Abstract
Genetic factors associated with COVID-19 disease outcomes are poorly understood. This study aimed to associate genetic variants in the SLC6A20, LZTFL1, CCR9, FYCO1, CXCR6, XCR1, and ABO genes with the risk of severe forms of COVID-19 in Amazonian Native Americans, and to compare the frequencies with continental populations. The study population was composed of 64 Amerindians from the Amazon region of northern Brazil. The difference in frequencies between the populations was analyzed using Fisher’s exact test, and the results were significant when p ≤ 0.05. We investigated 64 polymorphisms in 7 genes; we studied 47 genetic variants that were new or had impact predictions of high, moderate, or modifier. We identified 15 polymorphisms with moderate impact prediction in 4 genes (ABO, CXCR6, FYCO1, and SLC6A20). Among the variants analyzed, 18 showed significant differences in allele frequency in the NAM population when compared to others. We reported two new genetic variants with modifier impact in the Amazonian population that could be studied to validate the possible associations with COVID-19 outcomes. The genomic profile of Amazonian Native Americans may be associated with protection from severe forms of COVID-19. This work provides genomic data that may help forthcoming studies to improve COVID-19 outcomes.
1. Introduction
The coronavirus disease 2019 (COVID-19) outbreak started when a few patients were hospitalized with acute respiratory distress syndrome in December 2019. At the end of January 2020, a total of 1975 COVID-19 cases were confirmed in China, with a total of 56 deaths [1]. The new infection spread worldwide, and the World Health Organization (WHO) declared COVID-19 a pandemic on 11 March 2020 [2]. Globally, the number of confirmed cases of COVID-19 has reached almost 386,548,962, including 5,705,754 deaths as of 6 February 2022 [3]
All individuals are susceptible to COVID-19 infection; however, the severity of the disease varies significantly between individuals and populations. There are many host, viral, and environmental factors contributing to the COVID-19 phenotype [4]; however, the genetic factors associated with COVID-19 disease outcomes are poorly understood. The discovery of human genetic factors associated with this disease’s severity would be invaluable in identifying high-risk groups, and would enable the stratification of individuals according to risk in order to guide personalized prevention and therapeutics [5].
Infectious diseases continue to disproportionately affect indigenous peoples and admixture populations with Amerindian ancestry [6,7,8,9,10], and COVID-19 has reached indigenous communities [11,12,13,14,15,16]. This population has a particular genetic vulnerability to infection due to different frequencies of alleles in immune system genes [17]. Their high genetic homozygosity has been suggested to be a consequence of a serial founder effect, compounded by successive generations of inbreeding [18]. This genetic factor may result in a significant loss of diversity and have consequences on health and performance [8].
In genome-wide association studies (GWASs), the identification of potential genetic factors associated with the development of COVID-19 has been investigated. The first GWAS analysis with 1980 patients with COVID-19 identified two loci associated with the most severe forms of COVID-19: one locus was 3p21.31, which includes the genes SLC6A20, LZTFL1, CCR9, FYCO1, CXCR6, and XCR1, while the other was 9q34.21, including the ABO blood group. These results might explain the heterogeneity of the disease [19].
The CXCR6, CCR9, and XCR1 genes are chemokine receptors, and directly participate in the functioning of cells of the immune system and the expression of interleukins. Other selected genes have more heterogeneous actions. The LZTFL1 gene plays a role in intracellular signaling actions. The FYCO1 gene is involved in the transport of autophagic vesicles, the SLC6A20 gene activates virus adhesion co-receptors in the cell, and the ABO gene is related to glycosylation of the H antigen for the formation of blood group variability [19].
Subsequently, Shelton et al. identified a strong association between blood type and COVID-19 diagnosis. Moreover, variants on chromosome 3p21.31 were strongly associated with COVID-19 outcome severity [20]. The present study investigated genetic variants in the SLC6A20, LZTFL1, CCR9, FYCO1, CXCR6, XCR1, and ABO genes that are potentially related to severe forms of COVID-19 in Amazonian Native American populations, and compared their frequencies with continental populations.
2. Materials and Methods
2.1. Study and Reference Populations
All participants in the study and their ethnic group leaders signed written informed consent. The recruitment period was before the COVID-19 pandemic, from September 2017 to December 2018. The study was approved by the National Committee for Ethics in Research (CONEP) and the Research Ethics Committee of the UFPA Tropical Medicine Center under CAAE number 20654313.6.0000.5172, and by the Research Ethics Committee of the UFPA under project 123/98.
The Amazonian Native American (NAM) cohort was composed of 64 Amerindians from the Amazon region of northern Brazil. The NAM population was healthy and did not share family relationships. The genetic ancestry was obtained through a panel of 61 ancestry-informative markers (AIMs), which were used for estimating individual ancestry and admixture from three continents (European, African, and Amerindian) in three multiplex PCR reactions [7,21,22]. The amplicons were analyzed by electrophoresis using the ABI Prism 3130 sequencer and the GeneMapper ID v.3.2 software. The individual proportions were estimated using STRUCTURE v.2.3.3.
The allele frequencies of the NAM population were obtained directly by gene counting and compared with continental populations (available at http://www.1000genomes.org). The 1000 Genomes project dataset corresponds to full-length DNA sequences from 2504 human individuals that include 26 population groups clustered into 5 larger population groups (African: AFR; American: AMR; East Asian: EAS; European: EUR; and South Asian: SAS). These populations include 661 individuals of African (AFR), 347 of American (AMR), 504 of East Asian (EAS), 503 of European (EUR), and 489 of South Asian (SAS) descent.
