Tumor Suppressive Role of the PRELP Gene in Ovarian Clear Cell Carcinoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Clinical Materials
2.2. Database Analysis
2.3. Cell Culture
2.4. Reverse Transcription Polymerase Chain Reaction (RT-PCR)
2.5. Plasmids
2.6. Western Blotting
2.7. Cell Viability Assay
2.8. Soft Agar Colony Formation Assay
2.9. Chromatin Immunoprecipitation Followed by Sequencing (ChIP-Seq)
2.10. RNA-Seq
2.11. Bioinformatic and Statistical Analysis
3. Results
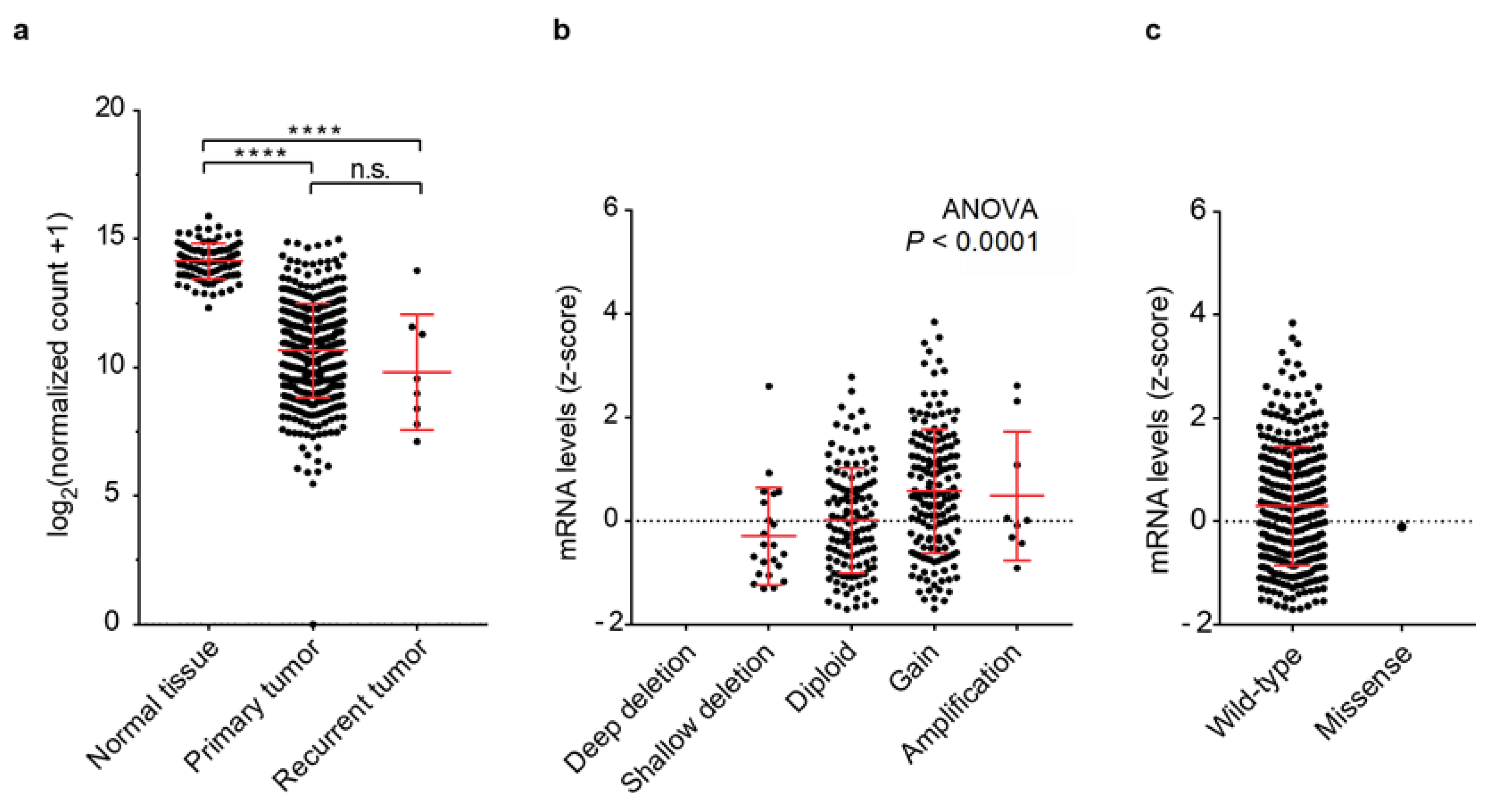
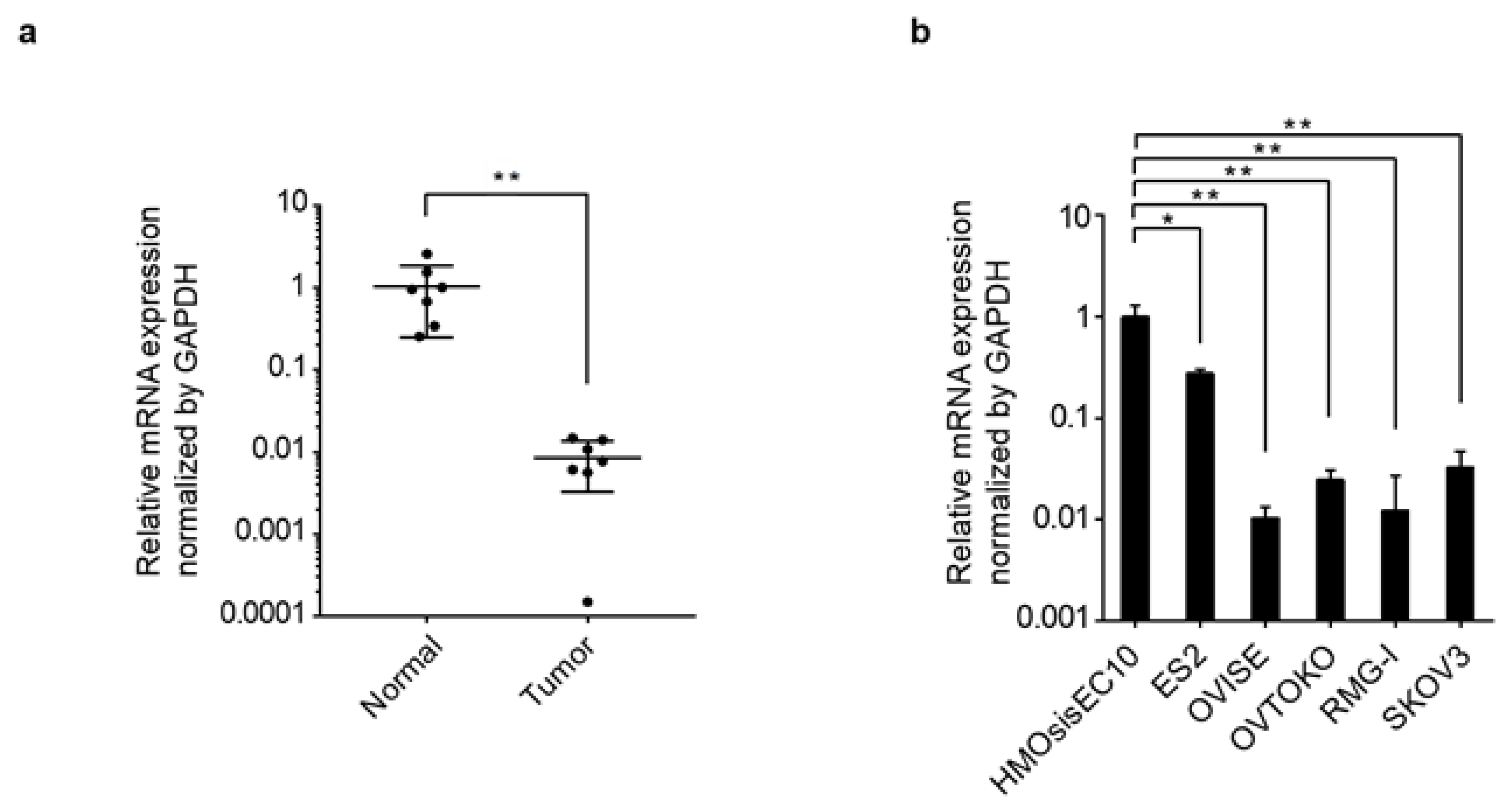
3.1. PRELP Gene Expression and Genomic Aberrations
3.2. Cell Viability upon PRELP Expression
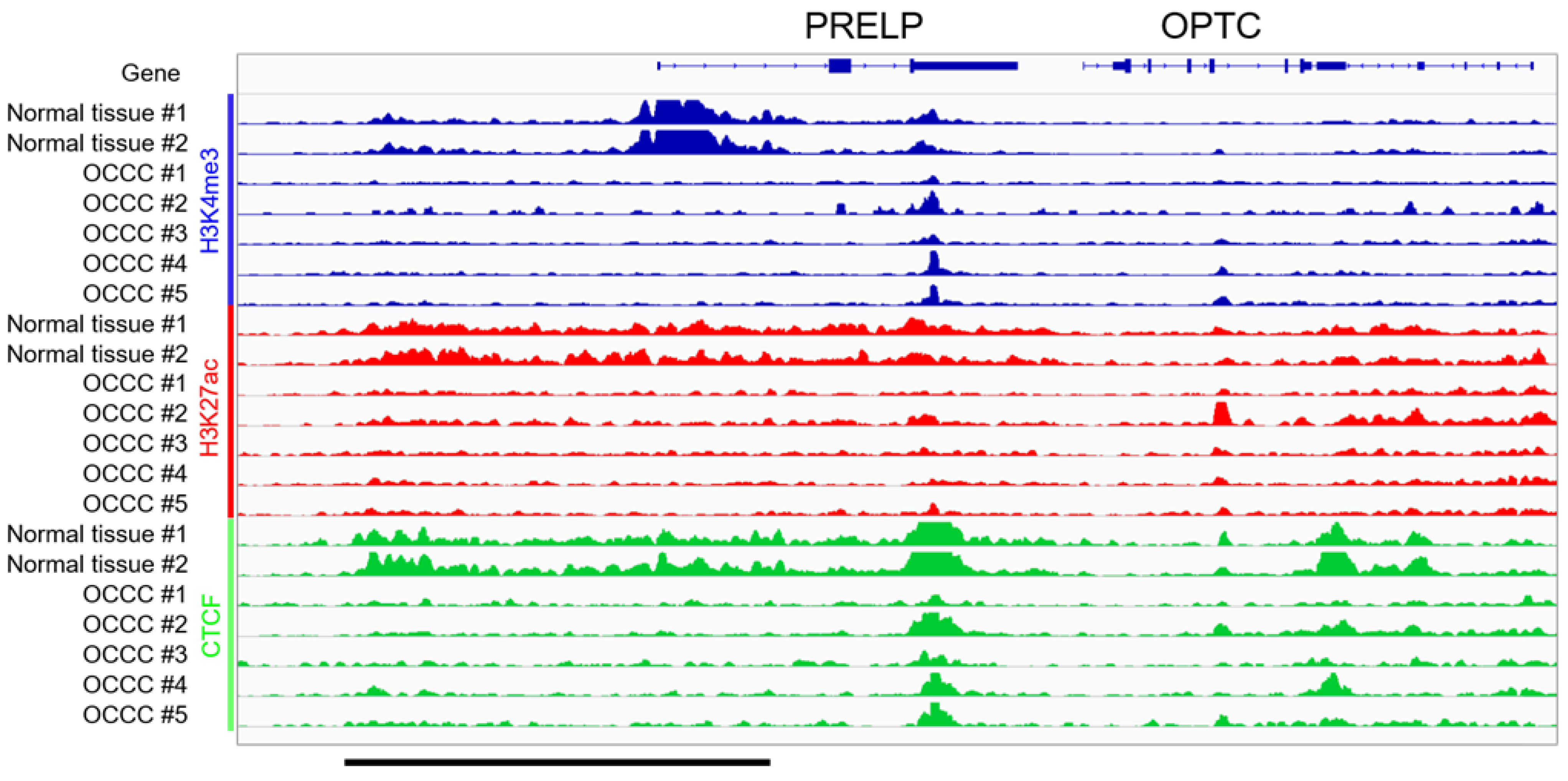
3.3. ChIP-Seq in Clinical Tissues of OCCC
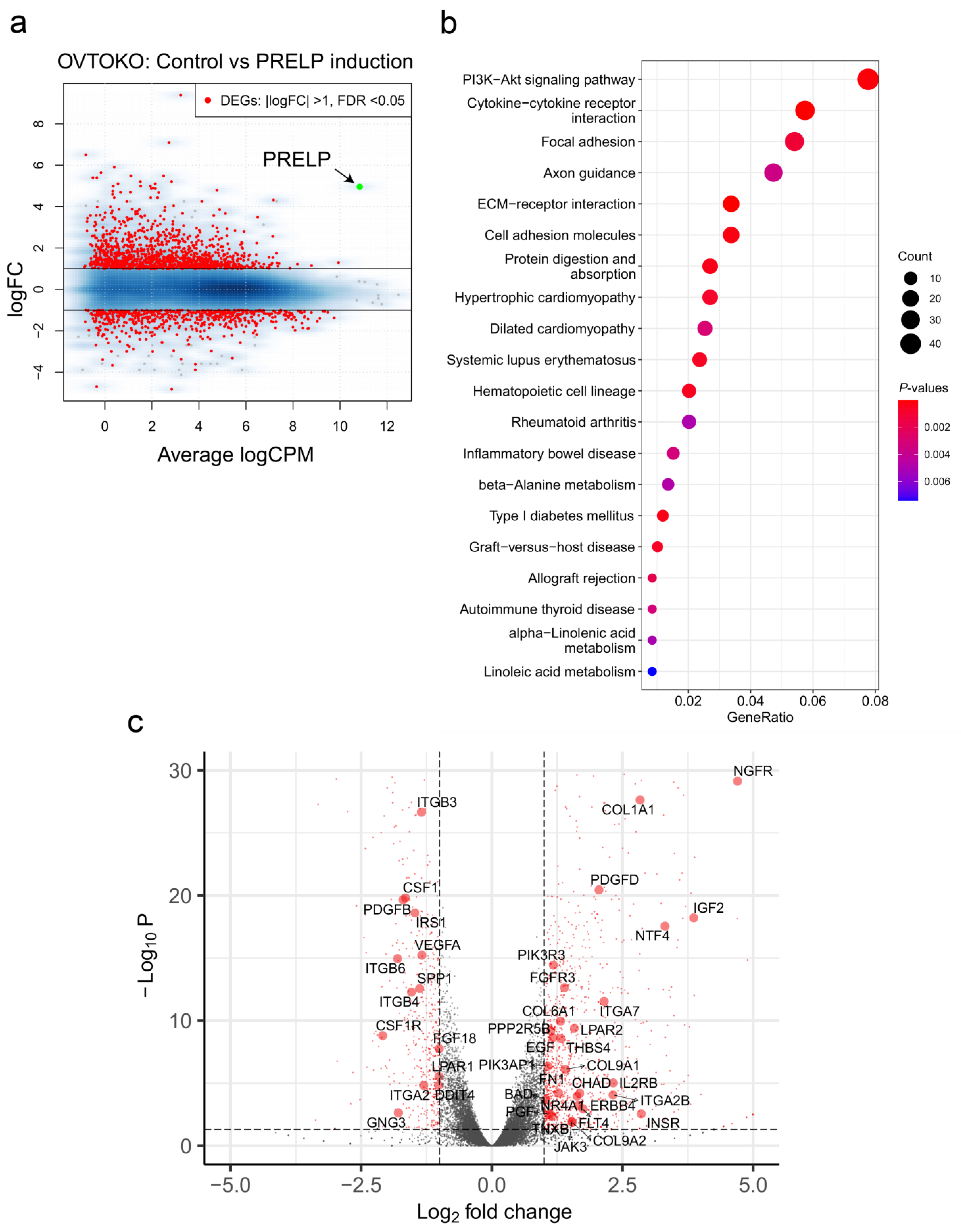
3.4. Gene Sets Altered by the Overexpression of PRELP in OCCC Cell Lines
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reid, B.M.; Permuth, J.B.; Sellers, T.A. Epidemiology of ovarian cancer: A review. Cancer Biol. Med. 2017, 14, 9–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matulonis, U.A.; Sood, A.K.; Fallowfield, L.; Howitt, B.E.; Sehouli, J.; Karlan, B.Y. Ovarian cancer. Nat. Rev. Dis. Primers 2016, 2, 16061. [Google Scholar] [CrossRef] [PubMed]
- Lisio, M.A.; Fu, L.; Goyeneche, A.; Gao, Z.H.; Telleria, C. High-grade serous ovarian cancer: Basic sciences, clinical and therapeutic standpoints. Int. J. Mol. Sci. 2019, 20, 952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, K.; Nakayama, K.; Ishikawa, N.; Ishikawa, M.; Sultana, R.; Kiyono, T.; Kyo, S. Reconstitution of high-grade serous ovarian carcinoma from primary fallopian tube secretory epithelial cells. Oncotarget 2018, 9, 12609–12619. [Google Scholar] [CrossRef] [Green Version]
