The Penn Medicine BioBank: Towards a Genomics-Enabled Learning Healthcare System to Accelerate Precision Medicine in a Diverse Population
Abstract
:1. Introduction
2. Materials and Methods
2.1. Planning and Development of PMBB
2.2. Patient-Participant Recruitment
2.3. Sample Collection
2.4. Genomic Data Generation
2.4.1. Genotyping and Imputation
2.4.2. Whole Exome Sequencing
2.5. Clinical Data and Clinical Informatics Core
2.6. Access to Data and Biospecimens
3. Results
3.1. Enrollment
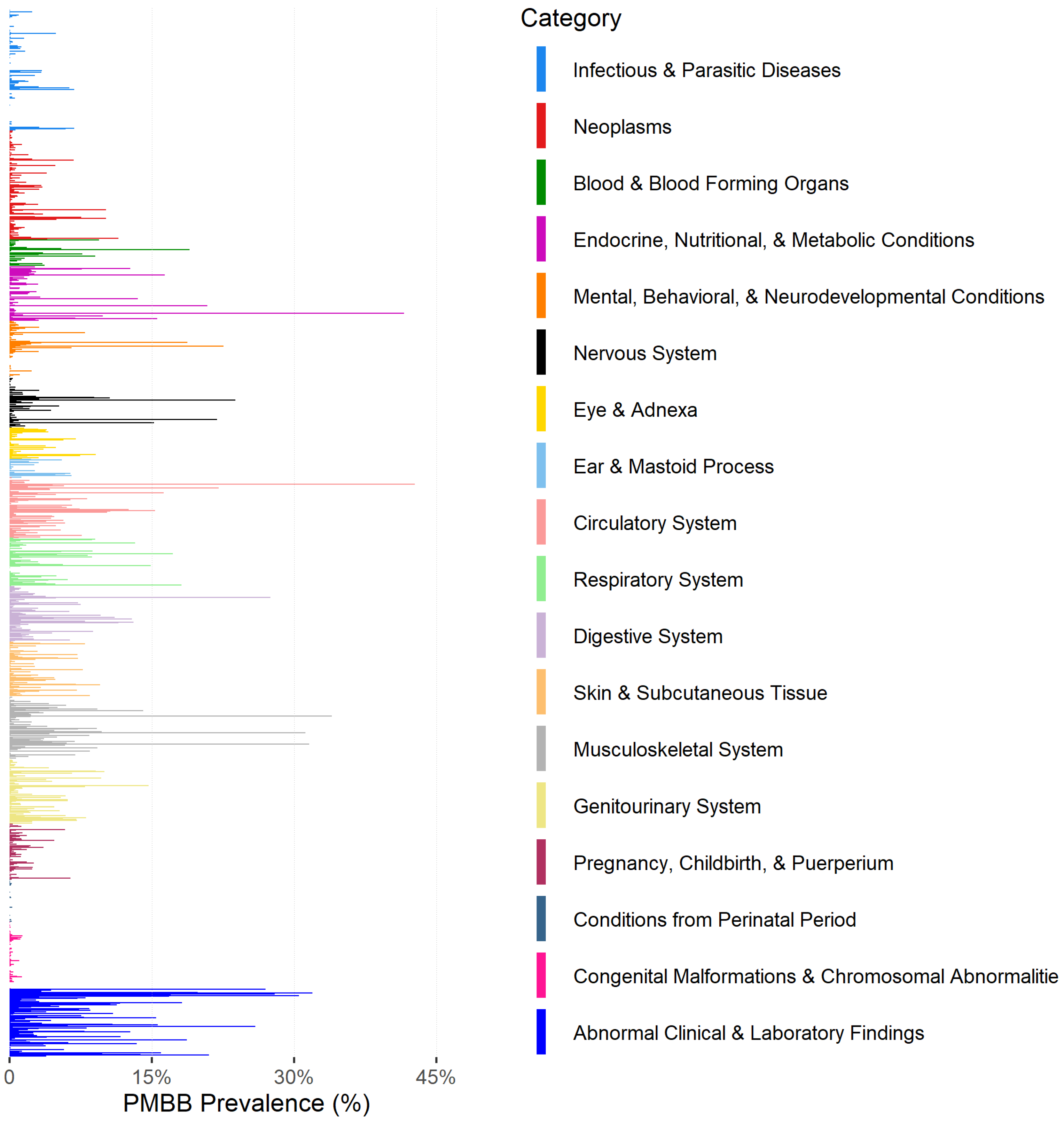
3.2. Clinical Data Availability
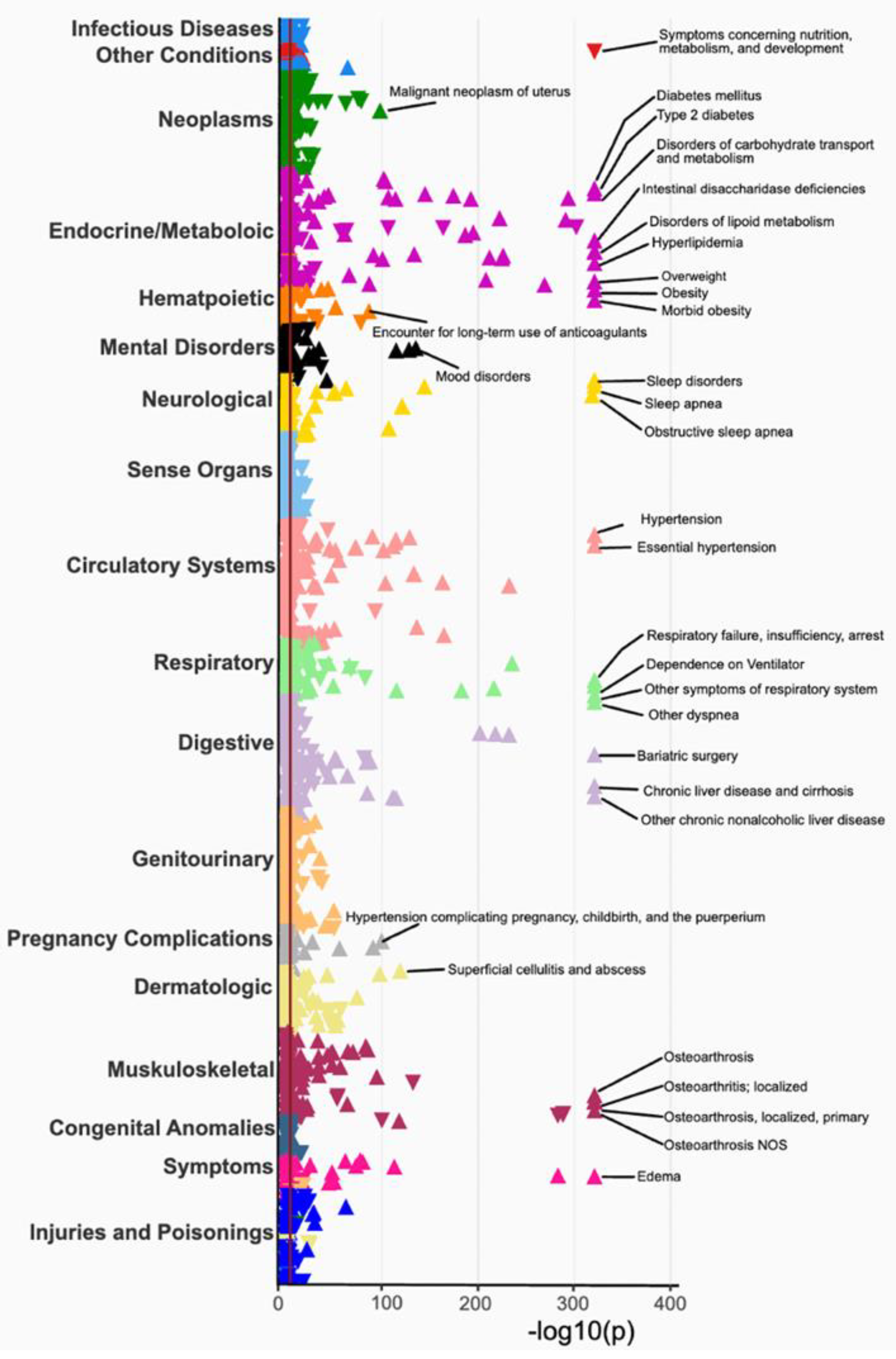
3.3. Phenome-Wide Association Study (PheWAS) of BMI
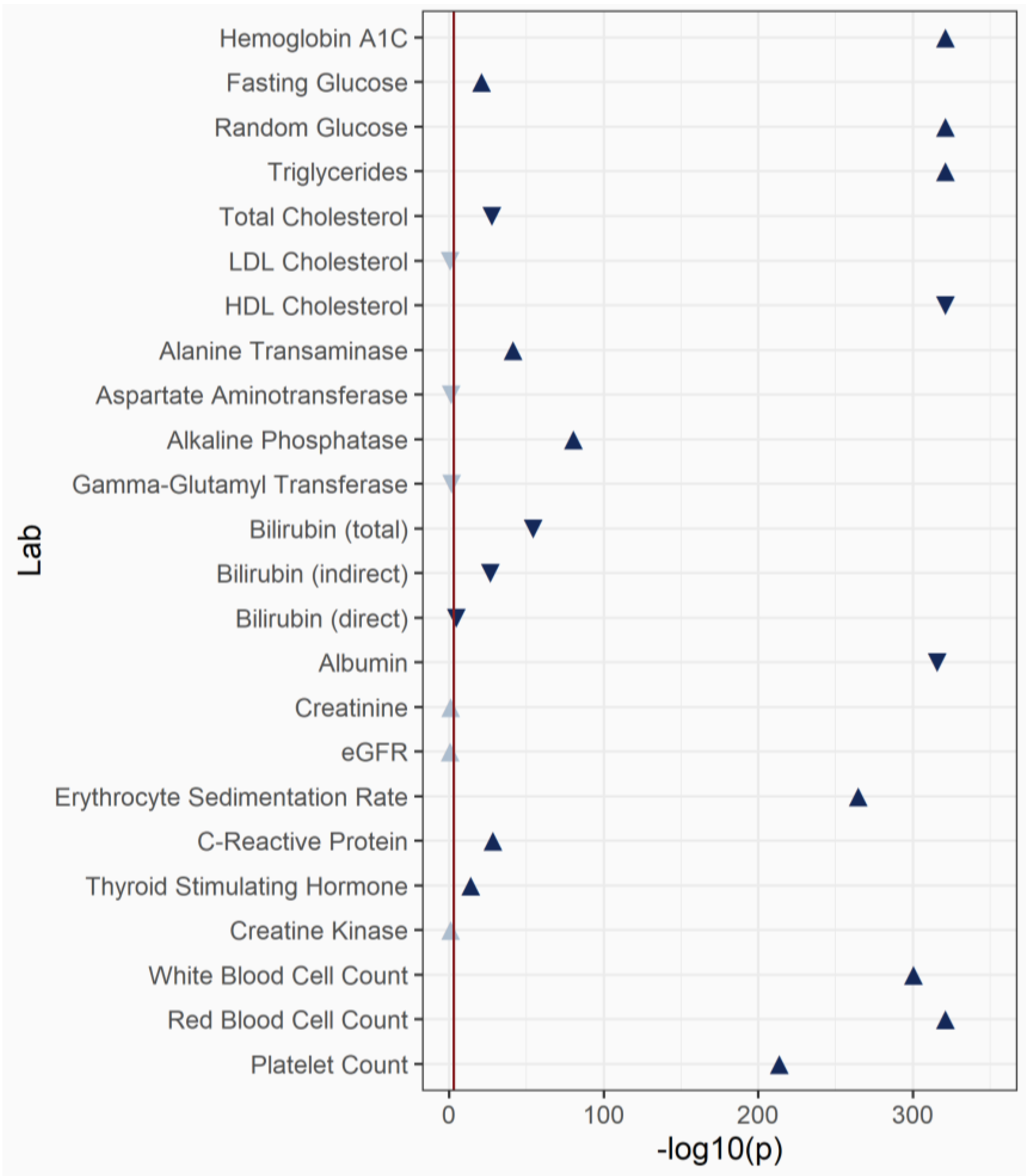
3.4. Laboratory-Wide Association Study (LabWAS) of BMI
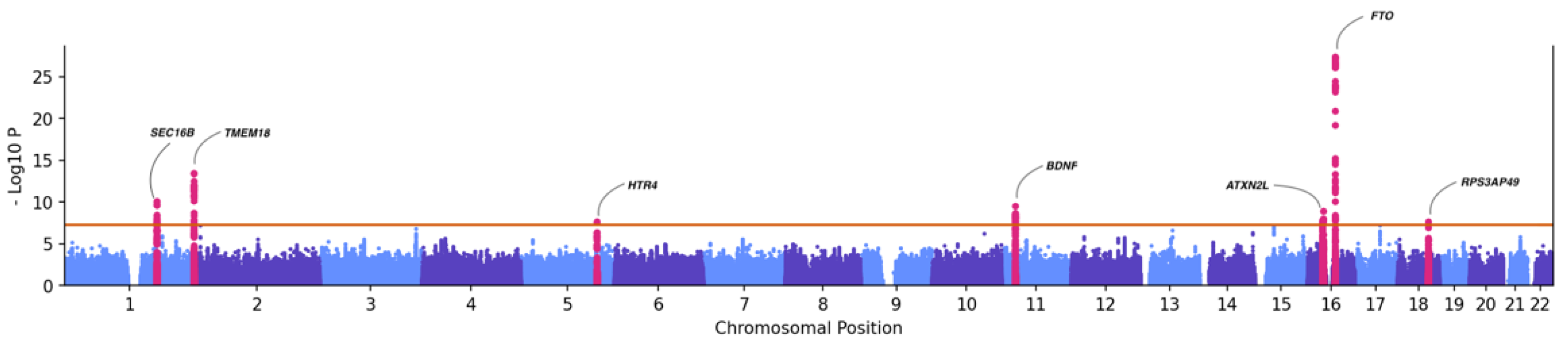
3.5. Genome-Wide Association (GWAS) with BMI
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Institute of Medicine. Genomics-Enabled Learning Health Care Systems: Gathering and Using Genomic Information to Improve Patient Care and Research: Workshop Summary; National Academies Press (US): Washington, DC, USA, 2015; ISBN 978-0-309-37112-4. [Google Scholar]
- Loh, P.-R.; Danecek, P.; Palamara, P.F.; Fuchsberger, C.; AReshef, Y.; Finucane, H.K.; Schoenherr, S.; Forer, L.; McCarthy, S.; Abecasis, G.R.; et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat. Genet. 2016, 48, 1443–1448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, S.; Forer, L.; Schönherr, S.; Sidore, C.; Locke, A.E.; Kwong, A.; Vrieze, S.I.; Chew, E.Y.; Levy, S.; McGue, M.; et al. Next-generation genotype imputation service and methods. Nat Genet. 2016, 48, 1284–1287. [Google Scholar] [CrossRef] [Green Version]
