA Integrity on the Success Rate of Tissue-Based next-Generation Sequencing: Lessons from Nationwide Cancer Genome Screening Project SCRUM-Japan GI-SCREEN. Pathol. Int. 2020, 70, 932–942. [Google Scholar] [CrossRef]
- Chen, H.; Luthra, R.; Goswami, R.S.; Singh, R.R.; Roy-Chowdhuri, S. Analysis of Pre-Analytic Factors Affecting the Success of Clinical Next-Generation Sequencing of Solid Organ Malignancies. Cancers 2015, 7, 1699–1715. [Google Scholar] [CrossRef] [PubMed]
- Kanagal-Shamanna, R.; Portier, B.P.; Singh, R.R.; Routbort, M.J.; Aldape, K.D.; Handal, B.A.; Rahimi, H.; Reddy, N.G.; Barkoh, B.A.; Mishra, B.M.; et al. Next-Generation Sequencing-Based Multi-Gene Mutation Profiling of Solid Tumors Using Fine Needle Aspiration Samples: Promises and Challenges for Routine Clinical Diagnostics. Mod. Pathol. 2014, 27, 314–327. [Google Scholar] [CrossRef] [Green Version]
- Fassan, M. Molecular Diagnostics in Pathology: Time for a Next-Generation Pathologist? Arch. Pathol. Lab. Med. 2018, 142, 313–320. [Google Scholar] [CrossRef] [Green Version]
- Dufraing, K.; De Hertogh, G.; Tack, V.; Keppens, C.; Dequeker, E.M.C.; van Krieken, J.H. External Quality Assessment Identifies Training Needs to Determine the Neoplastic Cell Content for Biomarker Testing. J. Mol. Diagn. 2018, 20, 455–464. [Google Scholar] [CrossRef] [Green Version]
- Witt, B.L. Rapid On Site Evaluation (ROSE): A Pathologists’ Perspective. Tech. Vasc. Interv. Radiol. 2021, 24, 100767. [Google Scholar] [CrossRef]
- Akahane, T.; Yamaguchi, T.; Kato, Y.; Yokoyama, S.; Hamada, T.; Nishida, Y.; Higashi, M.; Nishihara, H.; Suzuki, S.; Ueno, S.; et al. Comprehensive Validation of Liquid-Based Cytology Specimens for next-Generation Sequencing in Cancer Genome Analysis. PLoS ONE 2019, 14, e0217724. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, J.P.; Zhou, Y.; Jakubowski, M.A.; Wang, Z.; Brainard, J.A.; Klein, R.D.; Farver, C.F.; Almeida, F.A.; Cheng, Y.-W. Next-Generation Sequencing of Liquid-Based Cytology Non-Small Cell Lung Cancer Samples. Cancer Cytopathol. 2017, 125, 178–187. [Google Scholar] [CrossRef] [Green Version]
- Bellevicine, C.; Malapelle, U.; de Luca, C.; Iaccarino, A.; Troncone, G. EGFR Analysis: Current Evidence and Future Directions. Diagn. Cytopathol. 2014, 42, 984–992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vigliar, E.; Malapelle, U.; de Luca, C.; Bellevicine, C.; Troncone, G. Challenges and Opportunities of next-Generation Sequencing: A Cytopathologist’s Perspective. Cytopathology 2015, 26, 271–283. [Google Scholar] [CrossRef]
- Scarpa, A.; Sikora, K.; Fassan, M.; Rachiglio, A.M.; Cappellesso, R.; Antonello, D.; Amato, E.; Mafficini, A.; Lambiase, M.; Esposito, C.; et al. Molecular Typing of Lung Adenocarcinoma on Cytological Samples Using a Multigene next Generation Sequencing Panel. PLoS ONE 2013, 8, e80478. [Google Scholar] [CrossRef] [Green Version]
- Gleeson, F.C.; Kipp, B.R.; Levy, M.J.; Voss, J.S.; Campion, M.B.; Minot, D.M.; Tu, Z.J.; Klee, E.W.; Lazaridis, K.N.; Kerr, S.E. Lung Cancer Adrenal Gland Metastasis: Optimal Fine-Needle Aspirate and Touch Preparation Smear Cellularity Characteristics for Successful Theranostic next-Generation Sequencing. Cancer Cytopathol. 2014, 122, 822–832. [Google Scholar] [CrossRef] [PubMed]
- Do, H.; Dobrovic, A. Dramatic Reduction of Sequence Artefacts from DNA Isolated from Formalin-Fixed Cancer Biopsies by Treatment with Uracil- DNA Glycosylase. Oncotarget 2012, 3, 546–558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yonekura, S.-I.; Nakamura, N.; Yonei, S.; Zhang-Akiyama, Q.-M. Generation, Biological Consequences and Repair Mechanisms of Cytosine Deamination in DNA. J. Radiat. Res. 2009, 50, 19–26. [Google Scholar] [CrossRef] [Green Version]