- Kyo, S.; Ishikawa, N.; Nakamura, K.; Nakayama, K. The fallopian tube as origin of ovarian cancer: Change of diagnostic and preventive strategies. Cancer Med. 2020, 9, 421–431. [Google Scholar] [CrossRef] [Green Version]
- Bowtell, D.D.; Böhm, S.; Ahmed, A.A.; Aspuria, P.J.; Bast, R.C., Jr.; Beral, V.; Berek, J.S.; Birrer, M.J.; Blagden, S.; Bookman, M.A.; et al. Rethinking ovarian cancer II: Reducing mortality from high-grade serous ovarian cancer. Nat. Rev. Cancer 2015, 15, 668–679. [Google Scholar] [CrossRef] [Green Version]
- Ledermann, J.; Harter, P.; Gourley, C.; Friedlander, M.; Vergote, I.; Rustin, G.; Scott, C.; Meier, W.; Shapira-Frommer, R.; Safra, T.; et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N. Engl. J. Med. 2012, 366, 1382–1392. [Google Scholar] [CrossRef] [Green Version]
- Scott, C.L.; Swisher, E.M.; Kaufmann, S.H. Poly (ADP-ribose) polymerase inhibitors: Recent advances and future development. J. Clin. Oncol. 2015, 33, 1397–1406. [Google Scholar] [CrossRef] [Green Version]
- Bai, H.; Cao, D.; Yuan, F.; Sha, G.; Yang, J.; Chen, J.; Wang, Y.; Zhang, Z.; Shen, K. Prognostic value of endometriosis in patients with stage I ovarian clear cell carcinoma: Experiences at three academic institutions. Gynecol. Oncol. 2016, 143, 526–531. [Google Scholar] [CrossRef]