- Price, A.L.; Patterson, N.J.; Plenge, R.M.; Weinblatt, M.E.; Shadick, N.A.; Reich, D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006, 38, 904–909. [Google Scholar] [CrossRef] [PubMed]
- Klann, J.G.; Joss, M.A.H.; Embree, K.; Murphy, S.N. Data model harmonization for the All Of Us Research Program: Transforming i2b2 data into the OMOP common data model. PLoS ONE 2019, 14, e0212463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDonald, C.J.; Huff, S.M.; Suico, J.G.; Hill, G.; Leavelle, D.; Aller, R.; Forrey, A.; Mercer, K.; DeMoor, G.; Hook, J.; et al. LOINC, a Universal Standard for Identifying Laboratory Observations: A 5-Year Update. Clin. Chem. 2003, 49, 624–633. [Google Scholar] [CrossRef] [Green Version]
- Nelson, S.J.; Zeng, K.; Kilbourne, J.; Powell, T.; Moore, R. Normalized names for clinical drugs: RxNorm at 6 years. J. Am. Med. Inform. Assoc. 2011, 18, 441–448. [Google Scholar] [CrossRef] [Green Version]
- Wu, P.; Gifford, A.; Meng, X.; Li, X.; Campbell, H.; Varley, T.; Zhao, J.; Carroll, R.; Bastarache, L.; Denny, J.C.; et al. Mapping ICD-10 and ICD-10-CM Codes to Phecodes: Workflow Development and Initial Evaluation. JMIR Med. Inform. 2019, 7, e14325. [Google Scholar] [CrossRef]
- Green, E.D.; Gunter, C.; Biesecker, L.G.; Di Francesco, V.; Easter, C.L.; Feingold, E.A.; Felsenfeld, A.L.; Kaufman, D.J.; Ostrander, E.A.; Pavan, W.J.; et al. Strategic vision for improving human health at The Forefront of Genomics. Nature 2020, 586, 683–692. [Google Scholar] [CrossRef]
