Investigation of Molecular Features Involved in Clinical Responses and Survival in Advanced Endometrial Carcinoma Treated by Hormone Therapy
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Tumor Samples
2.3. Targeted Next-Generation Sequencing (tNGS)
2.4. Array Comparative Genomic Hybridization (aCGH)
2.5. Statistical Analysis
3. Results
3.1. Patients and Samples Characteristics
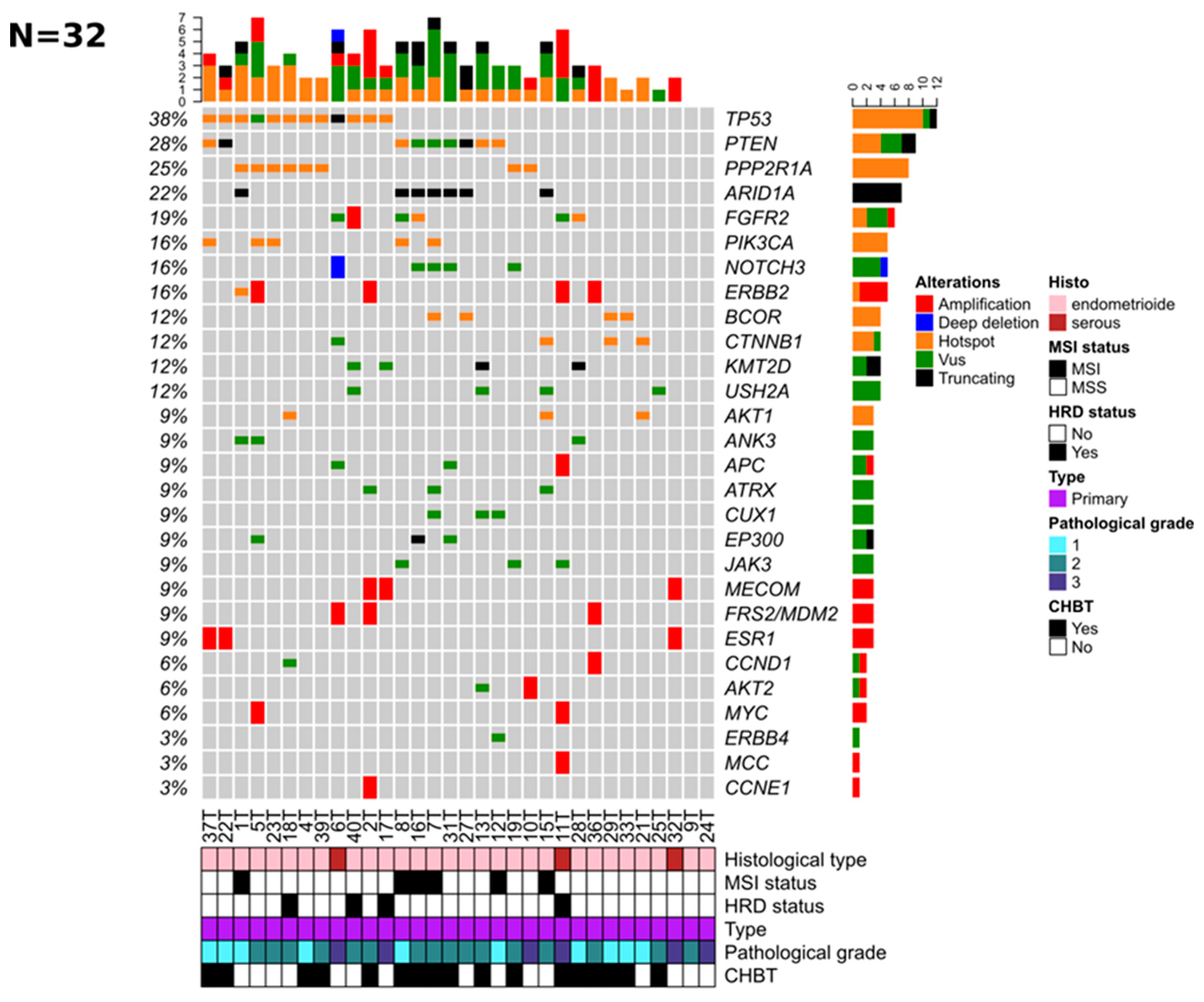
3.2. Genomic Alterations of Tumor Samples
3.3. Clinical Benefit to HT (CBHT)
3.4. Durable Clinical Response to HT (LRHT) and Overall Survival
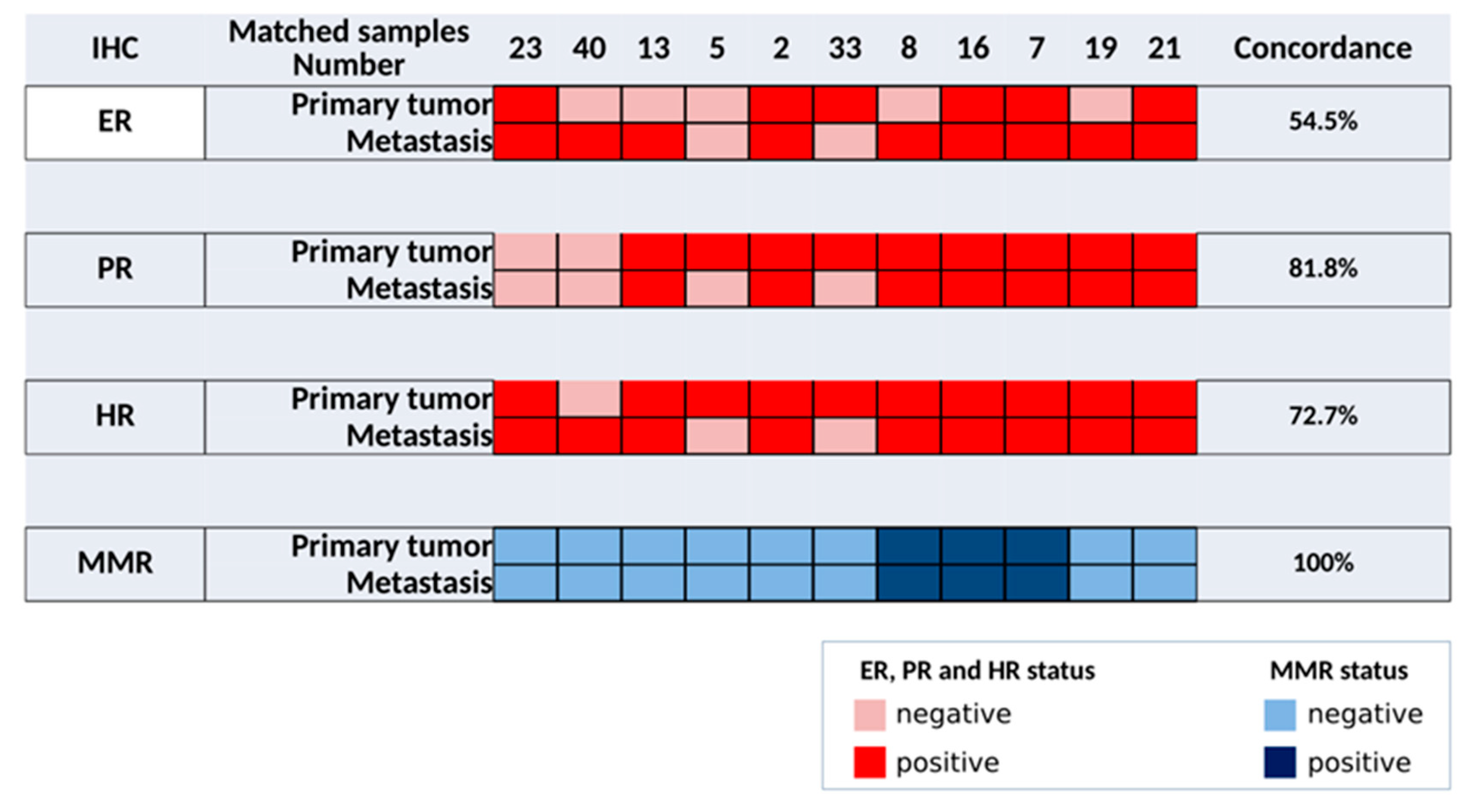
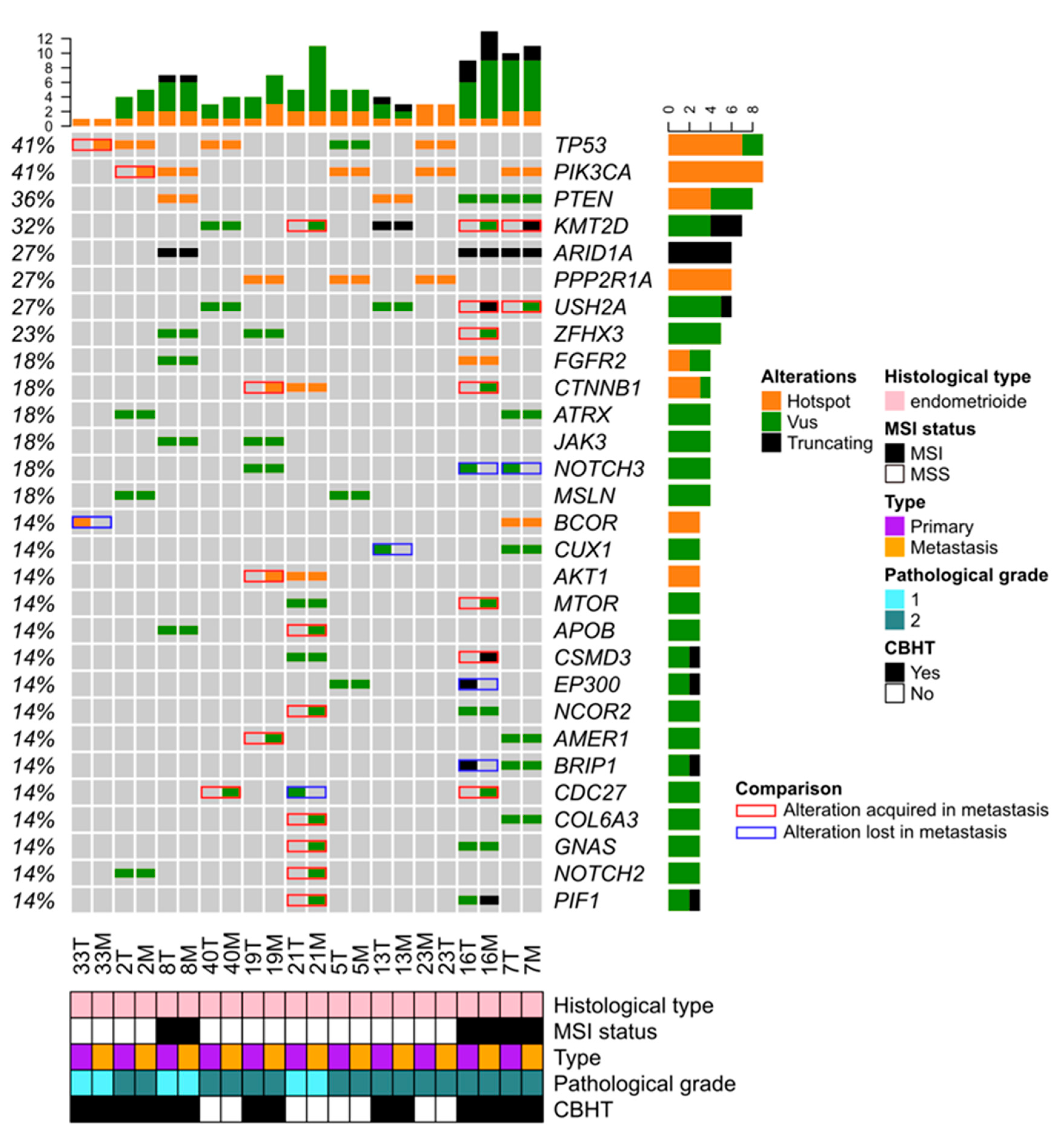
3.5. Comparison of Paired Primary and Metastatic EC Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Bricou, A.; Bendifallah, S.; Daix-Moreux, M.; Ouldamer, L.; Lavoue, V.; Benbara, A.; Huchon, C.; Canlorbe, G.; Raimond, E.; Coutant, C.; et al. A Proposal for a Classification for Recurrent Endometrial Cancer: Analysis of a French Multicenter Database From the FRANCOGYN Study Group. Int. J. Gynecol. Cancer 2018, 28, 1278–1284. [Google Scholar] [CrossRef] [PubMed]
- Concin, N.; Matias-Guiu, X.; Vergote, I.; Cibula, D.; Mirza, M.R.; Marnitz, S.; Ledermann, J.; Bosse, T.; Chargari, C.; Fagotti, A.; et al. ESGO/ESTRO/ESP guidelines for the management of patients with endometrial carcinoma. Int. J. Gynecol. Cancer 2021, 31, 12–39. [Google Scholar] [CrossRef] [PubMed]
- Thigpen, J.T.; Brady, M.F.; Alvarez, R.D.; Adelson, M.D.; Homesley, H.D.; Manetta, A.; Soper, J.T.; Given, F.T. Oral medroxyprogesterone acetate in the treatment of advanced or recurrent endometrial carcinoma: A dose-response study by the Gynecologic Oncology Group. J. Clin. Oncol. 1999, 17, 1736–1744. [Google Scholar] [CrossRef]
- Pautier, P.; Vergote, I.; Joly, F.; Melichar, B.; Kutarska, E.; Hall, E.; Lisyanskaya, A.; Reed, N.; Oaknin, A.; Ostapenko, V.; et al. A Phase 2, Randomized, Open-Label Study of Irosustat Versus Megestrol Acetate in Advanced Endometrial Cancer. Int. J. Gynecol. Cancer 2017, 27, 258–266. [Google Scholar] [CrossRef]
- Yamazawa, K.; Hirai, M.; Fujito, A.; Nishi, H.; Terauchi, F.; Ishikura, H.; Shozu, M.; Isaka, K. Fertility-preserving treatment with progestin, and pathological criteria to predict responses, in young women with endometrial cancer. Hum. Reprod. 2007, 22, 1953–1958. [Google Scholar] [CrossRef]
- Qin, Y.; Yu, Z.; Yang, J.; Cao, D.; Yu, M.; Wang, Y.; Shen, K. Oral Progestin Treatment for Early-Stage Endometrial Cancer: A Systematic Review and Meta-analysis. Int. J. Gynecol. Cancer 2016, 26, 1081–1091. [Google Scholar] [CrossRef]
- Derbyshire, A.; Ryan, N.; Crosbie, E. Biomarkers needed to predict progestin response in endometrial cancer. BJOG Int. J. Obs. Gynaecol. 2017, 124, 1584. [Google Scholar] [CrossRef]
- Ethier, J.-L.; Desautels, D.N.; Amir, E.; MacKay, H. Is hormonal therapy effective in advanced endometrial cancer? A systematic review and meta-analysis. Gynecol. Oncol. 2017, 147, 158–166. [Google Scholar] [CrossRef]
