Deep-ADCA: Development and Validation of Deep Learning Model for Automated Diagnosis Code Assignment Using Clinical Notes in Electronic Medical Records
Abstract
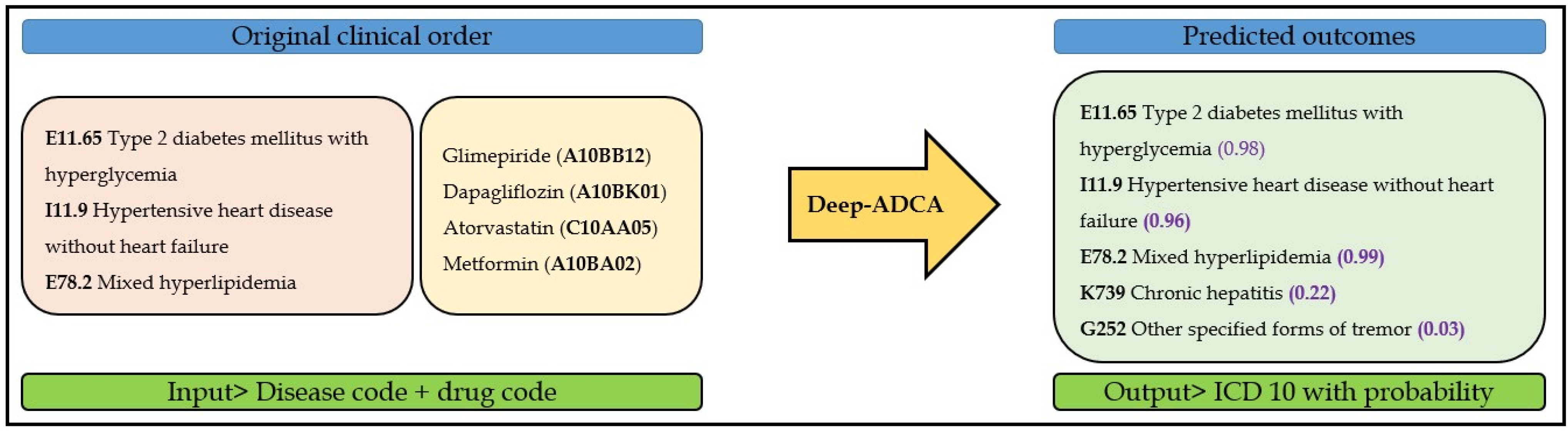
:1. Introduction
2. Methods
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rae, K.; Britt, H.; Orchard, J.; Finch, C. Classifying sports medicine diagnoses: A comparison of the International classification of diseases 10-Australian modification (ICD-10-AM) and the Orchard sports injury classification system (OSICS-8). Br. J. Sports Med. 2005, 39, 907–911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stein, B.D.; Bautista, A.; Schumock, G.T.; Lee, T.A.; Charbeneau, J.T.; Lauderdale, D.S.; Krishnan, J.A. The validity of International Classification of Diseases, Ninth Revision, Clinical Modification diagnosis codes for identifying patients hospitalized for COPD exacerbations. Chest 2012, 141, 87–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shortliffe, E.H.; Cimino, J. Computer Applications in Health Care and Biomedicine; Springer: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Woods, S. Counting Death and Disease: International Classification of Death and Diseases. DttP 2011, 39, 12. [Google Scholar]
- Bowker, G.C. The history of information infrastructures: The case of the international classification of diseases. Inf. Process. Manag. 1996, 32, 49–61. [Google Scholar] [CrossRef]
- Lindholm, V. Designing and Assessing an Interactive Sunburst Diagram for ICD; UPPSALA University: Uppsala, Sweden, 2020. [Google Scholar]
- Biruk, E.; Habtamu, T.; Taye, G.; Ayele, W.; Tassew, B.; Nega, A.; Sisay, A. Improving the Quality of Clinical Coding through Mapping of National Classification of Diseases (NCoD) and International Classification of Disease (ICD-10). Ethiop. J. Health Dev. 2021, 35, 59–65. [Google Scholar]
- Subotin, M.; Davis, A. A system for predicting ICD-10-PCS codes from electronic health records. In Proceedings of the BioNLP 2014, Baltimore, MD, USA, 26–27 June 2014; pp. 59–67.