- Park, J.; Levin, M.G.; Haggerty, C.M.; Hartzel, D.N.; Judy, R.; Kember, R.L.; Reza, N.; Ritchie, M.D.; Owens, A.T.; Damrauer, S.M.; et al. A genome-first approach to aggregating rare genetic variants in LMNA for association with electronic health record phenotypes. Genet. Med. 2020, 22, 102–111. [Google Scholar] [CrossRef]
- Park, J.; Packard, E.A.; Levin, M.G.; Judy, R.L.; Regeneron Genetics Center; Damrauer, S.M.; Day, S.M.; Ritchie, M.D.; Rader, D.J. A genome-first approach to rare variants in hypertrophic cardiomyopathy genes MYBPC3 and MYH7 in a medical biobank. Hum. Mol. Genet. 2022, 31, 827–837. [Google Scholar] [CrossRef]
- Damrauer, S.M.; Hardie, K.; Kember, R.L.; Judy, R.; Birtwell, D.; Williams, H.; Rader, D.J.; Pyeritz, R.E. FBN1 Coding Variants and Nonsyndromic Aortic Disease. Circ. Genom. Precis. Med. 2019, 12, e002454. [Google Scholar] [CrossRef]
- Wang, L.; Desai, H.; Verma, S.S.; Le, A.; Hausler, R.; Verma, A.; Judy, R.; Doucette, A.; Gabriel, P.E.; Nathanson, K.L.; et al. Performance of polygenic risk scores for cancer prediction in a racially diverse academic biobank. Genet. Med. 2022, 24, 601–609. [Google Scholar] [CrossRef]
- Kember, R.L.; Levin, M.G.; Cousminer, D.L.; Tsao, N.; Judy, R.; Schur, G.M.; Lubitz, S.A.; Ellinor, P.T.; McCormack, S.E.; Grant, S.F.A.; et al. Genetically Determined Birthweight Associates with Atrial Fibrillation: A Mendelian Randomization Study. Circ. Genom. Precis. Med. 2020, 13, e002553. [Google Scholar] [CrossRef]
- Zhang, C.; Verma, A.; Feng, Y.; Melo, M.C.R.; McQuillan, M.; Hansen, M.; Lucas, A.; Park, J.; Ranciaro, A.; Thompson, S.; et al. Impact of natural selection on global patterns of genetic variation and association with clinical phenotypes at genes involved in SARS-CoV-2 infection. Proc. Natl. Acad. Sci. USA 2022, 119, e2123000119. [Google Scholar] [CrossRef]