For the samples with European, East Asian, and South Asian ancestry, populations across the geographic range had ~1% FST; populations from Africa were related to the Yoruba and, therefore, not a comprehensive representation of Africa; for populations in the Americas, the samples were from two populations with primarily African and European ancestry—people with African Ancestry in the southwest USA (ASW), and those of Afro-Caribbean descent in Barbados (ACB)—and four populations (people with Mexican Ancestry in Los Angeles, CA, USA (MXL), Colombians in Medellin, Colombia (CLM), Puerto Ricans in Puerto Rico (PUR), and Peruvians in Lima, Peru (PEL)) with a wide range of European, African, and Indigenous American ancestry were chosen to represent the wide variation in ancestry proportions observed in North, Central, and South America.
2.2. Extraction of DNA and Preparation of the Exome Library
The DNA extraction was performed via the phenol–chloroform method [23]. The quantification and integrity of genetic material were analyzed using a NanoDrop 8000 spectrophotometer (Thermo Fisher Scientific Inc., Wilmington, DE, USA) and electrophoresis in 2% agarose gel, respectively.
The exome libraries were prepared using the Nextera Rapid Capture Exome (Illumina®, San Diego, CA, USA) and SureSelect Human All Exon V6 (Agilent, Santa Clara, CA, USA) kits. The sequencing reactions were run on the NextSeq 500® platform (Illumina®, San Diego, CA, USA) using the NextSeq 500 High-Output v2 300 cycle kit (Illumina®, San Diego, CA, USA).
2.3. Bioinformatic Analysis
The quality of the FASTQ reads was analyzed (FastQC v.0.11—http://www.bioinformatics.babraham.ac.uk/projects/fastqc/; accessed on 20 January 2022), and the samples were filtered to eliminate low-quality readings (fastx_tools v.0.13—http://hannonlab.cshl.edu/fastx_toolkit/; accessed on 20 January 2022). The sequences were mapped and aligned with the reference genome (GRCh38) using the BWA v.0.7 tool (http://bio-bwa.sourceforge.net/; accessed on 20 January 2022). Following this alignment with the reference genome, the file was indexed and sorted (SAMtools v.1.2—http://sourceforge.net/projects/samtools/; accessed on 20 January 2022). Subsequently, the alignment was processed for duplicate PCR removal (Picard Tools v.1.129—http://broadinstitute.github.io/picard/; accessed on 20 January 2022), mapping quality recalibration, and local realignment (GATK v.3.2—https://www.broadinstitute.org/gatk/; accessed on 20 January 2022). The results were processed in order to determine the variants from the reference genome (GATK v.3.2). The analysis of the variant annotations was carried out using the ViVa1 (Viewer of Variants) software developed by the Federal University of Rio Grande do Norte (UFRN)’s bioinformatics team. The databases and their versions used for variant annotations were SnpEff v.4.3.T, Ensembl Variant Effect Predictor (Ensembl release 99), and ClinVar (v.2018-10). For in silico prediction of pathogenicity, we used SIFT (v.6.2.1), PolyPhen-2 (v.2.2), LRT (November 2009), Mutation Assessor (v.3.0), Mutation Taster (v. 2.0), FATHMM (v.2.3), PROVEAN (v.1.1.3), MetaSVM (v1.0), M-CAP (v1.4), and FATHMM-MKL (http://fathmm.biocompute.org.uk/about.html; accessed on 20 January 2022). More information about bioinformatic analyses is described in the works of Rodrigues et al. (2020) and Ribeiro-dos-Santos et al. (2020) [22,24].
2.4. Statistical Analyses
The difference in frequencies between the populations was analyzed using Fisher’s exact test, and the results were significant when p ≤ 0.05. The interpopulation variability of the polymorphisms was assessed using Wright’s fixation index (FST). These analyses were performed using RStudio v.4.1.0.
2.5. Selection of Variants
Seven genes (SLC6A20, LZTFL1, CCR9, FYCO1, CXCR6, XCR1, and ABO) used in recent GWAS studies were selected. These studies identified genes at the loci 3p21.31 and 9q34.21 as being likely related to disease severity [19,20,25]. For subsequent analyses, the selection of variants was based on three main criteria: (a) a minimum of 10 reads of coverage (fastx_tools v.0.13-http://hannonlab.cshl.edu/fastx_toolkit/; accessed on 20 January 2022), (b) the variant should have an allele frequency described in all continental populations from the 1000 Genomes Project Consortium [10], and (c) the variant impact should be either modifier, moderate, or high, according to SnpEff classification (https://pcingola.github.io/SnpEff/; accessed on 20 January 2022)—a program that predicts coding effects such as synonymous or non-synonymous amino acid replacement, start codon gains or losses, stop codon gains or losses, and/or frameshifts. Predicted effects concerned protein-coding genes [26]. A total of 64 variants were found in the ABO, CCR9, CXCR6, FYCO1, LZTFL1, XCR1, and SLC6A20 genes, and are described in Supplementary Table S1. The analyses were directed to 38 variants that met all specifications of the selection criteria. The function of these genes is summarized in Table 1.

Table 1.
Function of the SLC6A20, LZTFL1, CCR9, CXCR6, XCR1, FYCO1, and ABO genes.
