Patient-Derived Organoids of Colorectal Cancer: A Useful Tool for Personalized Medicine
Abstract
1. Introduction
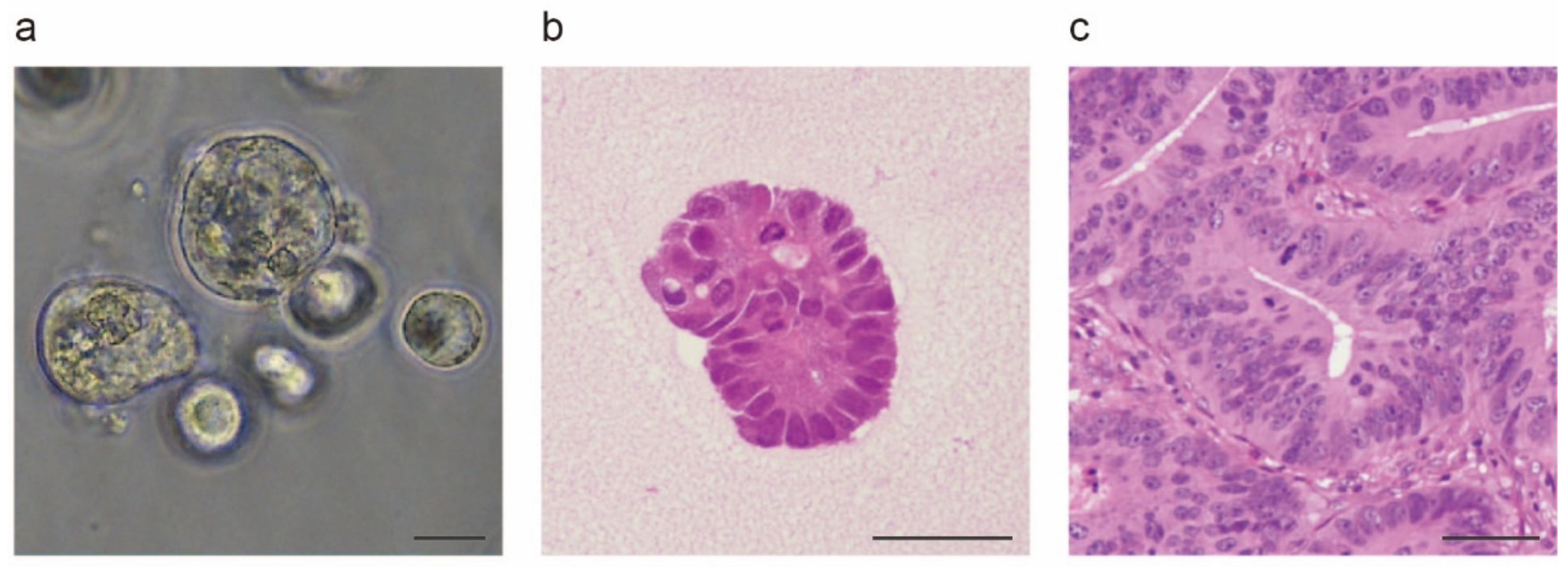
2. In Vitro Models of Colorectal Cancer: From Two to Three Dimensions
3. Patient-Derived Organoids as Predictive Biomarkers in Cancer Therapy
4. Patient-Derived Organoids in Precision Medicine
5. Challenges and Prospects for the Clinical Application of Patient-Derived Organoid-Based Therapy
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dekker, E.; Tanis, P.J.; Vleugels, J.L.A.; Kasi, P.M.; Wallace, M.B. Colorectal Cancer. Lancet 2019, 394, 1467–1480. [Google Scholar] [CrossRef]
- Keum, N.; Giovannucci, E. Global Burden of Colorectal Cancer: Emerging Trends, Risk Factors and Prevention Strategies. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 713–732. [Google Scholar] [CrossRef] [PubMed]
- Rawla, P.; Sunkara, T.; Barsouk, A. Epidemiology of Colorectal Cancer: Incidence, Mortality, Survival, and Risk Factors. Gastroenterol. Rev. Gastroenterol. 2019, 14, 89–103. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Network Comprehensive Molecular Characterization of Human Colon and Rectal Cancer. Nature 2012, 487, 330–337. [CrossRef]
- Guinney, J.; Dienstmann, R.; Wang, X.; de Reyniès, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The Consensus Molecular Subtypes of Colorectal Cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Stange, D.E.; Ferrante, M.; Vries, R.G.J.; Van Es, J.H.; Van den Brink, S.; Van Houdt, W.J.; Pronk, A.; Van Gorp, J.; Siersema, P.D.; et al. Long-Term Expansion of Epithelial Organoids from Human Colon, Adenoma, Adenocarcinoma, and Barrett’s Epithelium. Gastroenterology 2011, 141, 1762–1772. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, D.; Eide, P.W.; Eilertsen, I.A.; Danielsen, S.A.; Eknæs, M.; Hektoen, M.; Lind, G.E.; Lothe, R.A. Epigenetic and Genetic Features of 24 Colon Cancer Cell Lines. Oncogenesis 2013, 2, e71. [Google Scholar] [CrossRef]
- Berg, K.C.G.; Eide, P.W.; Eilertsen, I.A.; Johannessen, B.; Bruun, J.; Danielsen, S.A.; Bjørnslett, M.; Meza-Zepeda, L.A.; Eknæs, M.; Lind, G.E.; et al. Multi-Omics of 34 Colorectal Cancer Cell Lines—A Resource for Biomedical Studies. Mol. Cancer 2017, 16, 116. [Google Scholar] [CrossRef]
- Marian, B. In Vitro Models for the Identification and Characterization of Tumor-Promoting and Protective Factors for Colon Carcinogenesis. Food Chem. Toxicol. 2002, 40, 1099–1104. [Google Scholar] [CrossRef]
- Leibovitz, A.; Stinson, J.C.; McCombs, W.B.; McCoy, C.E.; Mazur, K.C.; Mabry, N.D. Classification of Human Colorectal Adenocarcinoma Cell Lines. Cancer Res. 1976, 36, 4562–4569. [Google Scholar]
- Fogh, J.; Fogh, J.M.; Orfeo, T. One Hundred and Twenty-Seven Cultured Human Tumor Cell Lines Producing Tumors in Nude Mice. J. Natl. Cancer Inst. 1977, 59, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Brattain, M.G.; Brattain, D.E.; Fine, W.D.; Khaled, F.M.; Marks, M.E.; Kimball, P.M.; Arcolano, L.A.; Danbury, B.H. Initiation and Characterization of Cultures of Human Colonic Carcinoma with Different Biological Characteristics Utilizing Feeder Layers of Confluent Fibroblasts. Oncodevelopmental Biol. Med. 1981, 2, 355–366. [Google Scholar]
- Kataoka, H.; Nabeshima, K.; Komada, N.; Koono, M. New Human Colorectal Carcinoma Cell Lines That Secrete Proteinase Inhibitors in Vitro. Virchows Arch. B 1989, 57, 157. [Google Scholar] [CrossRef] [PubMed]
- Bian, X.; Cao, F.; Wang, X.; Hou, Y.; Zhao, H.; Liu, Y. Establishment and Characterization of a New Human Colon Cancer Cell Line, PUMC-CRC1. Sci. Rep. 2021, 11, 13122. [Google Scholar] [CrossRef] [PubMed]
- Linnekamp, J.F.; van Hooff, S.R.; Prasetyanti, P.R.; Kandimalla, R.; Buikhuisen, J.Y.; Fessler, E.; Ramesh, P.; Lee, K.A.S.T.; Bochove, G.G.W.; de Jong, J.H.; et al. Consensus Molecular Subtypes of Colorectal Cancer Are Recapitulated in in Vitro and in Vivo Models. Cell Death Differ. 2018, 25, 616–633. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.R.; Drabkowski, D.; Hay, R.J.; Macy, M.; Peterson, W. WiDr Is a Derivative of Another Colon Adenocarcinoma Cell Line, HT-29. Cancer Genet. Cytogenet. 1987, 27, 125–134. [Google Scholar] [CrossRef]
- Chen, T.R.; Dorotinsky, C.S.; McGuire, L.J.; Macy, M.L.; Hay, R.J. DLD-1 and HCT-15 Cell Lines Derived Separately from Colorectal Carcinomas Have Totally Different Chromosome Changes but the Same Genetic Origin. Cancer Genet. Cytogenet. 1995, 81, 103–108. [Google Scholar] [CrossRef]