- Bajaj, A.; Ihegword, A.; Qiu, C.; Small, A.M.; Wei, W.-Q.; Bastarache, L.; Feng, Q.; Kember, R.L.; Risman, M.; Bloom, R.D.; et al. Phenome-wide association analysis suggests the APOL1 linked disease spectrum primarily drives kidney-specific pathways. Kidney Int. 2020, 97, 1032–1041. [Google Scholar] [CrossRef]
- Shefchek, K.A.; Harris, N.L.; Gargano, M.; Matentzoglu, N.; Unni, D.; Brush, M.; Keith, D.; Conlin, T.; Vasilevsky, N.; Zhang, X.A.; et al. The Monarch Initiative in 2019: An integrative data and analytic platform connecting phenotypes to genotypes across species. Nucleic Acids Res. 2020, 48, D704–D715. [Google Scholar] [CrossRef] [Green Version]
- Verma, S.S.; Chung, W.K.; Dudek, S.; Williamson, J.L.; Verma, A.; Robinson, S.; Rader, D.J.; Reilly, M.P.; Sengupta, S.; FitzGerald, G.A.; et al. Research on COVID-19 through patient-reported data: A survey for observational studies in the COVID-19 pandemic. J. Clin. Transl. Sci. 2021, 5, e17. [Google Scholar] [CrossRef]
- Drivas, T.G.; Lucas, A.; Zhang, X.; Ritchie, M.D. Mendelian pathway analysis of laboratory traits reveals distinct roles for ciliary subcompartments in common disease pathogenesis. Am. J. Hum. Genet. 2021, 108, 482–501. [Google Scholar] [CrossRef]
- Damrauer, S.M.; Chaudhary, K.; Cho, J.H.; Liang, L.W.; Argulian, E.; Chan, L.; Dobbyn, A.; Guerraty, M.A.; Judy, R.; Kay, J.; et al. Association of the V122I Hereditary Transthyretin Amyloidosis Genetic Variant With Heart Failure Among Individuals of African or Hispanic/Latino Ancestry. JAMA 2019, 322, 2191. [Google Scholar] [CrossRef]