- Park, J.Y.; Kim, D.Y.; Suh, D.S.; Kim, J.H.; Kim, Y.M.; Kim, Y.T.; Nam, J.H. Significance of ovarian endometriosis on the prognosis of ovarian clear cell carcinoma. Int. J. Gynecol. Cancer 2018, 28, 11–18. [Google Scholar] [CrossRef]
- Gadducci, A.; Multinu, F.; Cosio, S.; Carinelli, S.; Ghioni, M.; Aletti, G.D. Clear cell carcinoma of the ovary: Epidemiology, pathological and biological features, treatment options and clinical outcomes. Gynecol. Oncol. 2021, 162, 741–750. [Google Scholar] [CrossRef] [PubMed]
- Chao, A.; Lai, C.H.; Wang, T.H.; Jung, S.M.; Lee, Y.S.; Chang, W.Y.; Yang, L.Y.; Ku, F.C.; Huang, H.J.; Chao, A.S.; et al. Genomic scar signatures associated with homologous recombination deficiency predict adverse clinical outcomes in patients with ovarian clear cell carcinoma. J. Mol. Med. 2018, 96, 527–536. [Google Scholar] [CrossRef] [PubMed]
- Dellett, M.; Hu, W.; Papadaki, V.; Ohnuma, S. Small leucine rich proteoglycan family regulates multiple signalling pathways in neural development and maintenance. Dev. Growth Differ. 2012, 54, 327–340. [Google Scholar] [CrossRef]
- Schaefer, L.; Iozzo, R.V. Biological functions of the small leucine-rich proteoglycans: From genetics to signal transduction. J. Biol. Chem. 2008, 283, 21305–21309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hocking, A.M.; Shinomura, T.; McQuillan, D.J. Leucine-rich repeat glycoproteins of the extracellular matrix. Matrix Biol. J. Int. Soc. Matrix Biol. 1998, 17, 1–19. [Google Scholar] [CrossRef]