- Tan, D.S.; Rothermundt, C.; Thomas, K.; Bancroft, E.; Eeles, R.; Shanley, S.; Ardern-Jones, A.; Norman, A.; Kaye, S.B.; Gore, M.E. “BRCAness” syndrome in ovarian cancer: A case-control study describing the clinical features and outcome of patients with epithelial ovarian cancer associated with BRCA1 and BRCA2 mutations. J. Clin. Oncol. 2008, 26, 5530–5536. [Google Scholar] [CrossRef]
- Ledermann, J.; Harter, P.; Gourley, C.; Friedlander, M.; Vergote, I.; Rustin, G.; Scott, C.L.; Meier, W.; Shapira-Frommer, R.; Safra, T.; et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: A preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol. 2014, 15, 852–861. [Google Scholar] [CrossRef]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network; Kandoth, C.; Schultz, N.; Cherniack, A.D.; Akbani, R.; Liu, Y.; Hui, S.; Robertson, A.G.; Pashtan, I.; Shen, R.; et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013, 497, 67–73. [Google Scholar] [PubMed]
- Talhouk, A.; McConechy, M.K.; Leung, S.; Li-Chang, H.H.; Kwon, J.S.; Melnyk, N.; Yang, W.; Senz, J.; Boyd, N.F.; Karnezis, A.N.; et al. A clinically applicable molecular-based classification for endometrial cancers. Br. J. Cancer 2015, 113, 299–310. [Google Scholar] [CrossRef]
- Talhouk, A.; McConechy, M.K.; Leung, S.; Yang, W.; Lum, A.; Snez, J.; Boyd, N.; Pike, J.; Anglesio, M.; Kwon, J.S.; et al. Confirmation of ProMisE: A simple, genomics-based clinical classifier for endometrial cancer. Cancer 2017, 123, 802–813. [Google Scholar] [CrossRef]
- Yen, T.-T.; Wang, T.-L.; Fader, A.N.; Shih, I.-M.; Gaillard, S. Molecular Classification and Emerging Targeted Therapy in Endometrial Cancer. Int. J. Gynecol. Pathol. 2020, 39, 26–35. [Google Scholar] [CrossRef]
- Goodfellow, P.J.; Billingsley, C.C.; Lankes, H.A.; Ali, S.; Cohn, D.E.; Broaddus, R.J.; Ramirez, N.; Pritchard, C.C.; Hampel, H.; Chassen, A.S.; et al. Combined Microsatellite Instability, MLH1 Methylation Analysis, and Immunohistochemistry for Lynch Syndrome Screening in Endometrial Cancers From GOG210: An NRG Oncology and Gynecologic Oncology Group Study. J. Clin. Oncol. 2015, 33, 4301–4308. [Google Scholar] [CrossRef]
- Bertucci, F.; Finetti, P.; Guille, A.; Adélaïde, J.; Garnier, S.; Carbuccia, N.; Monneur, A.; Charafe-Jauffret, E.; Goncalves, A.; Viens, P.; et al. Comparative genomic analysis of primary tumors and metastases in breast cancer. Oncotarget 2016, 7, 27208–27219. [Google Scholar] [CrossRef]
- Soumerai, T.E.; Donoghue, M.T.A.; Bandlamudi, C.; Srinivasan, P.; Chang, M.T.; Zamarin, D.; Cadoo, K.A.; Grisham, R.N.; O’Cearbhaill, R.E.; Tew, W.P.; et al. Clinical Utility of Prospective Molecular Characterization in Advanced Endometrial Cancer. Clin. Cancer Res. 2018, 24, 5939–5947. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Koboldt, D.C.; Zhang, Q.; Larson, D.E.; Shen, D.; McLellan, M.D.; Lin, L.; Miller, C.A.; Mardis, E.R.; Ding, L.; Wilson, R.K. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012, 22, 568–576. [Google Scholar] [CrossRef] [PubMed]
- DePristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; Del Angel, G.; Rivas, M.A.; Hanna, M.; et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011, 43, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Ye, K.; Schulz, M.H.; Long, Q.; Apweiler, R.; Ning, Z. Pindel: A pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 2009, 25, 2865–2871. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Thorvaldsdóttir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef]