- Hinoue, T.; Weisenberger, D.J.; Pan, F.; Campan, M.; Kim, M.; Young, J.; Whitehall, V.L.; Leggett, B.A.; Laird, P.W. Analysis of the Association between CIMP and BRAFV600E in Colorectal Cancer by DNA Methylation Profiling. PLoS ONE 2009, 4, e8357. [Google Scholar] [CrossRef]
- Breslin, S.; O’Driscoll, L. The Relevance of Using 3D Cell Cultures, in Addition to 2D Monolayer Cultures, When Evaluating Breast Cancer Drug Sensitivity and Resistance. Oncotarget 2016, 7, 45745–45756. [Google Scholar] [CrossRef]
- Riedl, A.; Schlederer, M.; Pudelko, K.; Stadler, M.; Walter, S.; Unterleuthner, D.; Unger, C.; Kramer, N.; Hengstschläger, M.; Kenner, L.; et al. Comparison of Cancer Cells in 2D vs 3D Culture Reveals Differences in AKT-MTOR-S6K Signaling and Drug Responses. J. Cell Sci. 2017, 130, 203–218. [Google Scholar]
- Rombouts, C.; De Spiegeleer, M.; Van Meulebroek, L.; Vanhaecke, L.; De Vos, W.H. Comprehensive Polar Metabolomics and Lipidomics Profiling Discriminates the Transformed from the Non-Transformed State in Colon Tissue and Cell Lines. Sci. Rep. 2021, 11, 17249. [Google Scholar] [CrossRef] [PubMed]
- Debnath, J.; Brugge, J.S. Modelling Glandular Epithelial Cancers in Three-Dimensional Cultures. Nat. Rev. Cancer 2005, 5, 675–688. [Google Scholar] [CrossRef] [PubMed]
- Smalley, K.S.M.; Lioni, M.; Herlyn, M. Life Ins’t Flat: Taking Cancer Biology to the next Dimension. In Vitro Cell. Dev. Biol.-Anim. 2006, 42, 242–247. [Google Scholar] [CrossRef]
- Yamada, K.M.; Cukierman, E. Modeling Tissue Morphogenesis and Cancer in 3D. Cell 2007, 130, 601–610. [Google Scholar] [CrossRef]
- Hirschhaeuser, F.; Menne, H.; Dittfeld, C.; West, J.; Mueller-Klieser, W.; Kunz-Schughart, L.A. Multicellular Tumor Spheroids: An Underestimated Tool Is Catching up Again. J. Biotechnol. 2010, 148, 3–15. [Google Scholar] [CrossRef]
- Booth, C.; O’Shea, J.A.; Potten, C.S. Maintenance of Functional Stem Cells in Isolated and Cultured Adult Intestinal Epithelium. Exp. Cell Res. 1999, 249, 359–366. [Google Scholar] [CrossRef]
- Hearn, C.J.; Young, H.M.; Ciampoli, D.; Lomax, A.E.G.; Newgreen, D. Catenary Cultures of Embryonic Gastrointestinal Tract Support Organ Morphogenesis, Motility, Neural Crest Cell Migration, and Cell Differentiation. Dev. Dyn. 1999, 214, 239–247. [Google Scholar] [CrossRef]
- Abud, H.E.; Watson, N.; Heath, J.K. Growth of Intestinal Epithelium in Organ Culture Is Dependent on EGF Signalling. Exp. Cell Res. 2005, 303, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 Stem Cells Build Crypt-Villus Structures in Vitro without a Mesenchymal Niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef]
- Sasaki, T.; Giltay, R.; Talts, U.; Timpl, R.; Talts, J.F. Expression and Distribution of Laminin A1 and A2 Chains in Embryonic and Adult Mouse Tissues: An Immunochemical Approach. Exp. Cell Res. 2002, 275, 185–199. [Google Scholar] [CrossRef]
- Dignass, A.U.; Sturm, A. Peptide Growth Factors in the Intestine. Eur. J. Gastroenterol. Hepatol. 2001, 13, 763–770. [Google Scholar] [CrossRef]
- Haramis, A.-P.G.; Begthel, H.; Van Den Born, M.; Van Es, J.; Jonkheer, S.; Offerhaus, G.J.A.; Clevers, H. De Novo Crypt Formation and Juvenile Polyposis on BMP Inhibition in Mouse Intestine. Science 2004, 303, 1684–1686. [Google Scholar] [CrossRef] [PubMed]
- de Lau, W.; Barker, N.; Low, T.Y.; Koo, B.-K.; Li, V.S.W.; Teunissen, H.; Kujala, P.; Haegebarth, A.; Peters, P.J.; van de Wetering, M.; et al. Lgr5 Homologues Associate with Wnt Receptors and Mediate R-Spondin Signalling. Nature 2011, 476, 293–297. [Google Scholar] [CrossRef]
- Ootani, A.; Li, X.; Sangiorgi, E.; Ho, Q.T.; Ueno, H.; Toda, S.; Sugihara, H.; Fujimoto, K.; Weissman, I.L.; Capecchi, M.R.; et al. Sustained in Vitro Intestinal Epithelial Culture within a Wnt-Dependent Stem Cell Niche. Nat. Med. 2009, 15, 701–706. [Google Scholar] [CrossRef] [PubMed]
- Weiswald, L.-B.; Richon, S.; Validire, P.; Briffod, M.; Lai-Kuen, R.; Cordelières, F.P.; Bertrand, F.; Dargere, D.; Massonnet, G.; Marangoni, E.; et al. Newly Characterised Ex Vivo Colospheres as a Three-Dimensional Colon Cancer Cell Model of Tumour Aggressiveness. Br. J. Cancer 2009, 101, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Weiswald, L.-B.; Richon, S.; Massonnet, G.; Guinebretière, J.-M.; Vacher, S.; Laurendeau, I.; Cottu, P.; Marangoni, E.; Nemati, F.; Validire, P.; et al. A Short-Term Colorectal Cancer Sphere Culture as a Relevant Tool for Human Cancer Biology Investigation. Br. J. Cancer 2013, 108, 1720–1731. [Google Scholar] [CrossRef][Green Version]