- Vogel, K.G.; Paulsson, M.; Heinegård, D. Specific inhibition of type I and type II collagen fibrillogenesis by the small proteoglycan of tendon. Biochem. J. 1984, 223, 587–597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papadaki, V.; Asada, K.; Watson, J.K.; Tamura, T.; Leung, A.; Hopkins, J.; Dellett, M.; Sasai, N.; Davaapil, H.; Nik-Zainal, S.; et al. Two Secreted Proteoglycans, Activators of urothelial cell-cell adhesion, negatively contribute to bladder cancer initiation and progression. Cancers 2020, 12, 3362. [Google Scholar] [CrossRef] [PubMed]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
- Network, T.C.G.A.R. Integrated genomic analyses of ovarian carcinoma. Nature 2011, 474, 609–615. [Google Scholar] [CrossRef] [Green Version]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Machino, H.; Kaneko, S.; Komatsu, M.; Ikawa, N.; Asada, K.; Nakato, R.; Shozu, K.; Dozen, A.; Sone, K.; Yoshida, H.; et al. The metabolic stress-activated checkpoint LKB1-MARK3 axis acts as a tumor suppressor in high-grade serous ovarian carcinoma. Commun. Biol. 2022, 5, 39. [Google Scholar] [CrossRef] [PubMed]
- Shozu, K.; Kaneko, S.; Shinkai, N.; Dozen, A.; Kosuge, H.; Nakakido, M.; Machino, H.; Takasawa, K.; Asada, K.; Komatsu, M.; et al. Repression of the PRELP gene is relieved by histone deacetylase inhibitors through acetylation of histone H2B lysine 5 in bladder cancer. Clin. Epigenet. 2022, 14, 147. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.M.; Nakayama, K.; Shanta, K.; Razia, S.; Ishikawa, M.; Ishibashi, T.; Yamashita, H.; Sato, S.; Iida, K.; Kanno, K.; et al. Establishment of a novel in vitro model of endometriosis with oncogenic KRAS and PIK3CA mutations for understanding the underlying biology and molecular pathogenesis. Cancers 2021, 13, 3174. [Google Scholar] [CrossRef]