3. Results
In our study, we identified 64 polymorphisms distributed in 7 genes: 14 of them from the ABO gene, 3 from the CCR9 gene, 1 from the CXCR6 gene, 1 from the XCR1 gene, 11 from the SLC6A20 gene, 4 from the LZTFL1 gene, and 30 from the FYCO1 gene (Supplementary Table S1). Among these 64 variants, only 47 were new variants or had impact prediction by SnpEff of high, moderate, or modifier. A total of 10 SNPs were located in the ABO gene, 2 in the CCR9 gene, 1 in the CXCR6 gene, 21 in the FYCO1 gene, 4 in the LZTFL1 gene, and 8 in the SLC6A20 gene.
These variants are described in Table 2, which contains characteristics including SNP ID, chromosomal region, nucleotide change, SnpEff software impact prediction, and allele frequency for the NAM population and the five continental populations present in the 1000 Genomes platform (AFR, AMR, EAS, EUR, and SAS).

Table 2.
Description of the variants with predicted high, moderate, or modifier impact, and new variants for the genes ABO, CCR9, CXCR6, FYCO1, LZTFL1, and SLC6A20.
We identified three new variants in the Amazonian Native American population: One of the variants was located in the ABO gene at position 133262062, with base exchange C > A, in the intronic region, with an allele frequency of 0.016. The second variant was identified in the FYCO1 gene at position 45959401, with base exchange G > A, in the exonic region, with low impact predicted by SnpEff and an allele frequency of 0.018. The third variant was identified in the LZTFL1 gene at position 45827235, with a TCTG > T deletion, in the intronic region, and with an allele frequency of 0.016.
Among the polymorphisms analyzed, eight showed no variant allele frequency in Amerindian populations: two of these were in the ABO gene (rs200932155 and rs8176721), one in the CCR9 gene (rs17764980), two in the FYCO1 gene (rs3796376 and rs13069079), one in the LZTFL1 gene (rs1129183), and two in the SLC6A20 gene (rs61731475 and rs139429025).
Regarding the classification based on impact forecast by SnpEff, the polymorphism in the ABO gene (rs55727303) presented a high impact with a significant difference in all of the correlations. We identified 15 polymorphisms with moderate impact prediction in 4 genes, distributed in 4 polymorphisms in the ABO gene (rs8176748, rs8176740, rs8176720, and rs1053878), 1 in the CXCR6 gene (rs2234355), 8 in the FYCO1 gene (rs3796375, rs35678722, rs113517878, rs4683158, rs13079478, rs13059238, rs33910087, and rs3733097), and 2 in the SLC6A20 gene (rs140440513 and rs17279437).
Additionally, 38 variants had allele frequency in all populations—a necessary requirement for comparative analysis between the populations studied using Fisher’s exact test (Table 2). The remaining variants were excluded because they had no frequency description in the 1000 Genomes Project.
Among the variants analyzed, 18 showed significant differences of the NAM population when compared to all other continental populations (AFR, AMR, EAS, EUR, and SAS): 7 belonging to the ABO gene (rs55727303, rs8176748 rs8176740, rs8176720, rs2073824, rs559723, and rs616154), 7 from the FYCO1 gene (rs3733100, rs3796375, rs35678722, rs113517878, rs1532071, rs3733097, and rs751552), 2 from the SLC6A20 gene (rs2252547 and rs140440513), 1 from the LZTFL1 gene (rs141398338), and 1 from the CCR9 gene (rs147314165). The remaining 20 variants that were not significant in any comparisons are described in Table 3.

Table 3.
Comparison of the allele frequencies between the NAM population and the continental populations (AFR, AMR, EUR, EAS, and SAS).
Figure 1 represents the genomic differences between the studied populations in multidimensional scaling (MDS), based on the fixation index (FST) of polymorphisms. We can observe the presence of a genetically similar core set composed of the AMR, EUR, and SAS components, while the other groups (NAM, EAS, and AFR) are at extreme points of the graph.
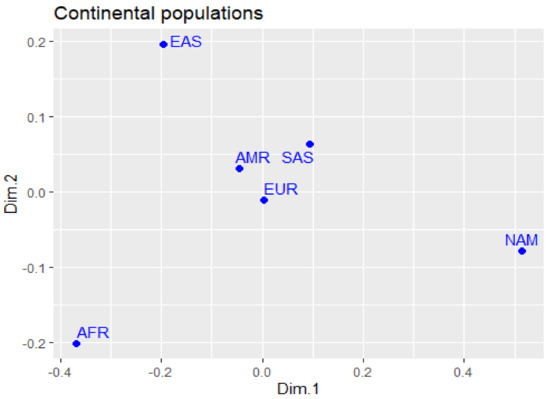
Figure 1.
Differences in allele frequencies of the variants studied in the continental populations and the Native American population, plotted in MDS.
4. Discussion
COVID-19 presents a new threat to the health of Native Amerindians living remotely. In the Amazon region, it is estimated that there are around 78 Amerindian tribes living in isolation [44]. The Amerindian people belong to a vulnerable population, who lack immunity to many infectious diseases [8]. In Brazilian territory, there were 59,574 cases and 871 deaths recorded in Amerindians as of 6 February 2022 [16].
The occurrence of numerous stochastic events—such as geographic isolation, inbreeding, and genetic drift—may contribute to genetic differentiation in Amazonian Native American populations [45,46,47,48,49]. These variables influence the formation of indigenous peoples, their ethnic structure and, consequently, their genomic patterns, with different allele frequencies from other continental populations [50,51].