- Bertucci, F.; Rypens, C.; Finetti, P.; Guille, A.; Adelaide, J.; Monneur, A.; Carbuccia, N.; Garnier, S.; Dirix, P.; Goncalves, A.; et al. NOTCH and DNA repair pathways are more frequently targeted by genomic alterations in inflammatory than in non-inflammatory breast cancers. Mol. Oncol. 2020, 14, 504–519. [Google Scholar] [CrossRef]
- Adélaïde, J.; Finetti, P.; Bekhouche, I.; Repellini, L.; Geneix, J.; Sircoulomb, F.; Charafe-Jauffret, E.; Cervera, N.; Desplans, J.; Parzy, D.; et al. Integrated Profiling of Basal and Luminal Breast Cancers. Cancer Res. 2007, 67, 11565–11575. [Google Scholar] [CrossRef]
- Abkevich, V.; Timms, K.M.; Hennessy, B.T.; Potter, J.; Carey, M.S.; Meyer, L.A.; Smith-McCune, K.; Broaddus, R.; Lu, K.H.; Chen, J.; et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br. J. Cancer 2012, 107, 1776–1782. [Google Scholar] [CrossRef]
- Knijnenburg, T.A.; Wang, L.; Zimmermann, M.T.; Chambwe, N.; Gao, G.F.; Cherniack, A.D.; Fan, H.; Shen, H.; Way, G.P.; Greene, C.S.; et al. Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell Rep. 2018, 23, 239–254.e6. [Google Scholar] [CrossRef]
- Sanchez-Vega, F.; Mina, M.; Armenia, J.; Chatila, W.K.; Luna, A.; La, K.C.; Dimitriadoy, S.; Liu, D.L.; Kantheti, H.S.; Saghafinia, S.; et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018, 173, 321–337.e10. [Google Scholar] [CrossRef]
- Kaplan, E.L.; Meier, P. Nonparametric Estimation from Incomplete Observations. J. Am. Stat. Assoc. 1958, 53, 457–481. [Google Scholar] [CrossRef]
- Kokka, F.; Brockbank, E.; Oram, D.; Gallagher, C.; Bryant, A. Hormonal therapy in advanced or recurrent endometrial cancer. Cochrane Database Syst. Rev. 2010, 12, CD007926. [Google Scholar] [CrossRef] [PubMed]
- Carlson, M.J.; Thiel, K.W.; Leslie, K.K. Past, present, and future of hormonal therapy in recurrent endometrial cancer. Int. J. Women’s Health 2014, 6, 429–435. [Google Scholar]
- Jerzak, K.J.; Duska, L.; MacKay, H.J. Endocrine therapy in endometrial cancer: An old dog with new tricks. Gynecol. Oncol. 2019, 153, 175–183. [Google Scholar] [CrossRef]
- van Weelden, W.J.; Massuger, L.F.; Pijnenborg, J.M.A.; Romano, A. Anti-estrogen Treatment in Endometrial Cancer: A Systematic Review. Front. Oncol. 2019, 9, 359. [Google Scholar] [CrossRef]
- Zehir, A.; Benayed, R.; Shah, R.H.; Syed, A.; Middha, S.; Kim, H.R.; Srinivasan, P.; Gao, J.; Chakravarty, D.; Devlin, S.M.; et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 2017, 23, 703–713. [Google Scholar] [CrossRef]
- Dou, Y.; Kawaler, E.A.; Zhou, D.C.; Gritsenko, M.A.; Huang, C.; Blumenberg, L.; Karpova, A.; Petyuk, V.A.; Savage, S.R.; Satpathy, S.; et al. Proteogenomic Characterization of Endometrial Carcinoma. Cell 2020, 180, 729–748.e26. [Google Scholar] [CrossRef]
- Watanabe, T.; Nanamiya, H.; Kojima, M.; Nomura, S.; Furukawa, S.; Soeda, S.; Tanaka, D.; Isogai, T.; Imai, J.-I.; Watanabe, S.; et al. Clinical relevance of oncogenic driver mutations identified in endometrial carcinoma. Transl. Oncol. 2021, 14, 101010. [Google Scholar] [CrossRef]