- Williams, C.; Pontén, F.; Moberg, C.; Söderkvist, P.; Uhlén, M.; Pontén, J.; Sitbon, G.; Lundeberg, J. A High Frequency of Sequence Alterations Is due to Formalin Fixation of Archival Specimens. Am. J. Pathol. 1999, 155, 1467–1471. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.; Mosier, S.; Gocke, C.D.; Lin, M.-T.; Eshleman, J.R. Cytosine Deamination Is a Major Cause of Baseline Noise in next-Generation Sequencing. Mol. Diagn. Ther. 2014, 18, 587–593. [Google Scholar] [CrossRef] [Green Version]
- McCall, C.M.; Mosier, S.; Thiess, M.; Debeljak, M.; Pallavajjala, A.; Beierl, K.; Deak, K.L.; Datto, M.B.; Gocke, C.D.; Lin, M.-T.; et al. False Positives in Multiplex PCR-Based next-Generation Sequencing Have Unique Signatures. J. Mol. Diagn. 2014, 16, 541–549. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Z.; Liebers, M.; Zhelyazkova, B.; Cao, Y.; Panditi, D.; Lynch, K.D.; Chen, J.; Robinson, H.E.; Shim, H.S.; Chmielecki, J.; et al. Anchored Multiplex PCR for Targeted next-Generation Sequencing. Nat. Med. 2014, 20, 1479–1484. [Google Scholar] [CrossRef]
- Mutz, K.-O.; Heilkenbrinker, A.; Lönne, M.; Walter, J.-G.; Stahl, F. Transcriptome Analysis Using next-Generation Sequencing. Curr. Opin. Biotechnol. 2013, 24, 22–30. [Google Scholar] [CrossRef]
- Penault-Llorca, F.; Rudzinski, E.R.; Sepulveda, A.R. Testing Algorithm for Identification of Patients with TRK Fusion Cancer. J. Clin. Pathol. 2019, 72, 460–467. [Google Scholar] [CrossRef] [PubMed]
- Davies, K.D.; Le, A.T.; Sheren, J.; Nijmeh, H.; Gowan, K.; Jones, K.L.; Varella-Garcia, M.; Aisner, D.L.; Doebele, R.C. Comparison of Molecular Testing Modalities for Detection of ROS1 Rearrangements in a Cohort of Positive Patient Samples. J. Thorac. Oncol. 2018, 13, 1474–1482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beadling, C.; Wald, A.I.; Warrick, A.; Neff, T.L.; Zhong, S.; Nikiforov, Y.E.; Corless, C.L.; Nikiforova, M.N. A Multiplexed Amplicon Approach for Detecting Gene Fusions by Next-Generation Sequencing. J. Mol. Diagn. 2016, 18, 165–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davies, K.D.; Lomboy, A.; Lawrence, C.A.; Yourshaw, M.; Bocsi, G.T.; Camidge, D.R.; Aisner, D.L. DNA-Based versus RNA-Based Detection of MET Exon 14 Skipping Events in Lung Cancer. J. Thorac. Oncol. 2019, 14, 737–741. [Google Scholar] [CrossRef] [Green Version]
- Ludyga, N.; Grünwald, B.; Azimzadeh, O.; Englert, S.; Höfler, H.; Tapio, S.; Aubele, M. Nucleic Acids from Long-Term Preserved FFPE Tissues Are Suitable for Downstream Analyses. Virchows Arch. 2012, 460, 131–140. [Google Scholar] [CrossRef]
- Murphy, D.A.; Ely, H.A.; Shoemaker, R.; Boomer, A.; Culver, B.P.; Hoskins, I.; Haimes, J.D.; Walters, R.D.; Fernandez, D.; Stahl, J.A.; et al. Detecting Gene Rearrangements in Patient Populations Through a 2-Step Diagnostic Test Comprised of Rapid IHC Enrichment Followed by Sensitive Next-Generation Sequencing. Appl. Immunohistochem. Mol. Morphol. 2017, 25, 513–523. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Mashock, M.; Tong, Z.; Mu, X.; Chen, H.; Zhou, X.; Zhang, H.; Zhao, G.; Liu, B.; Li, X. Changing Technologies of RNA Sequencing and Their Applications in Clinical Oncology. Front. Oncol. 2020, 10, 447. [Google Scholar] [CrossRef] [Green Version]
- Dupont, C.; Armant, D.R.; Brenner, C.A. Epigenetics: Definition, Mechanisms and Clinical Perspective. Semin. Reprod. Med. 2009, 27, 351–357. [Google Scholar] [CrossRef] [Green Version]
- Arora, I.; Tollefsbol, T.O. Computational Methods and next-Generation Sequencing Approaches to Analyze Epigenetics Data: Profiling of Methods and Applications. Methods 2021, 187, 92–103. [Google Scholar] [CrossRef] [PubMed]
- Nava, M.; Dutta, P.; Farias-Eisner, R.; Vadgama, J.V.; Wu, Y. Utilization of NGS Technologies to Investigate Transcriptomic and Epigenomic Mechanisms in Trastuzumab Resistance. Sci. Rep. 2019, 9, 5141. [Google Scholar] [CrossRef] [PubMed]