- Banerji, A.; Lai, K.H.; Li, Y.; Saff, R.R.; Camargo, C.A., Jr.; Blumenthal, K.G.; Zhou, L. Natural language processing combined with ICD-9-CM codes as a novel method to study the epidemiology of allergic drug reactions. J. Allergy Clin. Immunol. Pract. 2020, 8, 1032–1038.e1. [Google Scholar] [CrossRef] [PubMed]
- Farkas, R.; Szarvas, G. Automatic construction of rule-based ICD-9-CM coding systems. In BMC Bioinformatics; Springer: Berlin/Heidelberg, Germany, 2008; pp. 1–9. [Google Scholar]
- Goldstein, I.; Arzumtsyan, A.; Uzuner, Ö. Three approaches to automatic assignment of ICD-9-CM codes to radiology reports. In AMIA Annual Symposium Proceedings; American Medical Informatics Association: Bethesda, MD, USA, 2007; p. 279. [Google Scholar]
- Zhang, D.; He, D.; Zhao, S.; Li, L. Enhancing automatic icd-9-cm code assignment for medical texts with pubmed. In BioNLP 2017; Association for Computational Linguistics: Vancouver, BC, Canada, 2017; pp. 263–271. [Google Scholar]
- Yang, L.; Ke nny, E.M.; Ng, T.L.J.; Yang, Y.; Smyth, B.; Dong, R. Generating plausible counterfactual explanations for deep transformers in financial text classification. arXiv 2020, arXiv:201012512. [Google Scholar]
- Melville, P.; Gryc, W.; Lawrence, R.D. Sentiment analysis of blogs by combining lexical knowledge with text classification. In Proceedings of the 15th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Paris, France, 28 June–1 July 2009; pp. 1275–1284. [Google Scholar]
- Slater, L.T.; Karwath, A.; Williams, J.A.; Russell, S.; Makepeace, S.; Carberry, A.; Gkoutos, G.V. Towards similarity-based differential diagnostics for common diseases. Comput. Biol. Med. 2021, 133, 104360. [Google Scholar] [CrossRef] [PubMed]
- Kavuluru, R.; Rios, A.; Lu, Y. An empirical evaluation of supervised learning approaches in assigning diagnosis codes to electronic medical records. Artif. Intell. Med. 2015, 65, 155–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, H.; Xie, P.; Hu, Z.; Zhang, M.; Xing, E.P. An explainable CNN approach for medical codes prediction from clinical text. arXiv 2017, arXiv:171104075. [Google Scholar]
- Xie, P.; Xing, E. A neural architecture for automated ICD coding. In Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), Melbourne, Australia, 15–20 July 2018; pp. 1066–1076. [Google Scholar]
- Huang, J.; Osorio, C.; Sy, L.W. An empirical evaluation of deep learning for ICD-9 code assignment using MIMIC-III clinical notes. Comput. Methods Programs Biomed. 2019, 177, 141–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, M.; Li, M.; Fei, Z.; Yu, Y.; Pan, Y.; Wang, J. Automatic ICD-9 coding via deep transfer learning. Neurocomputing 2019, 324, 43–50. [Google Scholar] [CrossRef]
- Samonte, M.J.C.; Gerardo, B.D.; Fajardo, A.C.; Medina, R.P. ICD-9 tagging of clinical notes using topical word embedding. In Proceedings of the 2018 International Conference on Internet and e-Business, Singapore, 25–27 April 2018; pp. 118–123. [Google Scholar]
- Hsu, C.-C.; Chang, P.-C.; Chang, A. Multi-label classification of ICD coding using deep learning. 2020 International Symposium on Community-Centric Systems (CcS), Tokyo, Japan, 23–26 September 2020; IEEE: Piscataway Township, NJ, USA, 2020; pp. 1–6. [Google Scholar]
- Gangavarapu, T.; Krishnan, G.S.; Kamath, S.; Jeganathan, J. FarSight: Long-term disease prediction using unstructured clinical nursing notes. IEEE Trans. Emerg. Top. Comput. 2022, 9, 1151–1169. [Google Scholar] [CrossRef]