- Spratt, D.E.; Chan, T.; Waldron, L.; Speers, C.; Feng, F.Y.; Ogunwobi, O.; Osborne, J.R. Racial/Ethnic Disparities in Genomic Sequencing. JAMA Oncol. 2016, 2, 1070–1074. [Google Scholar] [CrossRef]
- Ignatiadis, M.; Sotiriou, C. Luminal breast cancer: From biology to treatment. Nat. Rev. Clin. Oncol. 2013, 10, 494–506. [Google Scholar] [CrossRef]
- Baxter, E.; Windloch, K.; Kelly, G.; Lee, J.S.; Gannon, F.; Brennan, D.J. Molecular basis of distinct oestrogen responses in endometrial and breast cancer. Endocr. Relat. Cancer 2019, 26, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Holst, F.; Hoivik, E.A.; Gibson, W.J.; Taylor-Weiner, A.; Schumacher, S.E.; Asmann, Y.W.; Grossmann, P.; Trovik, J.; Necela, B.M.; Thompson, E.A.; et al. Recurrent hormone-binding domain truncated ESR1 amplifications in primary endometrial cancers suggest their implication in hormone independent growth. Sci. Rep. 2016, 6, 25521. [Google Scholar] [CrossRef] [PubMed]
- Backes, F.J.; Walker, C.J.; Goodfellow, P.J.; Hade, E.M.; Agarwal, G.; Mutch, D.; Cohn, D.E.; Suarez, A.A. Estrogen receptor-α as a predictive biomarker in endometrioid endometrial cancer. Gynecol. Oncol. 2016, 141, 312–317. [Google Scholar] [CrossRef] [PubMed]
- Gibson, W.J.; Hoivik, E.A.; Halle, M.K.; Taylor-Weiner, A.; Cherniack, A.D.; Berg, A.; Holst, F.; Zack, T.I.; Werner, H.M.J.; Staby, K.M.; et al. The genomic landscape and evolution of endometrial carcinoma progression and abdominopelvic metastasis. Nat. Genet. 2016, 48, 848–855. [Google Scholar] [CrossRef]
- Blanchard, Z.; Vahrenkamp, J.M.; Berrett, K.C.; Arnesen, S.; Gertz, J. Estrogen-independent molecular actions of mutant estrogen receptor 1 in endometrial cancer. Genome Res. 2019, 29, 1429–1441. [Google Scholar] [CrossRef]
- Odagiri, T.; Watari, H.; Hosaka, M.; Mitamura, T.; Konno, Y.; Kato, T.; Kobayashi, N.; Sudo, S.; Takeda, M.; Kaneuchi, M.; et al. Multivariate survival analysis of the patients with recurrent endometrial cancer. J. Gynecol. Oncol. 2011, 22, 3–8. [Google Scholar] [CrossRef]
- Bertucci, F.; Ng, C.K.Y.; Patsouris, A.; Droin, N.; Piscuoglio, S.; Carbuccia, N.; Soria, J.C.; Dien, A.T.; Adnani, Y.; Kamal, M.; et al. Genomic characterization of metastatic breast cancers. Nature 2019, 569, 560–564. [Google Scholar] [CrossRef]
- Tyran, M.; Carbuccia, N.; Garnier, S.; Guille, A.; Adelaïde, J.; Finetti, P.; Touzlian, J.; Viens, P.; Tallet, A.; Goncalves, A.; et al. A Comparison of DNA Mutation and Copy Number Profiles of Primary Breast Cancers and Paired Brain Metastases for Identifying Clinically Relevant Genetic Alterations in Brain Metastases. Cancers 2019, 11, 665. [Google Scholar] [CrossRef]
- Lee, J.-M.; Nair, J.; Zimmer, A.; Lipkowitz, S.; Annunziata, C.M.; Merino, M.J.; Swisher, P.E.M.; Harrell, M.I.; Trepel, J.B.; Lee, M.-J.; et al. Prexasertib, a cell cycle checkpoint kinase 1 and 2 inhibitor, in BRCA wild-type recurrent high-grade serous ovarian cancer: A first-in-class proof-of-concept phase 2 study. Lancet Oncol. 2018, 19, 207–215. [Google Scholar] [CrossRef]