- Jeppesen, M.; Hagel, G.; Glenthoj, A.; Vainer, B.; Ibsen, P.; Harling, H.; Thastrup, O.; Jørgensen, L.N.; Thastrup, J. Short-Term Spheroid Culture of Primary Colorectal Cancer Cells as an in Vitro Model for Personalizing Cancer Medicine. PLoS ONE 2017, 12, e0183074. [Google Scholar] [CrossRef]
- Kondo, J.; Endo, H.; Okuyama, H.; Ishikawa, O.; Iishi, H.; Tsujii, M.; Ohue, M.; Inoue, M. Retaining Cell–Cell Contact Enables Preparation and Culture of Spheroids Composed of Pure Primary Cancer Cells from Colorectal Cancer. Proc. Natl. Acad. Sci. USA 2011, 108, 6235–6240. [Google Scholar] [CrossRef]
- Matano, M.; Date, S.; Shimokawa, M.; Takano, A.; Fujii, M.; Ohta, Y.; Watanabe, T.; Kanai, T.; Sato, T. Modeling Colorectal Cancer Using CRISPR-Cas9–Mediated Engineering of Human Intestinal Organoids. Nat. Med. 2015, 21, 256–262. [Google Scholar] [CrossRef]
- Abu-Remaileh, M.; Bender, S.; Raddatz, G.; Ansari, I.; Cohen, D.; Gutekunst, J.; Musch, T.; Linhart, H.; Breiling, A.; Pikarsky, E.; et al. Chronic Inflammation Induces a Novel Epigenetic Program That Is Conserved in Intestinal Adenomas and in Colorectal Cancer. Cancer Res. 2015, 75, 2120–2130. [Google Scholar] [CrossRef]
- Fumagalli, A.; Drost, J.; Suijkerbuijk, S.J.E.; van Boxtel, R.; de Ligt, J.; Offerhaus, G.J.; Begthel, H.; Beerling, E.; Tan, E.H.; Sansom, O.J.; et al. Genetic Dissection of Colorectal Cancer Progression by Orthotopic Transplantation of Engineered Cancer Organoids. Proc. Natl. Acad. Sci. USA 2017, 114, E2357–E2364. [Google Scholar] [CrossRef] [PubMed]
- Lannagan, T.R.M.; Lee, Y.K.; Wang, T.; Roper, J.; Bettington, M.L.; Fennell, L.; Vrbanac, L.; Jonavicius, L.; Somashekar, R.; Gieniec, K.; et al. Genetic Editing of Colonic Organoids Provides a Molecularly Distinct and Orthotopic Preclinical Model of Serrated Carcinogenesis. Gut 2019, 68, 684–692. [Google Scholar] [CrossRef] [PubMed]
- Takeda, H.; Kataoka, S.; Nakayama, M.; Ali, M.A.E.; Oshima, H.; Yamamoto, D.; Park, J.-W.; Takegami, Y.; An, T.; Jenkins, N.A.; et al. CRISPR-Cas9-Mediated Gene Knockout in Intestinal Tumor Organoids Provides Functional Validation for Colorectal Cancer Driver Genes. Proc. Natl. Acad. Sci. USA 2019, 116, 15635–15644. [Google Scholar] [CrossRef]
- Kawasaki, K.; Fujii, M.; Sugimoto, S.; Ishikawa, K.; Matano, M.; Ohta, Y.; Toshimitsu, K.; Takahashi, S.; Hosoe, N.; Sekine, S.; et al. Chromosome Engineering of Human Colon-Derived Organoids to Develop a Model of Traditional Serrated Adenoma. Gastroenterology 2020, 158, 638–651. [Google Scholar] [CrossRef] [PubMed]
- Weeber, F.; van de Wetering, M.; Hoogstraat, M.; Dijkstra, K.K.; Krijgsman, O.; Kuilman, T.; Hooijdonk, C.G.M.G.; van der Velden, D.L.; Peeper, D.S.; Cuppen, E.P.J.G.; et al. Preserved Genetic Diversity in Organoids Cultured from Biopsies of Human Colorectal Cancer Metastases. Proc. Natl. Acad. Sci. USA 2015, 112, 13308–13311. [Google Scholar] [CrossRef]
- Vellinga, T.T.; den Uil, S.; Rinkes, I.H.B.; Marvin, D.; Ponsioen, B.; Alvarez-Varela, A.; Fatrai, S.; Scheele, C.; Zwijnenburg, D.A.; Snippert, H.; et al. Collagen-Rich Stroma in Aggressive Colon Tumors Induces Mesenchymal Gene Expression and Tumor Cell Invasion. Oncogene 2016, 35, 5263–5271. [Google Scholar] [CrossRef]
- Okuyama, H.; Kondo, J.; Sato, Y.; Endo, H.; Nakajima, A.; Piulats, J.M.; Tomita, Y.; Fujiwara, T.; Itoh, Y.; Mizoguchi, A.; et al. Dynamic Change of Polarity in Primary Cultured Spheroids of Human Colorectal Adenocarcinoma and Its Role in Metastasis. Am. J. Pathol. 2016, 186, 899–911. [Google Scholar] [CrossRef] [PubMed]
- O’Rourke, K.P.; Loizou, E.; Livshits, G.; Schatoff, E.M.; Baslan, T.; Manchado, E.; Simon, J.; Romesser, P.B.; Leach, B.; Han, T.; et al. Transplantation of Engineered Organoids Enables Rapid Generation of Metastatic Mouse Models of Colorectal Cancer. Nat. Biotechnol. 2017, 35, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Fumagalli, A.; Suijkerbuijk, S.J.E.; Begthel, H.; Beerling, E.; Oost, K.C.; Snippert, H.J.; van Rheenen, J.; Drost, J. A Surgical Orthotopic Organoid Transplantation Approach in Mice to Visualize and Study Colorectal Cancer Progression. Nat. Protoc. 2018, 13, 235–247. [Google Scholar] [CrossRef]
- Ganesh, K.; Basnet, H.; Kaygusuz, Y.; Laughney, A.M.; He, L.; Sharma, R.; O’Rourke, K.P.; Reuter, V.P.; Huang, Y.-H.; Turkekul, M.; et al. L1CAM Defines the Regenerative Origin of Metastasis-Initiating Cells in Colorectal Cancer. Nat. Cancer 2020, 1, 28–45. [Google Scholar] [CrossRef]