- Singaravelan, A.; Hsieh, C.-H.; Liao, Y.-K.; Hsu, J.L. Predicting ICD-9 Codes Using Self-Report of Patients. Appl. Sci. 2021, 11, 10046. [Google Scholar] [CrossRef]







| Characteristics | Number (%) | |
|---|---|---|
| Total Number of Patient | ||
| Male | 2212 | |
| Female | 2804 | |
| Age in year, mean (SD), year | 60.76 (18.38) | |
| Total number of clinical notes | All departments | 21,953 |
| Cardiology | 3668 | |
| Neurology | 2762 | |
| Nephrology | 5789 | |
| Metabolism | 3707 | |
| Psychiatry | 6027 | |
| Department | Test Cases | No. of ICD-10 Codes | No. of Drugs | Precision | Recall | F-Measure |
|---|---|---|---|---|---|---|
| Cardiology | 284 | 148 | 145 | 0.69 | 0.89 | 0.78 |
| Metabolism | 307 | 155 | 136 | 0.64 | 0.91 | 0.75 |
| Psychiatry | 475 | 193 | 128 | 0.50 | 0.87 | 0.64 |
| Nephrology | 432 | 277 | 221 | 0.48 | 0.84 | 0.62 |
| Neurology | 282 | 358 | 177 | 0.50 | 0.78 | 0.61 |
| Study | Approach | Dataset | Input | Target | Performance |
|---|---|---|---|---|---|
| Xie et al. [18] | Deep learning | MIMIC-III | Diagnosis description | 2833 ICD-9 codes | Sensitivity: 0.29 Specificity: 0.33 |
| Huang et al. [19] | Deep learning | MIMIC-III | Discharge summary | 10 ICD-9 codes and 10 blocks | F1 score: Full code-0.69, ICD-9 block-0.72 |
| Zeng et al. [20] | Deep learning | MIMIC-III | Discharge summary | 6984 ICD-9 codes | F1 score-0.42 |
| Samonte et al. [21] | Deep learning | MIMIC-III | Discharge summary | 10 ICD-9 codes | Recall: 0.62, F1-score: 0.67 |
| Hsu et al. [22] | Deep learning | MIMIC-III | Discharge summary | Chapters (19), 50 and 100 ICD-9 codes | Micro F1 score: 0.76 Full code: 0.57 top-50; 0.51-top-10 |
| Gangavarapu et al. [23] | Deep learning | MIMIC-III | Nursing notes | 19 Chapters | Accuracy- 0.83 |
| Singaravelan et al. [24] | Deep learning | Medical Center | Subjective component | 1871 ICD-19 codes | Recall score: Chapter-0.57, block—0.49, Three-digit code-0.43, Full code—0.45 |
| Our study | Depp learning | Medical Center | Clinical notes | 1131 ICD-10 codes | Precision: 0.50~0.69 Recall: 0.78~0.89 F1 score: 0.61~0.78 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Masud, J.H.B.; Shun, C.; Kuo, C.-C.; Islam, M.M.; Yeh, C.-Y.; Yang, H.-C.; Lin, M.-C. Deep-ADCA: Development and Validation of Deep Learning Model for Automated Diagnosis Code Assignment Using Clinical Notes in Electronic Medical Records. J. Pers. Med. 2022, 12, 707. https://doi.org/10.3390/jpm12050707
Masud JHB, Shun C, Kuo C-C, Islam MM, Yeh C-Y, Yang H-C, Lin M-C. Deep-ADCA: Development and Validation of Deep Learning Model for Automated Diagnosis Code Assignment Using Clinical Notes in Electronic Medical Records. Journal of Personalized Medicine. 2022; 12(5):707. https://doi.org/10.3390/jpm12050707
Chicago/Turabian StyleMasud, Jakir Hossain Bhuiyan, Chiang Shun, Chen-Cheng Kuo, Md. Mohaimenul Islam, Chih-Yang Yeh, Hsuan-Chia Yang, and Ming-Chin Lin. 2022. "Deep-ADCA: Development and Validation of Deep Learning Model for Automated Diagnosis Code Assignment Using Clinical Notes in Electronic Medical Records" Journal of Personalized Medicine 12, no. 5: 707. https://doi.org/10.3390/jpm12050707
APA StyleMasud, J. H. B., Shun, C., Kuo, C.-C., Islam, M. M., Yeh, C.-Y., Yang, H.-C., & Lin, M.-C. (2022). Deep-ADCA: Development and Validation of Deep Learning Model for Automated Diagnosis Code Assignment Using Clinical Notes in Electronic Medical Records. Journal of Personalized Medicine, 12(5), 707. https://doi.org/10.3390/jpm12050707