- Borowicz, S.; Van Scoyk, M.; Avasarala, S.; Karuppusamy Rathinam, M.K.; Tauler, J.; Bikkavilli, R.K.; Winn, R.A. The soft agar colony formation assay. J. Vis. Exp. 2014, 92, e51998. [Google Scholar] [CrossRef] [Green Version]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Ozawa, T.; Kaneko, S.; Szulzewsky, F.; Qiao, Z.; Takadera, M.; Narita, Y.; Kondo, T.; Holland, E.C.; Hamamoto, R.; Ichimura, K. C11orf95-RELA fusion drives aberrant gene expression through the unique epigenetic regulation for ependymoma formation. Acta Neuropathol. Commun. 2021, 9, 36. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The human genome browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef] [Green Version]
- Kubo, N.; Ishii, H.; Xiong, X.; Bianco, S.; Meitinger, F.; Hu, R.; Hocker, J.D.; Conte, M.; Gorkin, D.; Yu, M.; et al. Promoter-proximal CTCF binding promotes distal enhancer-dependent gene activation. Nat. Struct. Mol. Biol. 2021, 28, 152–161. [Google Scholar] [CrossRef]
- Yano, M.; Katoh, T.; Miyazawa, M.; Miyazawa, M.; Ogane, N.; Miwa, M.; Hasegawa, K.; Narahara, H.; Yasuda, M. Clinicopathological correlation of ARID1A status with HDAC6 and its related factors in ovarian clear cell carcinoma. Sci. Rep. 2019, 9, 2397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, S.; Tsuda, H.; Takano, M.; Tamai, S.; Matsubara, O. Loss of ARID1A protein expression occurs as an early event in ovarian clear-cell carcinoma development and frequently coexists with PIK3CA mutations. Mod. Pathol. 