The genomic differences and distinct sociodemographic and anthropological characteristics of Amerindian populations are related to epidemiological differences in respiratory and viral diseases—such as tuberculosis (TB), human immunodeficiency virus (HIV), human T-lymphotropic virus (HTLV), and human herpesvirus type 8 (HHV-8)—observed in comparisons between Amerindian and non-Amerindian populations. Studies suggest that genomic alterations in the immune response patterns and the parasite–host molecular interaction may be the causes of the observed differences [7,17,52].
GWA studies are widely used in the identification of genetic variants associated with complex and multifactorial diseases, such as cardiovascular, psychiatric, infectious, and numerous other diseases [53,54,55]. The first GWA study investigated the multigenic group of chromosome 3 in patients infected with COVID-19 [19]. The 3p21.31 locus has the XCR1, CCR9, CXCR6, SLC6A20, LZTFL1, and FYCO1 genes, in which rs11385942 (LZTFL1) and rs657152 (ABO) were significantly associated with severe forms of the disease [19].
In Shelton’s 2021 study, the variants related to disease severity and susceptibility were rs13078854 of the LZTFL1 gene and rs9411378 of the ABO gene [20]. These findings reinforce the importance of the 3p21.31 locus and the 9q34.21 locus—particularly with regard to the ABO and LZTFL1 genes; therefore, we focus our discussion on these two genes.
The association between the ABO blood group and COVID-19 infection and severity was studied. Blood type A might be more susceptible to COVID-19 infection, while blood type O might be less susceptible to this disease. [19]. In other epidemiological and genomic studies, susceptibility was also lower in O blood group patients [20,56,57]. The O allele is the most frequent blood type found in the Native American population [51,58]. In addition, the ABO blood group has previously been linked to susceptibility to other diseases, such as influenza, malaria, schistosomiasis, and SARS-CoV-2 [59].
Some proposed mechanisms for the association between ABO blood type and SARS-CoV-2 infection were investigated, as follows: (1) anti-A and/or anti-B antibodies play a role as viral neutralizing antibodies when binding to A and/or B antigens expressed on the viral envelope; (2) the SARS-CoV-2 S protein is bound by human anti-A antibodies, and prevents entry into the lung epithelium when blocking the interaction between the virus and ACE2R; (3) an increase in ACE-1 activity in group A individuals can cause predisposition to cardiovascular disease and lead to severe COVID-19; (4) ABH glycans in the SARS-CoV-2 S protein may modify cellular receptors of SARS-CoV-2 for ACE2R; (5) ABH glycans on target cells could serve as alternative, lower affinity receptors for the SARS-CoV-2 S protein, or could bind other viral envelope structures [42,60,61].
In our study, significant differences were found in the majority of polymorphisms in the ABO gene between the Amerindian populations and continental populations. We hypothesize that the differences in genomic profiles and the novel variants identified in the Native American population may influence the development of severe forms of COVID-19. However, further studies will be needed in COVID-19-positive individuals in this population in order to better understand the potential influence of these variants on this infection.
The LZTFL1 gene encodes the leucine zipper transcription factor-like 1, and its function is related to tumor-suppressor action and negative regulation of the hedgehog signaling pathways. In knockout zebrafish (Danio rerio) experimental models, impaired cell traffic in ciliary membranes, retinal degeneration, and obesity were observed [28]. In addition, this gene has high expression in lung tissues; however, the mechanisms directly related to SARS-CoV-2 infection remain unknown [25].
In our study, we found four variants related to the LZTFL1 gene, and only rs141398338 showed a significant difference in the NAM population when compared to the continental populations. There have been no reports in the literature on the association between rs141398338 and severe forms of COVID-19. New variants identified in the Native American population may influence the development of severe forms of COVID-19, and further human genetic studies need to be carried out in order to clarify this issue.
In addition, we identified three new variants in the Amazonian NAM population; these SNPs were located in the FYCO1, ABO, and LZTFL1 genes. The first has low clinical impact, while the following two have modifier impact. These mutations—especially the ones with modifier impact—could have important potential as markers of severe forms of COVID-19 in Amazonian indigenous populations, as well as intronic regional mutations that significantly influence gene expression levels [62]. Larger studies should be performed to confirm these new variants in patients diagnosed with COVID-19.
Genetic variants associated with severe COVID-19 indicated by the studies of Ellinghaus and Shelton et al. [19,20] were not found in the Amazonian Native American population. The allelic frequencies of the SNPs in the NAM group were lower than for any of the other groups in our study, showing that the Amazonian Native Americans have low genetic variability and a different genetic pool. Genetic variants present in the NAM population and low genetic variability could indicate a protective factor against severe COVID-19.
The limitation of our study was the small number of NAM individuals, who come from isolated and relatively small populations in the Amazonian region. This study is a preliminary severe COVID-19 study, and did not investigate individuals with COVID-19 infection. We collected blood samples from individuals before the COVID-19 pandemic. Our results may reveal important information and contribute to the assessment of individual risk for the development of this disease.