- Strelez, C.; Chilakala, S.; Ghaffarian, K.; Lau, R.; Spiller, E.; Ung, N.; Hixon, D.; Yoon, A.Y.; Sun, R.X.; Lenz, H.-J.; et al. Human Colorectal Cancer-on-Chip Model to Study the Microenvironmental Influence on Early Metastatic Spread. iScience 2021, 24, 102509. [Google Scholar] [CrossRef] [PubMed]
- Calon, A.; Lonardo, E.; Berenguer-Llergo, A.; Espinet, E.; Hernando-Momblona, X.; Iglesias, M.; Sevillano, M.; Palomo-Ponce, S.; Tauriello, D.V.F.; Byrom, D.; et al. Stromal Gene Expression Defines Poor-Prognosis Subtypes in Colorectal Cancer. Nat. Genet. 2015, 47, 320–329. [Google Scholar] [CrossRef]
- Kong, J.C.H.; Guerra, G.R.; Millen, R.M.; Roth, S.; Xu, H.; Neeson, P.J.; Darcy, P.K.; Kershaw, M.H.; Sampurno, S.; Malaterre, J.; et al. Tumor-Infiltrating Lymphocyte Function Predicts Response to Neoadjuvant Chemoradiotherapy in Locally Advanced Rectal Cancer. JCO Precis. Oncol. 2018, 2, 1–15. [Google Scholar] [CrossRef]
- Courau, T.; Bonnereau, J.; Chicoteau, J.; Bottois, H.; Remark, R.; Miranda, L.A.; Toubert, A.; Blery, M.; Aparicio, T.; Allez, M.; et al. Cocultures of Human Colorectal Tumor Spheroids with Immune Cells Reveal the Therapeutic Potential of MICA/B and NKG2A Targeting for Cancer Treatment. J. Immunother. Cancer 2019, 7, 74. [Google Scholar] [CrossRef] [PubMed]
- Nakano, M.; Kikushige, Y.; Miyawaki, K.; Kunisaki, Y.; Mizuno, S.; Takenaka, K.; Tamura, S.; Okumura, Y.; Ito, M.; Ariyama, H.; et al. Dedifferentiation Process Driven by TGF-Beta Signaling Enhances Stem Cell Properties in Human Colorectal Cancer. Oncogene 2019, 38, 780–793. [Google Scholar] [CrossRef]
- Schumacher, D.; Andrieux, G.; Boehnke, K.; Keil, M.; Silvestri, A.; Silvestrov, M.; Keilholz, U.; Haybaeck, J.; Erdmann, G.; Sachse, C.; et al. Heterogeneous Pathway Activation and Drug Response Modelled in Colorectal-Tumor-Derived 3D Cultures. PLoS Genet. 2019, 15, e1008076. [Google Scholar] [CrossRef] [PubMed]
- Holbeck, S.L.; Collins, J.M.; Doroshow, J.H. Analysis of Food and Drug Administration-Approved Anticancer Agents in the NCI60 Panel of Human Tumor Cell Lines. Mol. Cancer Ther. 2010, 9, 1451–1460. [Google Scholar] [CrossRef]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia Enables Predictive Modelling of Anticancer Drug Sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef]
- Seyhan, A.A. Lost in Translation: The Valley of Death across Preclinical and Clinical Divide—Identification of Problems and Overcoming Obstacles. Transl. Med. Commun. 2019, 4, 18. [Google Scholar] [CrossRef]
- Folkesson, E.; Niederdorfer, B.; Nakstad, V.T.; Thommesen, L.; Klinkenberg, G.; Lægreid, A.; Flobak, Å. High-Throughput Screening Reveals Higher Synergistic Effect of MEK Inhibitor Combinations in Colon Cancer Spheroids. Sci. Rep. 2020, 10, 11574. [Google Scholar] [CrossRef]
- Kawai, S.; Yamazaki, M.; Shibuya, K.; Yamazaki, M.; Fujii, E.; Nakano, K.; Suzuki, M. Three-Dimensional Culture Models Mimic Colon Cancer Heterogeneity Induced by Different Microenvironments. Sci. Rep. 2020, 10, 3156. [Google Scholar] [CrossRef] [PubMed]
- Durinikova, E.; Buzo, K.; Arena, S. Preclinical Models as Patients’ Avatars for Precision Medicine in Colorectal Cancer: Past and Future Challenges. J. Exp. Clin. Cancer Res. 2021, 40, 185. [Google Scholar] [CrossRef]
- Yan, H.H.N.; Siu, H.C.; Ho, S.L.; Yue, S.S.K.; Gao, Y.; Tsui, W.Y.; Chan, D.; Chan, A.S.; Wong, J.W.H.; Man, A.H.Y.; et al. Organoid Cultures of Early-Onset Colorectal Cancers Reveal Distinct and Rare Genetic Profiles. Gut 2020, 69, 2165–2179. [Google Scholar] [CrossRef] [PubMed]
- Codrich, M.; Dalla, E.; Mio, C.; Antoniali, G.; Malfatti, M.C.; Marzinotto, S.; Pierobon, M.; Baldelli, E.; Di Loreto, C.; Damante, G.; et al. Integrated Multi-Omics Analyses on Patient-Derived CRC Organoids Highlight Altered Molecular Pathways in Colorectal Cancer Progression Involving PTEN. J. Exp. Clin. Cancer Res. 2021, 40, 198. [Google Scholar] [CrossRef] [PubMed]
- Schütte, M.; Risch, T.; Abdavi-Azar, N.; Boehnke, K.; Schumacher, D.; Keil, M.; Yildiriman, R.; Jandrasits, C.; Borodina, T.; Amstislavskiy, V.; et al. Molecular Dissection of Colorectal Cancer in Pre-Clinical Models Identifies Biomarkers Predicting Sensitivity to EGFR Inhibitors. Nat. Commun. 2017, 8, 14262. [Google Scholar] [CrossRef]
- Vlachogiannis, G.; Hedayat, S.; Vatsiou, A.; Jamin, Y.; Fernández-Mateos, J.; Khan, K.; Lampis, A.; Eason, K.; Huntingford, I.; Burke, R.; et al. Patient-Derived Organoids Model Treatment Response of Metastatic Gastrointestinal Cancers. Science 2018, 359, 920–926. [Google Scholar] [CrossRef]