2012, 25, 615–624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuo, K.-T.; Mao, T.-L.; Jones, S.; Veras, E.; Ayhan, A.; Wang, T.-L.; Glas, R.; Slamon, D.; Velculescu, V.E.; Kuman, R.J.; et al. Frequent Activating Mutations of PIK3CA in Ovarian Clear Cell Carcinoma. Am. J. Pathol. 2009, 174, 1597–1601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiegand, K.C.; Shah, S.P.; Al-Agha, O.M.; Zhao, Y.; Tse, K.; Zeng, T.; Senz, J.; McConechy, M.K.; Anglesio, M.S.; Kalloger, S.E.; et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N. Engl. J. Med. 2010, 363, 1532–1543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katagiri, A.; Nakayama, K.; Rahman, M.T.; Rahman, M.; Katagiri, H.; Nakayama, N.; Ishikawa, M.; Ishibashi, T.; Iida, K.; Kobayashi, H.; et al. Loss of ARID1A expression is related to shorter progression-free survival and chemoresistance in ovarian clear cell carcinoma. Mod. Pathol. 2012, 25, 282–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yano, M.; Yasuda, M.; Sakaki, M.; Nagata, K.; Fujino, T.; Arai, E.; Hasebe, T.; Miyazawa, M.; Miyazawa, M.; Ogane, N.; et al. Association of histone deacetylase expression with histology and prognosis of ovarian cancer. Oncol. Lett. 2018, 15, 3524–3531. [Google Scholar] [CrossRef] [Green Version]
- Fukumoto, T.; Park, P.H.; Wu, S.; Fatkhutdinov, N.; Karakashev, S.; Nacarelli, T.; Kossenkov, A.V.; Speicher, D.W.; Jean, S.; Zhang, L.; et al. Repurposing Pan-HDAC inhibitors for ARID1A-mutated ovarian cancer. Cell Rep. 2018, 22, 3393–3400. [Google Scholar] [CrossRef] [Green Version]