5. Conclusions
Genetic variants associated with severe COVID-19 were not found in the Amazonian Native American population. The allele frequency for the candidate genes in the NAM group was significantly different from the frequencies observed in continental groups. This may provide a protective factor against severe COVID-19. We also identified two new genetic variants with modifier impact in the Amazonian population that could be studied in order to validate the possible associations with COVID-19 outcomes. This work contributes to the elucidation of the genomic profile of Amazonian Native Americans—an understudied population—by providing genomic data that may help forthcoming studies to improve COVID-19 outcomes. Future studies should be performed in this population to identify more genetic variants related to severe COVID-19.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jpm12040554/s1: Supplementary Table S1: Description of all variants for the genes ABO, CCR9, CXCR6, FYCO1, LZTFL1, SLC6A20, and XCR1 found in the 64 individuals sampled in the present study.
Author Contributions
Conceptualization, P.P.d.A., S.E.B.d.S. and N.P.C.d.S.; data curation, J.F.G.; formal analysis, J.C.G.R., S.J.d.S. and J.E.K.; funding acquisition, R.M.R.B., P.P.d.A. and S.E.B.d.S.; investigation, L.F.P., T.A.S., M.R.F. and S.E.B.d.S.; methodology, L.F.P., T.A.S., L.P.A.G., G.M.V., L.A.d.A., L.P.C.L., N.M.d.S., J.C.G.R., S.J.d.S., J.E.K. and A.M.R.-d.-S.; resources, A.M.R.-d.-S.; software, J.C.G.R., S.J.d.S. and J.E.K.; supervision, N.P.C.d.S.; visualization, R.d.C.C.C., A.L.d.A. and L.W.M.S.V.; writing—original draft preparation, L.F.P., T.A.S., D.F.d.V.B.L., M.R.F. and J.F.G.; writing—review and editing, D.F.d.V.B.L., M.R.F., R.M.R.B., P.P.d.A., Â.C.R.-d.-S., S.E.B.d.S. and N.P.C.d.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (http://www.cnpq.br), the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) (https://www.gov.br/capes/pt-br), the Pró-Reitoria de Pesquisa e Pós-Graduação da UFPA (PROPESP) (http://www. propesp.ufpa.br), and the Fundação Amazônia de Amparo a Estudos e Pesquisas (FAPESPA) (http://www.fapespa.pa.gov.br). This work is part of the Rede de Pesquisa em Genômica Populacional Humana (Biocomputacional-Protocol no. 3381/2013/CAPES).
Institutional Review Board Statement
This study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the National Committee for Ethics in Research (CONEP) and the Research Ethics Committee of the UFPA Tropical Medicine Center, under CAAE number 20654313.6.0000.5172.
Informed Consent Statement
All participants in the study and their ethnic group leaders signed written informed consent. Written informed consent was obtained from the patients to publish this paper.
Data Availability Statement
The data presented in this study are openly available on Figshare at https://doi.org/10.6084/m9.figshare.18728192.v1; accessed on 25 January 2022.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wang, W.; Tang, J.; Wei, F. Updated understanding of the outbreak of 2019 novel coronavirus (2019-nCoV) in Wuhan, China. J. Med. Virol. 2020, 92, 441–447. [Google Scholar] [CrossRef]
- Cucinotta, D.; Vanelli, M. WHO Declares COVID-19 a Pandemic. Acta Bio-Med. Atenei Parm. 2020, 91, 157–160. [Google Scholar]
- World Health Organization. WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int (accessed on 6 February 2022).
- Samadizadeh, S.; Masoudi, M.; Rastegar, M.; Salimi, V.; Shahbaz, M.B.; Tahamtan, A. COVID-19: Why does disease severity vary among individuals? Respir. Med. 2021, 180, 106356. [Google Scholar] [CrossRef] [PubMed]
- Anastassopoulou, C.; Gkizarioti, Z.; Patrinos, G.P.; Tsakris, A. Human genetic factors associated with susceptibility to SARS-CoV-2 infection and COVID-19 disease severity. Hum. Genom. 2020, 14, 40. [Google Scholar] [CrossRef] [PubMed]
- Mario-Vásquez, J.E.; Naranjo-González, C.A.; Montiel, J.; Zuluaga, L.M.; Vásquez, A.M.; Tobón-Castaño, A.; Bedoya, G.; Segura, C. Association of variants in IL1B, TLR9, TREM1, IL10RA, and CD3G and Native American ancestry on malaria susceptibility in Colombian populations. Infect. Genet. Evol. 2021, 87, 104675. [Google Scholar] [CrossRef]
- Leal, D.F.D.V.B.; Da Silva, M.N.S.; Fernandes, D.C.R.D.O.; Rodrigues, J.C.G.; Barros, M.C.D.C.; Pinto, P.D.D.C.; Pastana, L.F.; Da Silva, C.A.; Fernandes, M.R.; De Assumpção, P.P.; et al. Amerindian genetic ancestry as a risk factor for tuberculosis in an amazonian population. PLoS ONE 2020, 15, e0236033. [Google Scholar] [CrossRef] [PubMed]