- Francies, H.E.; Barthorpe, A.; McLaren-Douglas, A.; Barendt, W.J.; Garnett, M.J. Drug Sensitivity Assays of Human Cancer Organoid Cultures. Methods Mol. Biol. 2019, 1576, 339–351. [Google Scholar]
- Ganesh, K.; Wu, C.; O’Rourke, K.P.; Szeglin, B.C.; Zheng, Y.; Sauvé, C.-E.G.; Adileh, M.; Wasserman, I.; Marco, M.R.; Kim, A.S.; et al. A Rectal Cancer Organoid Platform to Study Individual Responses to Chemoradiation. Nat. Med. 2019, 25, 1607–1614. [Google Scholar] [CrossRef]
- van de Wetering, M.; Francies, H.E.; Francis, J.M.; Bounova, G.; Iorio, F.; Pronk, A.; van Houdt, W.; van Gorp, J.; Taylor-Weiner, A.; Kester, L.; et al. Prospective Derivation of a Living Organoid Biobank of Colorectal Cancer Patients. Cell 2015, 161, 933–945. [Google Scholar] [CrossRef]
- Fujii, M.; Shimokawa, M.; Date, S.; Takano, A.; Matano, M.; Nanki, K.; Ohta, Y.; Toshimitsu, K.; Nakazato, Y.; Kawasaki, K.; et al. A Colorectal Tumor Organoid Library Demonstrates Progressive Loss of Niche Factor Requirements during Tumorigenesis. Cell Stem Cell 2016, 18, 827–838. [Google Scholar] [CrossRef]
- Thai, A.A.; Solomon, B.J.; Sequist, L.V.; Gainor, J.F.; Heist, R.S. Lung Cancer. Lancet 2021, 398, 535–554. [Google Scholar] [CrossRef]
- Cerrito, M.G.; Grassilli, E. Identifying Novel Actionable Targets in Colon Cancer. Biomedicines 2021, 9, 579. [Google Scholar] [CrossRef]
- Ben-Hamo, R.; Jacob Berger, A.; Gavert, N.; Miller, M.; Pines, G.; Oren, R.; Pikarsky, E.; Benes, C.H.; Neuman, T.; Zwang, Y.; et al. Predicting and Affecting Response to Cancer Therapy Based on Pathway-Level Biomarkers. Nat. Commun. 2020, 11, 3296. [Google Scholar] [CrossRef] [PubMed]
- Hudziak, R.M.; Lewis, G.D.; Winget, M.; Fendly, B.M.; Shepard, H.M.; Ullrich, A. p185HER2 Monoclonal Antibody Has Antiproliferative Effects in Vitro and Sensitizes Human Breast Tumor Cells to Tumor Necrosis Factor. Mol. Cell. Biol. 1989, 3, 1165–1172. [Google Scholar]
- Slamon, D.J.; Leyland-Jones, B.; Shak, S.; Fuchs, H.; Paton, V.; Bajamonde, A.; Fleming, T.; Eiermann, W.; Wolter, J.; Pegram, M.; et al. Use of Chemotherapy plus a Monoclonal Antibody against HER2 for Metastatic Breast Cancer That Overexpresses HER2. N. Engl. J. Med. 2001, 344, 783–792. [Google Scholar] [CrossRef]
- Solit, D.B.; Garraway, L.A.; Pratilas, C.A.; Sawai, A.; Getz, G.; Basso, A.; Ye, Q.; Lobo, J.M.; She, Y.; Osman, I.; et al. BRAF Mutation Predicts Sensitivity to MEK Inhibition. Nature 2006, 439, 358–362. [Google Scholar] [CrossRef]
- McArthur, G.A.; Chapman, P.B.; Robert, C.; Larkin, J.; Haanen, J.B.; Dummer, R.; Ribas, A.; Hogg, D.; Hamid, O.; Ascierto, P.A.; et al. Safety and Efficacy of Vemurafenib in BRAF(V600E) and BRAF(V600K) Mutation-Positive Melanoma (BRIM-3): Extended Follow-up of a Phase 3, Randomised, Open-Label Study. Lancet Oncol. 2014, 15, 323–332. [Google Scholar] [CrossRef]
- Miao, D.; Margolis, C.A.; Vokes, N.I.; Liu, D.; Taylor-Weiner, A.; Wankowicz, S.M.; Adeegbe, D.; Keliher, D.; Schilling, B.; Tracy, A.; et al. Genomic Correlates of Response to Immune Checkpoint Blockade in Microsatellite-Stable Solid Tumors. Nat. Genet. 2018, 50, 1271–1281. [Google Scholar] [CrossRef]
- Hellmann, M.D.; Ciuleanu, T.-E.; Pluzanski, A.; Lee, J.S.; Otterson, G.A.; Audigier-Valette, C.; Minenza, E.; Linardou, H.; Burgers, S.; Salman, P.; et al. Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N. Engl. J. Med. 2018, 378, 2093–2104. [Google Scholar] [CrossRef]
- Tumeh, P.C.; Harview, C.L.; Yearley, J.H.; Shintaku, I.P.; Taylor, E.J.M.; Robert, L.; Chmielowski, B.; Spasic, M.; Henry, G.; Ciobanu, V.; et al. PD-1 Blockade Induces Responses by Inhibiting Adaptive Immune Resistance. Nature 2014, 515, 568–571. [Google Scholar] [CrossRef]
- Althammer, S.; Tan, T.H.; Spitzmüller, A.; Rognoni, L.; Wiestler, T.; Herz, T.; Widmaier, M.; Rebelatto, M.C.; Kaplon, H.; Damotte, D.; et al. Automated Image Analysis of NSCLC Biopsies to Predict Response to Anti-PD-L1 Therapy. J. Immunother. Cancer 2019, 7, 121. [Google Scholar] [CrossRef] [PubMed]
- Malone, E.R.; Oliva, M.; Sabatini, P.J.B.; Stockley, T.L.; Siu, L.L. Molecular Profiling for Precision Cancer Therapies. Genome Med. 2020, 12, 8. [Google Scholar] [CrossRef] [PubMed]
- Sveen, A.; Kopetz, S.; Lothe, R.A. Biomarker-Guided Therapy for Colorectal Cancer: Strength in Complexity. Nat. Rev. Clin. Oncol. 2020, 17, 11–32. [Google Scholar] [CrossRef] [PubMed]