- Oza, A.M.; Elit, L.; Tsao, M.; Kamel-Reid, S.; Biagi, J.; Provencher, D.M.; Gotlieb, W.H.; Hoskins, P.J.; Ghatage, P.; Tonkin, K.S.; et al. Phase II study of temsirolimus in women with recurrent or metastatic endometrial cancer: A trial of the NCIC Clinical Trials Group. J. Clin. Oncol. 2011, 29, 3278–3285. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, Z.; Feng, Y.; Fadare, O.; Wang, J.; Ai, Z.; Jin, H.; Gu, C.; Zheng, W. Aberrant survivin expression in endometrial hyperplasia: Another mechanism of progestin resistance. Mod. Pathol. 2009, 22, 699–708. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, Y.; Wang, Y.; Zhang, Z.; Park, J.Y.; Guo, D.; Liao, H.; Yi, X.; Zheng, Y.; Zhang, D.; Chambers, S.K.; et al. Mechanism of progestin resistance in endometrial precancer/cancer through NRF2-AKR1C1 pathway. Oncotarget 2016, 7, 10363–10372. [Google Scholar] [CrossRef] [PubMed]
- Fan, R.; Wang, Y.; Wang, Y.; Wei, L.; Zheng, W. Mechanism of progestin resistance in endometrial precancer/cancer through Nrf2-survivin pathway. Am. J. Transl. Res. 2017, 9, 1483–1491. [Google Scholar] [PubMed]
- Travaglino, A.; Raffone, A.; Saccone, G.; Insabato, L.; Mollo, A.; De Placido, G.; Zullo, F. Immunohistochemical predictive markers of response to conservative treatment of endometrial hyperplasia and early endometrial cancer: A systematic review. Acta Obstet. Gynecol. Scand. 2019, 98, 1086–1099. [Google Scholar] [CrossRef]
- Yang, B.; Hu, M.; Fu, Y.; Sun, D.; Zheng, W.; Liao, H.; Zhang, Z.; Chen, X. LASS2 mediates Nrf2-driven progestin resistance in endometrial cancer. Am. J. Transl. Res. 2021, 13, 1280–1289. [Google Scholar]
- Fleming, G.F.; Filiaci, V.L.; Marzullo, B.; Zaino, R.J.; Davidson, S.A.; Pearl, M.; Makker, V.; Burke, J.J.; Zweizig, S.L.; Van Le, L.; et al. Temsirolimus with or without megestrol acetate and tamoxifen for endometrial cancer: A gynecologic oncology group study. Gynecol. Oncol. 2014, 132, 585–592. [Google Scholar] [CrossRef]
- Ring, K.L.; Yates, M.S.; Schmandt, R.; Onstad, M.; Zhang, Q.; Celestino, J.; Kwan, S.Y.; Lu, K.H. Endometrial Cancers With Activating KRas Mutations Have Activated Estrogen Signaling and Paradoxical Response to MEK Inhibition. Int. J. Gynecol. Cancer 2017, 27, 854–862. [Google Scholar] [CrossRef]
- Bertucci, F.; Gonçalves, A.; Guille, A.; Adelaïde, J.; Garnier, S.; Carbuccia, N.; Billon, E.; Finetti, P.; Sfumato, P.; Monneur, A.; et al. Prospective high-throughput genome profiling of advanced cancers: Results of the PERMED-01 clinical trial. Genome Med. 2021, 13, 87. [Google Scholar] [CrossRef]
- Konstantinopoulos, P.A.; Luo, W.; Liu, J.F.; Gulhan, D.C.; Krasner, C.; Ishizuka, J.J.; Gockley, A.A.; Buss, M.; Growdon, W.B.; Crowe, H.; et al. Phase II Study of Avelumab in Patients With Mismatch Repair Deficient and Mismatch Repair Proficient Recurrent/Persistent Endometrial Cancer. J. Clin. Oncol. 2019, 37, 2786–2794. [Google Scholar] [CrossRef]
- Martin-Hirsch, P.P.; Bryant, A.; Keep, S.L.; Kitchener, H.C.; Lilford, R. Adjuvant progestagens for endometrial cancer. Cochrane Database Syst. Rev. 2011, 15, CD001040. [Google Scholar] [CrossRef]
- Tao, Y.; Liang, B. PTEN mutation: A potential prognostic factor associated with immune infiltration in endometrial carcinoma. Pathol. Res. Pract. 2020, 216, 152943. [Google Scholar] [CrossRef] [PubMed]
| Patient Characteristics | 38 Patients Included (Interquantile Range or %) | |