- Zhao, B.; Lin, J.; Rong, L.; Wu, S.; Deng, Z.; Fatkhutdinov, N.; Zundell, J.; Fukumoto, T.; Liu, Q.; Kossenkov, A.; et al. ARID1A promotes genomic stability through protecting telomere cohesion. Nat. Commun. 2019, 10, 4067. [Google Scholar] [CrossRef] [Green Version]
- Kosuge, H.; Nakakido, M.; Nagatoishi, S.; Fukuda, T.; Bando, Y.; Ohnuma, S.I.; Tsumoto, K. Proteomic identification and validation of novel interactions of the putative tumor suppressor PRELP with membrane proteins including IGFI-R and p75NTR. J. Biol. Chem. 2021, 296, 100278. [Google Scholar] [CrossRef]
- Farley, J.H.; Brady, W.E.; Fujiwara, K.; Nomura, H.; Yunokawa, M.; Tokunaga, H.; Saitou, M.; Gershenson, D.M. A phase II evaluation of temsirolimus in combination with carboplatin and paclitaxel followed by temsirolimus consolidation as first-line therapy in the treatment of stage III-IV clear cell carcinoma of the ovary. J. Clin. Oncol. 2016, 34, 5531. [Google Scholar] [CrossRef]
- Hong, R.; Gu, J.; Niu, G.; Hu, Z.; Zhang, X.; Song, T.; Han, S.; Hong, L.; Ke, C. PRELP has prognostic value and regulates cell proliferation and migration in hepatocellular carcinoma. J. Cancer 2020, 11, 6376–6389. [Google Scholar] [CrossRef] [PubMed]
- Rucci, N.; Capulli, M.; Ventura, L.; Angelucci, A.; Peruzzi, B.; Tillgren, V.; Muraca, M.; Heinegård, D.; Teti, A. Proline/arginine-rich end leucine-rich repeat protein N-terminus is a novel osteoclast antagonist that counteracts bone loss. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 2013, 28, 1912–1924. [Google Scholar] [CrossRef] [PubMed]