- Castro e Silva, M.A.; Ferraz, T.; Couto-Silva, C.M.; Lemes, R.B.; Nunes, K.; Comas, D.; Hünemeier, T. Population Histories and Genomic Diversity of South American Natives. Mol. Biol. Evol. 2021, 39, msab339. [Google Scholar] [CrossRef] [PubMed]
- Gilmour, B.; Alene, K.A.; Clements, A. The prevalence of tuberculosis and malaria in minority indigenous populations of South-East Asia and the Western Pacific Region: A systematic review and meta-analysis. Pathog. Glob. Health 2021, 1–19. [Google Scholar] [CrossRef]
- Pinto, P.; Salgado, C.; Santos, N.P.C.; Santos, S.; Ribeiro-dos-Santos, Â. Influence of Genetic Ancestry on INDEL Markers of NFKβ1, CASP8, PAR1, IL4 and CYP19A1 Genes in Leprosy Patients. PLoS Negl. Trop. Dis. 2015, 9, e0004050. [Google Scholar] [CrossRef]
- Tai, D.B.G.; Shah, A.; Doubeni, C.A.; Sia, I.G.; Wieland, M.L. The Disproportionate Impact of COVID-19 on Racial and Ethnic Minorities in the United States. Clin. Infect. Dis. 2021, 72, 703–706. [Google Scholar] [CrossRef]
- Da Cunha, A.A.; Corona, R.A.; Castilho-Martins, E.A. COVID-19 and race/color disparity: A brief analysis of the indigenous population in a state in the Brazilian Amazon. Einstein 2021, 19, eCE6734. [Google Scholar] [CrossRef]
- Stone, M.J.; Close, R.M.; Jentoft, C.K.; Pocock, K.; Lee-Gatewood, G.; Grow, B.I.; Parker, K.H.; Twarkins, A.; Nashio, J.T.; McAuley, J.B. High-Risk Outreach for COVID-19 Mortality Reduction in an Indigenous Community. Am. J. Public Health 2021, 111, 1939–1941. [Google Scholar] [CrossRef]
- Serrano-Coll, H.; Miller, H.; Rodríguez-Van, D.H.C.; Gastelbondo, B.; Novoa, W.; Oviedo, M.; Rivero, R.; Garay, E.; Mattar, S. High Prevalence of SARS-CoV-2 in an Indigenous Community of the Colombian Amazon Region. Trop. Med. Infect. Dis. 2021, 6, 191. [Google Scholar] [CrossRef]
- Cupertino, G.A.; Cupertino, M.D.C.; Gomes, A.P.; Braga, L.M.; Siqueira-Batista, R. COVID-19 and Brazilian Indigenous Populations. Am. J. Trop. Med. Hyg. 2020, 103, 609–612. [Google Scholar] [CrossRef]
- Secretaria de Saúde Indígena (SESAI). Boletim Epidemiológico 492, 2022, 1. Available online: https://www.gov.br/saude/pt-br/composicao/sesai (accessed on 30 November 2020).
- Lindenau, J.D.; Guimarães, L.S.P.; Friedrich, D.C.; Hurtado, A.M.; Hill, K.R.; Salzano, F.M.; Hutz, M.H. Cytokine gene polymorphisms are associated with susceptibility to tuberculosis in an Amerindian population. Int. J. Tuberc. Lung Dis. 2014, 18, 952–957. [Google Scholar] [CrossRef]
- Wang, S.; Lewis, C.M.; Jakobsson, M.; Ramachandran, S.; Ray, N.; Bedoya, G.; Rojas, W.; Parra-Marín, M.V.; A Molina, J.; Gallo, C.; et al. Genetic Variation and Population Structure in Native Americans. PLoS Genet. 2007, 3, e185. [Google Scholar] [CrossRef]
- Ellinghaus, D. Genomewide Association Study of Severe Covid-19 with Respiratory Failure. N. Engl. J. Med. 2020, 383, 1522–1534. [Google Scholar]
- Shelton, J.F.; Shastri, A.J.; Ye, C.; Weldon, C.H.; Filshtein-Sonmez, T.; Coker, D.; Symons, A.; Esparza-Gordillo, J.; Aslibekyan, S.; Auton, A.; et al. Trans-ancestry analysis reveals genetic and nongenetic associations with COVID-19 susceptibility and severity. Nat. Genet. 2021, 53, 801–808. [Google Scholar] [CrossRef]
- De Ramos, B.R.A.; D’Elia, M.P.B.; Amador, M.A.T.; Santos, N.P.C.; Santos, S.E.B.; da Castelli, E.C.; Witkin, S.S.; Miot, H.A.; Miot, L.D.B.; da Silva, M.G. Neither self-reported ethnicity nor declared family origin are reliable indicators of genomic ancestry. Genetica 2016, 144, 259–265. [Google Scholar] [CrossRef]
- Rodrigues, J.C.G.; de Souza, T.P.; Pastana, L.F.; Ribeiro dos Santos, A.M.; Fernandes, M.R.; Pinto, P.; Wanderley, A.V.; De Souza, S.J.; Kroll, J.E.; Pereira, A.L.; et al. Identification of NUDT15 gene variants in Amazonian Amerindians and admixed individuals from northern Brazil. PLoS ONE 2020, 15, e0231651. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Isolation of High-Molecular-Weight DNA Using Organic Solvents. Cold Spring Harb. Protoc. 2017, pdb.prot093450. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro-dos-Santos, A.M.; Vidal, A.F.; Vinasco-Sandoval, T.; Guerreiro, J.; Santos, S.; Ribeiro-dos-Santos, Â. Exome Sequencing of Native Populations from the Amazon Reveals Patterns on the Peopling of South America. Front. Genet. 2020, 11, 548507. [Google Scholar] [CrossRef] [PubMed]