|---|---|---|
| Age in years old at diagnosis, median (IQR) | 67 (61.25–74.50) | |
| Weight in kg at diagnosis, median (IQR) | 64 (56–75) | |
| Patients with high blood pressure n = 33 | 18 (47.4%) | |
| Patients with diabetes mellitus n = 32 | 5 (13.2%) | |
| Surgical route of the primary EC | ||
| Laparoscopy | 3 (7.9%) | |
| Open surgery | 31 (81.6%) | |
| Robotic surgery | 4 (10.5%) | |
| FIGO stage of the primary EC | ||
| 1A | 7 (18.4%) | |
| 1B | 8 (21.1%) | |
| 2 | 2 (5.3%) | |
| 3A | 7 (18.4%) | |
| 3B | 1 (2.6%) | |
| 3C | 8 (21.1%) | |
| 4A | 1 (2.6%) | |
| 4B | 4 (10.5%) | |
| Pathological grade of the primary EC | ||
| 1 | 11 (28.9%) | |
| 2 | 17 (44.7%) | |
| 3 | 10 (26.3%) | |
| Pathological type (centralized review) | ||
| HGSEC | 6 (15.8%) | |
| EEC | 32 (84.2%) | |
| Hormone therapy (metastatic EC) | ||
| Progestins | 27 (71.1%) | |
| AI | 7 (18.4%) | |
| Sequential AI—Progestins | 2 (5.3%) | |
| SERM | 2 (5.3%) | |
| Number of treatments before HT in the metastatic setting | ||
| 0 | 21 (54%) | |
| 1 | 11 (28.9%) | |
| 2 | 4 (10.5%) | |
| 3 | 1 (2.6%) | |
| 4 | 1 (2.6%) | |
| First-line chemotherapy (metastatic EC) (n = 25) | ||
| Carboplatin–Paclitaxel | 18 (72%) | |
| Carboplatin | 3 (12%) | |
| Carboplatin + Other drug | 1 (4%) | |
| Other drug | 3 (12%) |
| Variables | Univariate | Multivariate | ||
|---|---|---|---|---|
| OR [95% CI] | p-Value | OR [95% CI] | p-Value | |
| Age. * | 0.97 [0.89–1.05] | 4.63 × 10−1 | ||
| BMI > 30 (Yes vs. No) | 1.18 [1.03–1.42] | 4.31 × 10−2 | Inf [0–Inf] | 9.97 × 10−1 |
| Mitotic index * | 0.95 [0.90–0.99] | 4.49 × 10−1 | 0.72 [0.20–1.02] | 3.39 × 10−1 |
| PR (Pos vs. Neg) | 3.40 [0.86–14.69] | 8.62 × 10−1 | ||
| RE (Pos vs. Neg) | 2.67 [0.70–10.86] | 1.57 × 10−1 | ||
| Hormone receptors (ER or PR) (Pos vs. Neg) | 6.00 [1.15–46.35] | 4.74 × 10−2 | Inf [Inf–Inf] | 9.97 × 10−1 |
| Altered genome * | 0.61 [0.37–0.89] | 3.33 × 10−2 | 0.77 [NA–1.26] | 4.65 × 10−1 |
| MSI (MSS vs. MSI) | 1.46 [0.24–9.02] | 6.71 × 10−1 | ||
| Gene mutation (Mut vs. Wild) | ||||
| PTEN | 5.20 [1.03–39.83] | 6.56 × 10−2 | Inf [0–NA] | 9.97 × 10−1 |
| TP53 | 0.44 [0.10–1.84] | 2.65 × 10−1 | ||
| PPP2R1A | 0.40 [0.07–2.00] | 2.73 × 10−1 | ||
| ARID1A | 1.14 [0.21–6.79] | 8.76 × 10−1 | ||
| FGFR2 | 5.38 [0.74–110.60] | 1.47 × 10−1 | ||
| PIK3CA | 1.30 [0.19–11.05] | 7.91 × 10−1 | ||
| BCOR | 2.80 [0.32–60.38] | 3.96 × 10−1 | ||
| CTNNB1 | 0.24 [0.01–2.09] | 2.34 × 10−1 | ||
| KMT2D | 0.81 [0.09–7.54] | 8.46 × 10−1 | ||
| NOTCH3 | Inf [0–Inf] | 9.93 × 10−1 | ||
| USH2A | 0.81 [0.09–7.54] | 8.46 × 10−1 | ||
| AKT1 | 0 [NA–Inf] | 9.94 × 10−1 | ||
| ANK3 | 0.38 [0.02–4.42] | 4.52 × 10−1 | ||
| APC | 1.75 [0.15–40.07] | 6.62 × 10−1 | ||
| ATRX | 1.75 [0.15–40.07] | 6.62 × 10−1 | ||
| CUX1 | 1.75 [0.15–40.07] | 6.62 × 10−1 | ||
| EP300 | 1.75 [0.15–40.07] | 6.62 × 10−1 | ||
| JAK3 | Inf [0–Inf] | 9.94 × 10−1 | ||
| Oncogenic pathways alterations (Alt vs. non Alt) | ||||
| Cell Cycle | 2.00 [0.33–16.30] | 4.66 × 10−1 | ||
| HIPPO | 0.20 [0.03–1.08] | 7.99 × 10−2 | 12.23 [0–Inf] | 7.33 × 10−1 |
| MYC | 0.87 [0.09–8.07] | 8.94 × 10−1 | ||
| NOTCH | 3.93 [0.92–19.32] | 7.33 × 10−2 | 19.10 [0.14–3.9E06] | 3.96 × 10−1 |