- Nordlund, J. Advances in Whole Genome Methylomic Sequencing. In Epigenetics Methods; Tollefsbol, T., Ed.; Elsevier: Cambridge, MA, USA, 2020; pp. 213–233. [Google Scholar]
- Das, P.M.; Singal, R. DNA Methylation and Cancer. J. Clin. Oncol. 2004, 22, 4632–4642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darst, R.P.; Pardo, C.E.; Ai, L.; Brown, K.D.; Kladde, M.P. Bisulfite Sequencing of DNA. Curr. Protoc. Mol. Biol. 2010, 91, 7–9. [Google Scholar] [CrossRef] [Green Version]
- Bártová, E.; Krejcí, J.; Harnicarová, A.; Galiová, G.; Kozubek, S. Histone Modifications and Nuclear Architecture: A Review. J. Histochem. Cytochem. 2008, 56, 711–721. [Google Scholar] [CrossRef] [Green Version]
- Park, P.J. ChIP-Seq: Advantages and Challenges of a Maturing Technology. Nat. Rev. Genet. 2009, 10, 669–680. [Google Scholar] [CrossRef] [Green Version]
- Roy, S.; Coldren, C.; Karunamurthy, A.; Kip, N.S.; Klee, E.W.; Lincoln, S.E.; Leon, A.; Pullambhatla, M.; Temple-Smolkin, R.L.; Voelkerding, K.V.; et al. Standards and Guidelines for Validating Next-Generation Sequencing Bioinformatics Pipelines: A Joint Recommendation of the Association for Molecular Pathology and the College of American Pathologists. J. Mol. Diagn. 2018, 20, 4–27. [Google Scholar] [CrossRef] [Green Version]
- Ledergerber, C.; Dessimoz, C. Base-Calling for next-Generation Sequencing Platforms. Brief. Bioinform. 2011, 12, 489–497. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Agarwal, S. Ranvijay Fast and Memory Efficient Approach for Mapping NGS Reads to a Reference Genome. J. Bioinform. Comput. Biol. 2019, 17, 1950008. [Google Scholar] [CrossRef]
- Koboldt, D.C. Best Practices for Variant Calling in Clinical Sequencing. Genome Med. 2020, 12, 91. [Google Scholar] [CrossRef]
- Gullapalli, R.R.; Desai, K.V.; Santana-Santos, L.; Kant, J.A.; Becich, M.J. Next Generation Sequencing in Clinical Medicine: Challenges and Lessons for Pathology and Biomedical Informatics. J. Pathol. Inform. 2012, 3, 40. [Google Scholar] [CrossRef] [PubMed]
| Illumina MiSeq | Illumina HiSeq 2000 | Ion Torrent PGM | PacBio SMRT | Oxford Nanopore MinION | |
|---|---|---|---|---|---|
| Read Length | Up to 150 bases | Up to 150 bases | ~200 bases | Average 1500 bases | 13–20 kb |
| Paired-End | Yes | Yes | Yes | No | No |
| Reported Accuracy | Mostly >Q30 | Mostly >Q30 | Mostly Q20 | <Q10 | Mostly Q50 |
| Observed Raw Error Rate | 0.80% | 0.26% | 1.71% | 12.86% | 10.50% |
| Insert Size | Up to 700 bases | Up to 700 bases | Up to 250 bases | Up to 10 kb | Average of 331 bases |
| Run Time | 27 h | 11 days | 2 h | 2 h | 72 h |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cappello, F.; Angerilli, V.; Munari, G.; Ceccon, C.; Sabbadin, M.; Pagni, F.; Fusco, N.; Malapelle, U.; Fassan, M. FFPE-Based NGS Approaches into Clinical Practice: The Limits of Glory from a Pathologist Viewpoint. J. Pers. Med. 2022, 12, 750. https://doi.org/10.3390/jpm12050750
Cappello F, Angerilli V, Munari G, Ceccon C, Sabbadin M, Pagni F, Fusco N, Malapelle U, Fassan M. FFPE-Based NGS Approaches into Clinical Practice: The Limits of Glory from a Pathologist Viewpoint. Journal of Personalized Medicine. 2022; 12(5):750. https://doi.org/10.3390/jpm12050750
Chicago/Turabian StyleCappello, Filippo, Valentina Angerilli, Giada Munari, Carlotta Ceccon, Marianna Sabbadin, Fabio Pagni, Nicola Fusco, Umberto Malapelle, and Matteo Fassan. 2022. "FFPE-Based NGS Approaches into Clinical Practice: The Limits of Glory from a Pathologist Viewpoint" Journal of Personalized Medicine 12, no. 5: 750. https://doi.org/10.3390/jpm12050750
APA StyleCappello, F., Angerilli, V., Munari, G., Ceccon, C., Sabbadin, M., Pagni, F., Fusco, N., Malapelle, U., & Fassan, M. (2022). FFPE-Based NGS Approaches into Clinical Practice: The Limits of Glory from a Pathologist Viewpoint. Journal of Personalized Medicine, 12(5), 750. https://doi.org/10.3390/jpm12050750