- Ren, D.; Hua, Y.; Yu, B.; Ye, X.; He, Z.; Li, C.; Wang, J.; Mo, Y.; Wei, X.; Chen, Y.; et al. Predictive Biomarkers and Mechanisms Underlying Resistance to PD1/PD-L1 Blockade Cancer Immunotherapy. Mol. Cancer 2020, 19, 19. [Google Scholar] [CrossRef] [PubMed]
- Kern, S.E. Why Your New Cancer Biomarker May Never Work: Recurrent Patterns and Remarkable Diversity in Biomarker Failures. Cancer Res. 2012, 72, 6097–6101. [Google Scholar] [CrossRef]
- Ooft, S.N.; Weeber, F.; Dijkstra, K.K.; McLean, C.M.; Kaing, S.; van Werkhoven, E.; Schipper, L.; Hoes, L.; Vis, D.J.; van de Haar, J.; et al. Patient-Derived Organoids Can Predict Response to Chemotherapy in Metastatic Colorectal Cancer Patients. Sci. Transl. Med. 2019, 513, eaay2574. [Google Scholar] [CrossRef]
- Wang, T.; Pan, W.; Zheng, H.; Zheng, H.; Wang, Z.; Li, J.J.; Deng, C.; Yan, J. Accuracy of Using a Patient-Derived Tumor Organoid Culture Model to Predict the Response to Chemotherapy Regimens In Stage IV Colorectal Cancer: A Blinded Study. Dis. Colon Rectum 2021, 64, 833–850. [Google Scholar] [CrossRef]
- Yao, Y.; Xu, X.; Yang, L.; Zhu, J.; Wan, J.; Shen, L.; Xia, F.; Fu, G.; Deng, Y.; Pan, M.; et al. Patient-Derived Organoids Predict Chemoradiation Responses of Locally Advanced Rectal Cancer. Cell Stem Cell 2020, 26, 17–26. [Google Scholar] [CrossRef]
- Karakasheva, T.A.; Gabre, J.T.; Sachdeva, U.M.; Cruz-Acuña, R.; Lin, E.W.; DeMarshall, M.; Falk, G.W.; Ginsberg, G.G.; Yang, Z.; Kim, M.M.; et al. Patient-Derived Organoids as a Platform for Modeling a Patient’s Response to Chemoradiotherapy in Esophageal Cancer. Sci. Rep. 2021, 11, 21304. [Google Scholar] [CrossRef]
- Patel, U.B.; Taylor, F.; Blomqvist, L.; George, C.; Evans, H.; Tekkis, P.; Quirke, P.; Sebag-montefiore, D.; Moran, B.; Heald, R.; et al. Magnetic Resonance Imaging–Detected Tumor Response for Locally Advanced Rectal Cancer Predicts Survival Outcomes. MERCURY experience. J. Clin. Oncol. 2011, 28, 3753–3760. [Google Scholar] [CrossRef]
- Jin, G.; Liu, Y.; Zhang, J.; Bian, Z.; Yao, S.; Fei, B.; Zhou, L.; Yin, Y.; Huang, Z. A Panel of Serum Exosomal MicroRNAs as Predictive Markers for Chemoresistance in Advanced Colorectal Cancer. Cancer Chemother. Pharmacol. 2019, 84, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Xiao, Q.; Venkatachalam, N.; Hofheinz, R.-D.; Veldwijk, M.R.; Herskind, C.; Ebert, M.P.; Zhan, T. Predicting Response to Neoadjuvant Chemoradiotherapy in Rectal Cancer: From Biomarkers to Tumor Models. Ther. Adv. Med. Oncol. 2022, 14, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Ooft, S.N.; Weeber, F.; Schipper, L.; Dijkstra, K.K.; McLean, C.M.; Kaing, S.; van de Haar, J.; Prevoo, W.; van Werkhoven, E.; Snaebjornsson, P.; et al. Prospective Experimental Treatment of Colorectal Cancer Patients Based on Organoid Drug Responses. ESMO Open 2021, 6, 100103. [Google Scholar] [CrossRef] [PubMed]
- Friedman, A.A.; Letai, A.; Fisher, D.E.; Flaherty, K.T. Precision Medicine for Cancer with Next-Generation Functional Diagnostics. Nat. Rev. Cancer 2015, 15, 747–756. [Google Scholar] [CrossRef]
- Frampton, G.M.; Fichtenholtz, A.; Otto, G.A.; Wang, K.; Downing, S.R.; He, J.; Schnall-Levin, M.; White, J.; Sanford, E.M.; An, P.; et al. Development and Validation of a Clinical Cancer Genomic Profiling Test Based on Massively Parallel DNA Sequencing. Nat. Biotechnol. 2013, 31, 1023–1031. [Google Scholar] [CrossRef]
- Kohno, T. Implementation of “Clinical Sequencing” in Cancer Genome Medicine in Japan. Cancer Sci. 2018, 109, 507–512. [Google Scholar] [CrossRef]
- Saotome, K.; Chiyoda, T.; Aimono, E.; Nakamura, K.; Tanishima, S.; Nohara, S.; Okada, C.; Hayashi, H.; Kuroda, Y.; Nomura, H.; et al. Clinical Implications of Next-Generation Sequencing-Based Panel Tests for Malignant Ovarian Tumors. Cancer Med. 2020, 9, 7407–7417. [Google Scholar] [CrossRef]
- Stockley, T.L.; Oza, A.M.; Berman, H.K.; Leighl, N.B.; Knox, J.J.; Shepherd, F.A.; Chen, E.X.; Krzyzanowska, M.K.; Dhani, N.; Joshua, A.M.; et al. Molecular Profiling of Advanced Solid Tumors and Patient Outcomes with Genotype-Matched Clinical Trials: The Princess Margaret IMPACT/COMPACT Trial. Genome Med. 2016, 8, 109. [Google Scholar] [CrossRef]
- Zehir, A.; Benayed, R.; Shah, R.H.; Syed, A.; Middha, S.; Kim, H.R.; Srinivasan, P.; Gao, J.; Chakravarty, D.; Devlin, S.M.; et al. Mutational Landscape of Metastatic Cancer Revealed from Prospective Clinical Sequencing of 10,000 Patients. Nat. Med. 2017, 23, 703–713. [Google Scholar] [CrossRef]