- Asada, K.; Kaneko, S.; Takasawa, K.; Machino, H.; Takahashi, S.; Shinkai, N.; Shimoyama, R.; Komatsu, M.; Hamamoto, R. Integrated analysis of whole genome and epigenome data using machine learning technology: Toward the establishment of precision oncology. Front. Oncol. 2021, 11, 666937. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dozen, A.; Shozu, K.; Shinkai, N.; Ikawa, N.; Aoyama, R.; Machino, H.; Asada, K.; Yoshida, H.; Kato, T.; Hamamoto, R.; et al. Tumor Suppressive Role of the PRELP Gene in Ovarian Clear Cell Carcinoma. J. Pers. Med. 2022, 12, 1999. https://doi.org/10.3390/jpm12121999
Dozen A, Shozu K, Shinkai N, Ikawa N, Aoyama R, Machino H, Asada K, Yoshida H, Kato T, Hamamoto R, et al. Tumor Suppressive Role of the PRELP Gene in Ovarian Clear Cell Carcinoma. Journal of Personalized Medicine. 2022; 12(12):1999. https://doi.org/10.3390/jpm12121999
Chicago/Turabian StyleDozen, Ai, Kanto Shozu, Norio Shinkai, Noriko Ikawa, Rina Aoyama, Hidenori Machino, Ken Asada, Hiroshi Yoshida, Tomoyasu Kato, Ryuji Hamamoto, and et al. 2022. "Tumor Suppressive Role of the PRELP Gene in Ovarian Clear Cell Carcinoma" Journal of Personalized Medicine 12, no. 12: 1999. https://doi.org/10.3390/jpm12121999
APA StyleDozen, A., Shozu, K., Shinkai, N., Ikawa, N., Aoyama, R., Machino, H., Asada, K., Yoshida, H., Kato, T., Hamamoto, R., Kaneko, S., & Komatsu, M. (2022). Tumor Suppressive Role of the PRELP Gene in Ovarian Clear Cell Carcinoma. Journal of Personalized Medicine, 12(12), 1999. https://doi.org/10.3390/jpm12121999