- Lau-Min, K.S.; Asher, S.B.; Chen, J.; Domchek, S.M.; Feldman, M.; Joffe, S.; Landgraf, J.; Speare, V.; Varughese, L.A.; Tuteja, S.; et al. Real-world integration of genomic data into the electronic health record: The PennChart Genomics Initiative. Genet. Med. Off. J. Am. Coll. Med. Genet. 2021, 23, 603–605. [Google Scholar] [CrossRef]
- Lau-Min, K.S.; McKenna, D.; Asher, S.B.; Bardakjian, T.; Wollack, C.; Bleznuck, J.; Biros, D.; Anantharajah, A.; Clark, D.F.; Condit, C.; et al. Impact of integrating genomic data into the electronic health record on genetics care delivery. Genet. Med. 2022, 24, 2338–2350. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.S.; Keat, K.; Li, B.; Hoffecker, G.; Risman, M.; Regeneron Genetics Center; Sangkuhl, K.; Whirl-Carrillo, M.; Dudek, S.; Verma, A.; et al. Evaluating the frequency and the impact of pharmacogenetic alleles in an ancestrally diverse Biobank population. medRxiv 2022, medRxiv:2022.08.26.22279261. [Google Scholar] [CrossRef]
- Martin, A.R.; Kanai, M.; Kamatani, Y.; Okada, Y.; Neale, B.M.; Daly, M.J. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 2019, 51, 584–591. [Google Scholar] [CrossRef] [PubMed]
- Sirugo, G.; Williams, S.M.; Tishkoff, S.A. The Missing Diversity in Human Genetic Studies. Cell 2019, 177, 26–31. [Google Scholar] [CrossRef] [Green Version]
- Kember, R.L.; Merikangas, A.K.; Verma, S.S.; Verma, A.; Judy, R.; Regeneron Genetics Center; Damrauer, S.M.; Ritchie, M.D.; Rader, D.J.; Bućan, M. Polygenic Risk of Psychiatric Disorders Exhibits Cross-trait Associations in Electronic Health Record Data From European Ancestry Individuals. Biol. Psychiatry 2021, 89, 236–245. [Google Scholar] [CrossRef] [PubMed]