- Dalton, W.B.; Forde, P.M.; Kang, H.; Connolly, R.M.; Stearns, V.; Gocke, C.D.; Eshleman, J.R.; Axilbund, J.; Petry, D.; Geoghegan, C.; et al. Personalized Medicine in the Oncology Clinic: Implementation and Outcomes of the Johns Hopkins Molecular Tumor Board. JCO Precis. Oncol. 2017, 1, 1–19. [Google Scholar] [CrossRef]
- Sunami, K.; Ichikawa, H.; Kubo, T.; Kato, M.; Fujiwara, Y.; Shimomura, A.; Koyama, T.; Kakishima, H.; Kitami, M.; Matsushita, H.; et al. Feasibility and Utility of a Panel Testing for 114 Cancer-Associated Genes in a Clinical Setting: A Hospital-Based Study. Cancer Sci. 2019, 110, 1480–1490. [Google Scholar] [CrossRef] [PubMed]
- Pauli, C.; Hopkins, B.D.; Prandi, D.; Shaw, R.; Fedrizzi, T.; Sboner, A.; Sailer, V.; Augello, M.; Puca, L.; Rosati, R.; et al. Personalized In Vitro and In Vivo Cancer Models to Guide Precision Medicine. Cancer Discov. 2017, 7, 462–477. [Google Scholar] [CrossRef] [PubMed]
- Ashley, N.; Jones, M.; Ouaret, D.; Wilding, J.; Bodmer, W.F. Rapidly Derived Colorectal Cancer Cultures Recapitulate Parental Cancer Characteristics and Enable Personalized Therapeutic Assays. J. Pathol. 2014, 234, 34–45. [Google Scholar] [CrossRef]
- He, A.; Powell, S.; Kyle, M.; Rose, M.; Masmila, E.; Estrada, V.; Sicklick, J.K.; Molinolo, A.; Kaushal, S. Cryopreservation of Viable Human Tissues: Renewable Resource for Viable Tissue, Cell Lines, and Organoid Development. Biopreserv. Biobank. 2020, 18, 222–227. [Google Scholar] [CrossRef]
- Häfliger, J.; Morsy, Y.; Scharl, M.; Wawrzyniak, M. From Patient Material to New Discoveries: A Methodological Review and Guide for Intestinal Stem Cell Researchers. Stem Cell Rev. Rep. 2022, 18, 1309–1321. [Google Scholar] [CrossRef]
- Hughes, C.S.; Postovit, L.M.; Lajoie, G.A. Matrigel: A Complex Protein Mixture Required for Optimal Growth of Cell Culture. Proteomics 2010, 10, 1886–1890. [Google Scholar] [CrossRef]
- Aisenbrey, E.A.; Murphy, W.L. Synthetic Alternatives to Matrigel. Nat. Rev. Mater. 2020, 5, 539–551. [Google Scholar] [CrossRef] [PubMed]
- Kozlowski, M.T.; Crook, C.J.; Ku, H.T. Towards Organoid Culture without Matrigel. Commun. Biol. 2021, 4, 1387. [Google Scholar] [CrossRef]
- Jabaji, Z.; Brinkley, G.J.; Khalil, H.A.; Sears, C.M.; Lei, N.Y.; Lewis, M.; Stelzner, M.; Martín, M.G.; Dunn, J.C.Y. Type I Collagen as an Extracellular Matrix for the In Vitro Growth of Human Small Intestinal Epithelium. PLoS ONE 2014, 9, e107814. [Google Scholar] [CrossRef] [PubMed]
- Bergenheim, F.; Fregni, G.; Buchanan, C.F.; Riis, L.B.; Heulot, M.; Touati, J.; Seidelin, J.B.; Rizzi, S.C.; Nielsen, O.H. A Fully Defined 3D Matrix for Ex Vivo Expansion of Human Colonic Organoids from Biopsy Tissue. Biomaterials 2020, 262, 120248. [Google Scholar] [CrossRef] [PubMed]
- Praharaj, P.P.; Bhutia, S.K.; Nagrath, S.; Bitting, R.L.; Deep, G. Circulating Tumor Cell-Derived Organoids: Current Challenges and Promises in Medical Research and Precision Medicine. Biochim. Biophys. Acta Rev. Cancer 2018, 1869, 117–127. [Google Scholar] [CrossRef]
- Yang, C.; Xia, B.-R.; Jin, W.-L.; Lou, G. Circulating Tumor Cells in Precision Oncology: Clinical Applications in Liquid Biopsy and 3D Organoid Model. Cancer Cell Int. 2019, 19, 341. [Google Scholar] [CrossRef] [PubMed]
- Bidard, F.-C.; Kiavue, N.; Ychou, M.; Cabel, L.; Stern, M.-H.; Madic, J.; Saliou, A.; Rampanou, A.; Decraene, C.; Bouché, O.; et al. Circulating Tumor Cells and Circulating Tumor DNA Detection in Potentially Resectable Metastatic Colorectal Cancer: A Prospective Ancillary Study to the Unicancer Prodige-14 Trial. Cells 2019, 8, 516. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Vela, I.; Sboner, A.; Iaquinta, P.J.; Karthaus, W.R.; Gopalan, A.; Dowling, C.; Wanjala, J.N.; Undvall, E.A.; Arora, V.K.; et al. Organoid Cultures Derived from Patients with Advanced Prostate Cancer. Cell 2014, 159, 176–187. [Google Scholar] [CrossRef]
- Boehnke, K.; Iversen, P.W.; Schumacher, D.; Lallena, M.J.; Haro, R.; Amat, J.; Haybaeck, J.; Liebs, S.; Lange, M.; Schäfer, R.; et al. Assay Establishment and Validation of a High-Throughput Screening Platform for Three-Dimensional Patient-Derived Colon Cancer Organoid Cultures. J. Biomol. Screen. 2016, 21, 931–941. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.H.; Choi, D.-H.; Park, K.; Lee, S.; Kim, J.; Kim, H.; Jeong, H.-W.; Yang, J.H.; Kim, J.-A.; Chung, S.; et al. Drug Screening by Uniform Patient Derived Colorectal Cancer Hydro-Organoids. Biomaterials 2021, 276, 121004. [Google Scholar] [CrossRef] [PubMed]
- Mazrouei, R.; Velasco, V.; Esfandyarpour, R. 3D-Bioprinted All-Inclusive Bioanalytical Platforms for Cell Studies. Sci. Rep. 2020, 10, 14669. [Google Scholar] [CrossRef] [PubMed]