- Elhabyan, A.; Elyaacoub, S.; Sanad, E.; Abukhadra, A.; Elhabyan, A.; Dinu, V. The role of host genetics in susceptibility to severe viral infections in humans and insights into host genetics of severe COVID-19: A systematic review. Virus Res. 2020, 289, 198163. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Camargo, S.M.R.; Vuille-dit-Bille, R.N.; Meier, C.F.; Verrey, F. ACE2 and gut amino acid transport. Clin. Sci. 2020, 134, 2823–2833. [Google Scholar] [CrossRef]
- Wei, Q.; Chen, Y.; Gu, Y.F.; Zhao, W. Molecular Characterization and Functional Analysis of Leucine Zipper Transcription Factor like 1 in Zebrafish (Danio rerio). Front. Physiol. 2019, 10, 801. [Google Scholar] [CrossRef]
- Downes, D.J.; Cross, A.R.; Hua, P.; Roberts, N.; Schwessinger, R.; Cutler, A.J.; Munis, A.M.; Brown, J.; Mielczarek, O.; de Andrea, C.E.; et al. Identification of LZTFL1 as a candidate effector gene at a COVID-19 risk locus. Nat. Genet. 2021, 53, 1606–1615. [Google Scholar] [CrossRef]
- Wu, X.; Sun, M.; Yang, Z.; Lu, C.; Wang, Q.; Wang, H.; Deng, C.; Liu, Y.; Yang, Y. The Roles of CCR9/CCL25 in Inflammation and Inflammation-Associated Diseases. Front. Cell Dev. Biol. 2021, 9, 2244. [Google Scholar] [CrossRef]
- Pathak, M.; Lal, G. The Regulatory Function of CCR9+ Dendritic Cells in Inflammation and Autoimmunity. Front. Imunol. 2020, 2219. [Google Scholar]
- Liu, G.; Abas, O.; Strickland, A.B.; Chen, Y.; Shi, M. CXCR6+CD4+ T cells promote mortality during Trypanosoma brucei infection. PLoS Pathog. 2021, 17, e1009968. [Google Scholar] [CrossRef]
- Luoma, A.M.; Suo, S.; Williams, H.L.; Sharova, T.; Sullivan, K.; Manos, M.; Bowling, P.; Hodi, F.S.; Rahma, O.; Sullivan, R.J.; et al. Molecular Pathways of Colon Inflammation Induced by Cancer Immunotherapy. Cell 2020, 182, 655–671.e22. [Google Scholar] [CrossRef]
- Balan, S.; Saxena, M.; Bhardwaj, N. Chapter One—Dendritic cell subsets and locations. In International Review of Cell and Molecular Biology; Lhuillier, C., Galluzzi, L., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 1–68. Available online: https://www.sciencedirect.com/science/article/pii/S193764481930067X (accessed on 30 November 2020).
- Lei, Y.; Takahama, Y. XCL1 and XCR1 in the immune system. Microbes Infect. 2012, 14, 262–267. [Google Scholar] [CrossRef]
- Pankiv, S.; Alemu, E.A.; Brech, A.; Bruun, J.-A.; Lamark, T.; Overvatn, A.; Bjørkøy, G.; Johansen, T. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J. Cell Biol. 2010, 188, 253–269. [Google Scholar] [CrossRef]
- Kuhn, C.; Menke, M.; Senger, F.; Mack, C.; Dierck, F.; Hille, S.; Schmidt, I.; Brunke, G.; Brünger, P.; Schmiedel, N.; et al. FYCO1 Regulates Cardiomyocyte Autophagy and Pre-vents Heart Failure Due to Pressure Overload In Vivo. JACC Basic Transl. Sci. 2021, 6, 365–380. [Google Scholar] [CrossRef]
- Iqbal, H.; Khan, S.Y.; Zhou, L.; Irum, B.; Ali, M.; Ahmed, M.R.; Shahzad, M.; Ali, M.H.; Naeem, M.A.; Riazuddin, S.; et al. Mutations in FYCO1 identified in families with congenital cataracts. Mol. Vis. 2020, 26, 334–344. [Google Scholar]
- Rothwell, S.; Lilleker, J.B.; Lamb, J.A. Genetics in inclusion body myositis. Curr. Opin. Rheumatol. 2017, 29, 639. [Google Scholar] [CrossRef]
- Franchini, M.; Crestani, S.; Frattini, F.; Sissa, C.; Bonfanti, C. ABO blood group and von Willebrand factor: Biological implications. Clin. Chem. Lab. Med. (CCLM) 2014, 52, 1273–1276. [Google Scholar] [CrossRef]
- Breiman, A.; Ruvën-Clouet, N.; Le Pendu, J. Harnessing the natural anti-glycan immune response to limit the transmission of enveloped viruses such as SARS-CoV-2. PLoS Pathog. 2020, 16, e1008556. [Google Scholar] [CrossRef]
- Goel, R.; Bloch, E.M.; Pirenne, F.; Al-Riyami, A.Z.; Crowe, E.; Dau, L.; Land, K.; Townsend, M.; Jecko, T.; Rahimi-Levene, N.; et al. ABO blood group and COVID-19: A review on behalf of the ISBT COVID-19 working group. Vox Sang. 2021, 116, 849–861. [Google Scholar] [CrossRef]
- Murray, G.P.; Post, S.R.; Post, G.R. ABO blood group is a determinant of von Willebrand factor protein levels in human pulmonary endothelial cells. J. Clin. Pathol. 2020, 73, 347–349. [Google Scholar] [CrossRef]
- IBGE. Censo Brasileiro de 2010. Rio de Janeiro-RJ. 2012. Available online: https://biblioteca.ibge.gov.br/visualizacao/periodicos/552/cd_2010_agsn_if.pdf (accessed on 30 November 2020).