| PI3K | 0.71 [0.16–3.01] | 6.48 × 10−1 | ||
| RTK_RAS | 1.18 [0.23–6.11] | 8.38 × 10−1 | ||
| TP53 | 0.48 [0.11–1.95] | 3.08 × 10−1 | ||
| WNT | 1.50 [0.33–7.31] | 6.00 × 10−1 | ||
| BER | 2.00 [0.33–16.30] | 4.66 × 10−1 | ||
| NER | 0.53 [0.06–3.72] | 5.26 × 10−1 | ||
| FA | 0.62 [0.12–2.91] | 5.40 × 10−1 | ||
| HRD | 0.46 [0.09–2.08] | 3.20 × 10−1 | ||
| Other | 0.52 [0.11–2.27] | 3.91 × 10−1 | ||
| Variables | Univariate | Multivariate | ||
|---|---|---|---|---|
| OR [95% CI] | p-Value | OR [95% CI] | p-Value | |
| Age. * | 0.92 [0.79–1.03] | 1.62 × 10−1 | ||
| BMI > 30 (Yes vs. No) | 0.51 [0.02–4.01] | 5.74 × 10−1 | ||
| Mitotic index * | 0.99 [0.92–1.04] | 6.43 × 10−1 | ||
| PR (Pos vs. Neg) | Inf [0.00–NA] | 9.95 × 10−1 | ||
| RE (Pos vs. Neg) | 1.20 [0.20–9.61] | 8.46 × 10−1 | ||
| Hormone receptors (ER or PR) (Pos vs. Neg) | Inf [0.00–NA] | 9.94 × 10−1 | ||
| Altered genome * | 0.62 [0.19–1.03] | 2.52 × 10−1 | ||
| MSI (MSS vs. MSI) | Inf [0.00–NA] | 9.95 × 10−1 | ||
| Gene mutation (Mut vs. Wild) | ||||
| PTEN | 7.00 [1.10–60.51] | 4.75 × 10−2 | ||
| TP53 | 2.00 [0.31–12.85] | 4.48 × 10−1 | ||
| PPP2R1A | 1.75 [0.21–11.57] | 5.69 × 10−1 | ||
| ARID1A | 0.00 [NA–Inf] | 9.94 × 10−1 | ||
| FGFR2 | 0.88 [0.04–7.32] | 9.15 × 10−1 | ||
| PIK3CA | 1.15 [0.05–10.20] | 9.09 × 10−1 | ||
| BCOR | 0.00 [NA–Inf] | 9.96 × 10−1 | ||
| CTNNB1 | 0.00 [NA–Inf] | 9.96 × 10−1 | ||
| KMT2D | 1.60 [0.07–15.90] | 7.08 × 10−1 | ||
| NOTCH3 | 1.60 [0.07–15.90] | 7.08 × 10−1 | ||
| USH2A | 1.60 [0.07–15.90] | 7.08 × 10−1 | ||
| AKT1 | 0.00 [NA–Inf] | 9.94 × 10−1 | ||
| ANK3 | 0.00 [NA–Inf] | 9.94 × 10−1 | ||
| APC | 2.50 [0.10–31.67] | 4.87 × 10−1 | ||
| ATRX | 0.00 [NA–Inf] | 9.94 × 10−1 | ||
| CUX1 | 2.50 [0.10–31.67] | 4.87 × 10−1 | ||
| EP300 | 0.00 [NA–Inf] | 9.94 × 10−1 | ||
| JAK3 | 2.50 [0.10–31.67] | 4.87 × 10−1 | ||
| Oncogenic pathways alterations (Alt vs. non Alt) | ||||
| Cell Cycle | 0.00 [NA–Inf] | 9.95 × 10−1 | ||
| HIPPO | 0.71 [0.03–5.95] | 7.79 × 10−1 | ||
| MYC | 0.00 [NA–Inf] | 9.96 × 10−1 | ||
| NOTCH | 2.18 [0.31–18.73] | 4.32 × 10−1 | ||
| PI3K | Inf [0.00–NA] | 9.95 × 10−1 | ||
| RTK_RAS | 0.43 [0.06–3.82] | 4.08 × 10−1 | ||
| TP53 | 2.18 [0.31–18.73] | 4.32 × 10−1 | ||
| WNT | 0.50 [0.02–4.04] | 5.60 × 10−1 | ||
| BER | 1.10 [0.05–9.77] | 9.38 × 10−1 | ||
| NER | 1.44 [0.06–13.57] | 7.70 × 10−1 | ||
| FA | 0.59 [0.03–4.86] | 6.63 × 10−1 | ||
| HRD | 0.50 [0.02–4.04] | 5.60 × 10−1 | ||
| Other | 2.35 [0.29–49.50] | 4.71 × 10−1 | ||
| Variables | Univariate | Multivariate | ||
|---|---|---|---|---|
| OR [95% CI] | p-Value | OR [95% CI] | p-Value | |
| Age. * | 1.09 (1.03–1.15) | 1.50 × 10−3 | 1.09 [1.02–1.15] | 8.47× 10−3 |
| BMI > 30 (Yes vs. No) | 0.86 (0.34–2.17) | 7.50 × 10−1 | ||
| Mitotic index * | 1.04 (1.01–1.06) | 5.80 × 10−3 | 1.04 [1.00–1.08] | 3.73× 10−2 |
| PR (Pos vs. Neg) | 0.38 (0.18–0.80) | 1.00 × 10−2 | ||
| RE (Pos vs. Neg) | 0.79 (0.39–1.63) | 5.30 × 10−1 | ||
| Hormone receptors (ER or PR) (Pos vs. Neg) | 0.29 (0.13–0.67) | 3.60 × 10−3 | 0.21 [0.07–0.58] | 2.67× 10−3 |
| Altered genome * | 1.07 (1.01–1.14) | 2.90 × 10−2 | 0.98 [0.90–1.07] | 6.42 × 10−1 |
| MSI (MSS vs. MSI) | 0.53 (0.21–1.34) | 1.80 × 10−1 | ||