- Pinho, D.; Santos, D.; Vila, A.; Carvalho, S. Establishment of Colorectal Cancer Organoids in Microfluidic-Based System. Micromachines 2021, 12, 497. [Google Scholar] [CrossRef] [PubMed]
- Hirashita, Y.; Tsukamoto, Y.; Kudo, Y.; Kakisako, D.; Kurogi, S.; Hijiya, N.; Nakada, C.; Uchida, T.; Hirashita, T.; Hiratsuka, T.; et al. Early Response in Phosphorylation of Ribosomal Protein S6 Is Associated with Sensitivity to Trametinib in Colorectal Cancer Cells. Lab. Investig. 2021, 101, 1036–1047. [Google Scholar] [CrossRef]
- Waldman, A.D.; Fritz, J.M.; Lenardo, M.J. A Guide to Cancer Immunotherapy: From T Cell Basic Science to Clinical Practice. Nat. Rev. Immunol. 2020, 20, 651–668. [Google Scholar] [CrossRef]
- He, X.; Xu, C. Immune Checkpoint Signaling and Cancer Immunotherapy. Cell Res. 2020, 30, 660–669. [Google Scholar] [CrossRef] [PubMed]
- Balko, J.M.; Sosman, J.A. A Critical Need for Better Cancer Immunotherapy Models: Are Organotypic Tumor Spheroid Cultures the Answer? Cancer Discov. 2018, 8, 143–145. [Google Scholar] [CrossRef] [PubMed]
- Martín-Ruiz, A.; Fiuza-Luces, C.; Martínez-Martínez, E.; Arias, C.F.; Gutiérrez, L.; Ramírez, M.; Martín-Acosta, P.; Coronado, M.J.; Lucia, A.; Provencio, M. Effects of Anti-PD-1 Immunotherapy on Tumor Regression: Insights from a Patient-Derived Xenograft Model. Sci. Rep. 2020, 10, 7078. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.K.; Yun, N.H.; Jeong, Y.-L.; Park, J.; Doh, J.; Lee, W.Y.; Cho, Y.B. Establishment of Patient-Derived Organotypic Tumor Spheroid Models for Tumor Microenvironment Modeling. Cancer Med. 2021, 10, 5589–5598. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Higa, A.; Hiyama, G.; Tamura, H.; Hoshi, H.; Dobashi, Y.; Katahira, K.; Ishihara, H.; Takagi, K.; Goda, K.; et al. Construction of in Vitro Patient-derived Tumor Models to Evaluate Anticancer Agents and Cancer Immunotherapy. Oncol. Lett. 2021, 21, 406. [Google Scholar] [CrossRef]
- Hu, H.; Piotrowska, Z.; Hare, P.J.; Chen, H.; Mulvey, H.E.; Mayfield, A.; Noeen, S.; Kattermann, K.; Greenberg, M.; Williams, A.; et al. Three Subtypes of Lung Cancer Fibroblasts Define Distinct Therapeutic Paradigms. Cancer Cell 2021, 39, 1531–1547. [Google Scholar] [CrossRef]
- Cheng, N.; Bai, X.; Shu, Y.; Ahmad, O.; Shen, P. Targeting Tumor-Associated Macrophages as an Antitumor Strategy. Biochem. Pharmacol. 2021, 183, 114354. [Google Scholar] [CrossRef]
- Deng, L.; Jiang, N.; Zeng, J.; Wang, Y.; Cui, H. The Versatile Roles of Cancer-Associated Fibroblasts in Colorectal Cancer and Therapeutic Implications. Front. Cell Dev. Biol. 2021, 9, 733270. [Google Scholar] [CrossRef]
- Li, J.; Zhang, A.-H.; Wu, F.-F.; Wang, X.-J. Alterations in the Gut Microbiota and Their Metabolites in Colorectal Cancer: Recent Progress and Future Prospects. Front. Oncol. 2022, 12, 841552. [Google Scholar] [CrossRef]
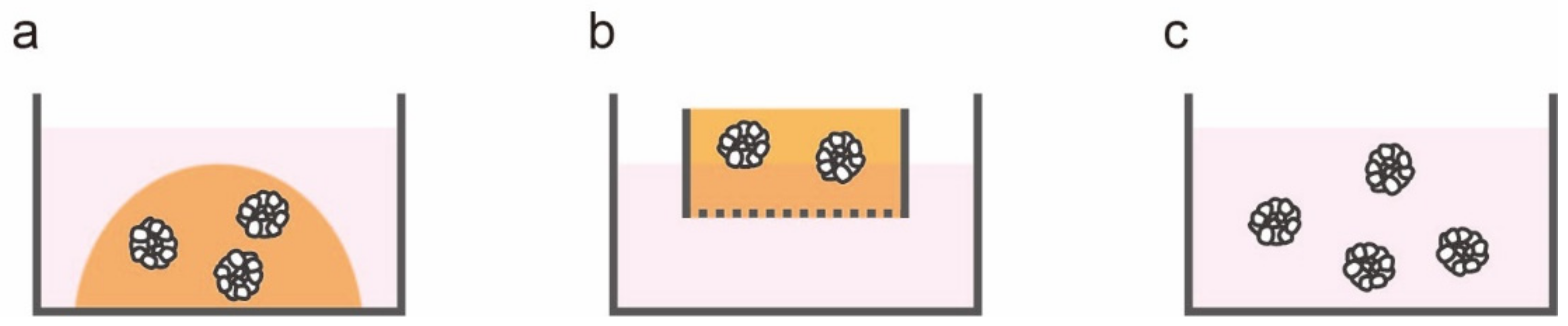

| Method | Procedure | Merit | Demerit |
|---|---|---|---|
| 3D culture models | |||
| Matrigel embedding [6] |
|
|
|
| Air-liquid interface [34] |
|
|
|
| Spheroid culture [38] |
|
|
|
| Other models | |||
| 2D culture |
|
|
|
| Patient- derived xenografts |
|
|
|
| Study | Enrolled Patients | Culture Success Rate | Outcome |
|---|---|---|---|
| Predictive biomarker | |||
| Ooft et al. (2019) [86] | Metastatic CRC | 63.5% (40/63) |
|
| Yao et al. (2020) [88] | Locally advanced rectal cancer | 85.7% (96/112) |
|
| Wang et al. (2021) [87] | Stage IV CRC | 80.2% (77/96) |
|
| Precision medicine | |||
| Ooft et al. (2021) [93] | Metastatic CRC | 57.4% (31/54) |
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kiwaki, T.; Kataoka, H. Patient-Derived Organoids of Colorectal Cancer: A Useful Tool for Personalized Medicine. J. Pers. Med. 2022, 12, 695. https://doi.org/10.3390/jpm12050695
Kiwaki T, Kataoka H. Patient-Derived Organoids of Colorectal Cancer: A Useful Tool for Personalized Medicine. Journal of Personalized Medicine. 2022; 12(5):695. https://doi.org/10.3390/jpm12050695
Chicago/Turabian StyleKiwaki, Takumi, and Hiroaki Kataoka. 2022. "Patient-Derived Organoids of Colorectal Cancer: A Useful Tool for Personalized Medicine" Journal of Personalized Medicine 12, no. 5: 695. https://doi.org/10.3390/jpm12050695
APA StyleKiwaki, T., & Kataoka, H. (2022). Patient-Derived Organoids of Colorectal Cancer: A Useful Tool for Personalized Medicine. Journal of Personalized Medicine, 12(5), 695. https://doi.org/10.3390/jpm12050695