- Salzano, F.M. Fatores dete rminísticos e estocásticos no processo microevolucionário humano. Actas V Congr. Latinoam. Genet. 1982, 1, 81–89. [Google Scholar]
- Amos, W.; Hoffman, J.I. Evidence that two main bottleneck events shaped modern human genetic diversity. Proc. Soc. Biol. Sci. 2010, 277, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Dobbin, E.A.F.; Medeiros, J.A.G.; Costa, M.S.C.R.; Rodrigues, J.C.G.; Guerreiro, J.F.; Kroll, J.E.; Souza, S.; de Assumpção, P.; Ribeiro-Dos-Santos, Â.; Santos, S.; et al. Identification of Variants (rs11571707, rs144848, and rs11571769) in the BRCA2 Gene Associated with Hereditary Breast Cancer in Indigenous Populations of the Brazilian Amazon. Genes 2021, 12, 142. [Google Scholar] [CrossRef] [PubMed]
- O’Fallon, B.D.; Fehren-Schmitz, L. Native Americans experienced a strong population bottleneck coincident with European contact. Proc. Natl. Acad. Sci. USA 2011, 108, 20444–20448. [Google Scholar] [CrossRef] [PubMed]
- De Brito Vargas, L.; Beltrame, M.H.; Ho, B.; Marin, W.M.; Dandekar, R.; Montero-Martín, G.; Fernández-Viña, M.A.; Hurtado, A.M.; Hill, K.R.; Tsuneto, L.T.; et al. Remarkably Low KIR and HLA Diversity in Amerindians Reveals Signatures of Strong Purifying Selection Shaping the Centromeric KIR Region. Mol. Biol. Evol. 2022, 39, msab298. [Google Scholar] [CrossRef]
- Battilana, J.; Fagundes, N.J.R.; Heller, A.H.; Goldani, A.; Freitas, L.B.; Tarazona-Santos, E.; Munkhbat, B.; Munkhtuvshin, N.; Krylov, M.; Benevolenskaia, L.; et al. Alu insertion polymorphisms in Native Americans and related Asian populations. Ann. Hum. Biol. 2006, 33, 142–160. [Google Scholar] [CrossRef]
- Estrada-Mena, B.; Estrada, F.J.; Ulloa-Arvizu, R.; Guido, M.; Méndez, R.; Coral, R.; Canto, T.; Granados, J.; Rubí-Castellanos, R.; Rangel-Villalobos, H.; et al. Blood group O alleles in Native Americans: Implications in the peopling of the Americas. Am. J. Phys. Anthropol. 2010, 142, 85–94. [Google Scholar] [CrossRef]
- Salzano, F.M. The Amerindian Microcosm: Anthropology, Comparative History, Ecology, Genetics and Evolution; Hutz, M.H., Bortolini, M.C., Eds.; Cambridge Scholars Publishing: Newcastle upon Tyne, UK, 2019; p. 607. [Google Scholar]
- Hart, A.B.; Kranzler, H.R. Alcohol Dependence Genetics: Lessons Learned from Genome-Wide Association Studies (GWAS) and Post-GWAS Analyses. Alcohol. Clin. Exp. Res. 2015, 39, 1312–1327. [Google Scholar] [CrossRef]
- Nikpay, M.; Goel, A.; Won, H.-H.; Hall, L.M.; Willenborg, C.; Kanoni, S.; Saleheen, D.; Kyriakou, T.; Nelson, C.P.; Hopewell, J.C.; et al. A comprehensive 1000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 2015, 47, 1121–1130. [Google Scholar]
- Ko, D.C.; Urban, T.J. Understanding Human Variation in Infectious Disease Susceptibility through Clinical and Cellular GWAS. PLoS Pathog. 2013, 9, e1003424. [Google Scholar] [CrossRef]
- Chen, T.; Wu, D.; Chen, H.; Yan, W.; Yang, D.; Chen, G.; Ma, K.; Xu, D.; Yu, H.; Wang, H.; et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: Retrospective study. BMJ 2020, 368, m1091. [Google Scholar] [CrossRef]
- Zhao, J.; Yang, Y.; Huang, H.; Li, D.; Gu, D.; Lu, X.; Zhang, Z.; Liu, L.; Liu, T.; Liu, Y.; et al. Relationship between the ABO Blood Group and the Coronavirus Disease 2019 (COVID-19) Susceptibility. Clin. Infect. Dis. 2021, 73, 328–331. [Google Scholar] [CrossRef]
- Halverson, M.S.; Bolnick, D.A. An ancient DNA test of a founder effect in native American ABO blood group frequencies. American J. Phys. Anthropol. 2008, 137, 342–347. [Google Scholar] [CrossRef]
- Fricke-Galindo, I.; Falfán-Valencia, R. Genetics Insight for COVID-19 Susceptibility and Severity: A Review. Front. Immunol. 2021, 12, 1057. [Google Scholar] [CrossRef]
- Cooling, L. Blood Groups in Infection and Host Susceptibility. Clin. Microbiol. Rev. 2015, 28, 801–870. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef]
- Rigau, M.; Juan, D.; Valencia, A.; Rico, D. Intronic CNVs and gene expression variation in human populations. PLoS Genet. 2019, 15, e1007902. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).