| Gene mutation (Mut vs. Wild) | ||||
| PTEN | 0.70 (0.30–1.60) | 3.90 × 10−1 | ||
| TP53 | 1.08 (0.49–2.39) | 8.40 × 10−1 | ||
| PPP2R1A | 0.83 (0.33–2.10) | 7.00 × 10−1 | ||
| ARID1A | 2.16 (0.86–5.43) | 1.00 × 10−1 | ||
| FGFR2 | 1.19 (0.45–3.16) | 7.30 × 10−1 | ||
| PIK3CA | 0.91 (0.31–2.67) | 8.70 × 10−1 | ||
| BCOR | 1.30 (0.44–3.82) | 6.40 × 10−1 | ||
| CTNNB1 | 1.17 (0.40–3.44) | 7.70 × 10−1 | ||
| KMT2D | 0.59 (0.14–2.49) | 4.70 × 10−1 | ||
| NOTCH3 | 0.78 (0.23–2.61) | 6.80 × 10−1 | ||
| USH2A | 0.73 (0.22–2.45) | 6.10 × 10−1 | ||
| AKT1 | 1.21 (0.36–4.04) | 7.60 × 10−1 | ||
| ANK3 | 1.15 (0.27–4.97) | 8.50 × 10−1 | ||
| APC | 1.40 (0.42–4.72) | 5.80 × 10−1 | ||
| ATRX | 1.50 (0.44–5.19) | 5.20 × 10−1 | ||
| CUX1 | 0.72 (0.17–3.06) | 6.60 × 10−1 | ||
| EP300 | 2.12 (0.60–7.49) | 2.40 × 10−1 | ||
| JAK3 | 0.59 (0.14–2.53) | 4.80 × 10−1 | ||
| Oncogenic pathways alterations (Alt vs. non Alt) | ||||
| Cell Cycle | 1.33 (0.44–4.00) | 6.10 × 10−1 | ||
| HIPPO | 1.29 (0.51–3.25) | 5.90 × 10−1 | ||
| MYC | 2.10 (0.71–6.23) | 1.80 × 10−1 | ||
| NOTCH | 0.72 (0.32–1.59) | 4.20 × 10−1 | ||
| PI3K | 0.63 (0.28–1.42) | 2.60 × 10−1 | ||
| RTK_RAS | 1.73 (0.68–4.42) | 2.50 × 10−1 | ||
| TP53 | 1.05 (0.48–2.31) | 9.00 × 10−1 | ||
| WNT | 1.11 (0.49–2.50) | 8.00 × 10−1 | ||
| BER | 0.97 (0.36–2.60) | 9.60 × 10−1 | ||
| NER | 1.23 (0.42–3.60) | 7.10 × 10−1 | ||
| FA | 1.37 (0.60–3.17) | 4.60 × 10−1 | ||
| HRD | 1.07 (0.46–2.46) | 8.80 × 10−1 | ||
| Other | 1.51 (0.65–3.49) | 3.40 × 10−1 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neron, M.; Guille, A.; Allegre, L.; Colombo, P.-E.; Leaha, C.; Adelaide, J.; Carbuccia, N.; Courtier, F.; Boissiere, F.; Crapez, E.; et al. Investigation of Molecular Features Involved in Clinical Responses and Survival in Advanced Endometrial Carcinoma Treated by Hormone Therapy. J. Pers. Med. 2022, 12, 655. https://doi.org/10.3390/jpm12050655
Neron M, Guille A, Allegre L, Colombo P-E, Leaha C, Adelaide J, Carbuccia N, Courtier F, Boissiere F, Crapez E, et al. Investigation of Molecular Features Involved in Clinical Responses and Survival in Advanced Endometrial Carcinoma Treated by Hormone Therapy. Journal of Personalized Medicine. 2022; 12(5):655. https://doi.org/10.3390/jpm12050655
Chicago/Turabian StyleNeron, Mathias, Arnaud Guille, Lucie Allegre, Pierre-Emmanuel Colombo, Cristina Leaha, José Adelaide, Nadine Carbuccia, Frédéric Courtier, Florence Boissiere, Evelyne Crapez, and et al. 2022. "Investigation of Molecular Features Involved in Clinical Responses and Survival in Advanced Endometrial Carcinoma Treated by Hormone Therapy" Journal of Personalized Medicine 12, no. 5: 655. https://doi.org/10.3390/jpm12050655
APA StyleNeron, M., Guille, A., Allegre, L., Colombo, P.-E., Leaha, C., Adelaide, J., Carbuccia, N., Courtier, F., Boissiere, F., Crapez, E., Fabbro, M., Gouy, S., Mamessier, E., Lambaudie, É., Birnbaum, D., Bertucci, F., & Chaffanet, M. (2022). Investigation of Molecular Features Involved in Clinical Responses and Survival in Advanced Endometrial Carcinoma Treated by Hormone Therapy. Journal of Personalized Medicine, 12(5), 655. https://doi.org/10.3390/jpm12050655