- Hartwell, E.E.; Merikangas, A.K.; Verma, S.S.; Ritchie, M.D.; Regeneron Genetics Center; Kranzler, H.R.; Kember, R.L. Genetic liability for substance use associated with medical comorbidities in electronic health records of African- and European-ancestry individuals. Addict. Biol. 2022, 27, e13099. [Google Scholar] [CrossRef]
- Alanio, C.; Verma, A.; Mathew, D.; Gouma, S.; Liang, G.; Dunn, T.; Oldridge, D.A.; Weaver, J.; Kuri-Cervantes, L.; Pampena, M.B.; et al. Cytomegalovirus Latent Infection is Associated with an Increased Risk of COVID-19-Related Hospitalization. J. Infect. Dis. 2022, 226, 463–473. [Google Scholar] [CrossRef]
- Banday, A.R.; Stanifer, M.L.; Florez-Vargas, O.; Onabajo, O.O.; Papenberg, B.W.; Zahoor, M.A.; Mirabello, L.; Ring, T.J.; Lee, C.-H.; Albert, P.S.; et al. Genetic regulation of OAS1 nonsense-mediated decay underlies association with COVID-19 hospitalization in patients of European and African ancestries. Nat. Genet. 2022, 54, 1103–1116. [Google Scholar] [CrossRef]
- Anderson, E.M.; Goodwin, E.C.; Verma, A.; Arevalo, C.P.; Bolton, M.J.; Weirick, M.E.; Gouma, S.; McAllister, C.M.; Christensen, S.R.; Weaver, J.; et al. Seasonal human coronavirus antibodies are boosted upon SARS-CoV-2 infection but not associated with protection. Cell 2021, 184, 1858–1864. [Google Scholar] [CrossRef]
- Flannery, D.D.; Gouma, S.; Dhudasia, M.B.; Mukhopadhyay, S.; Pfeifer, M.R.; Woodford, E.C.; Gerber, J.S.; Arevalo, C.P.; Bolton, M.J.; Weirick, M.E.; et al. SARS-CoV-2 seroprevalence among parturient women in Philadelphia. Sci. Immunol. 2020, 5, eabd5709. [Google Scholar] [CrossRef]
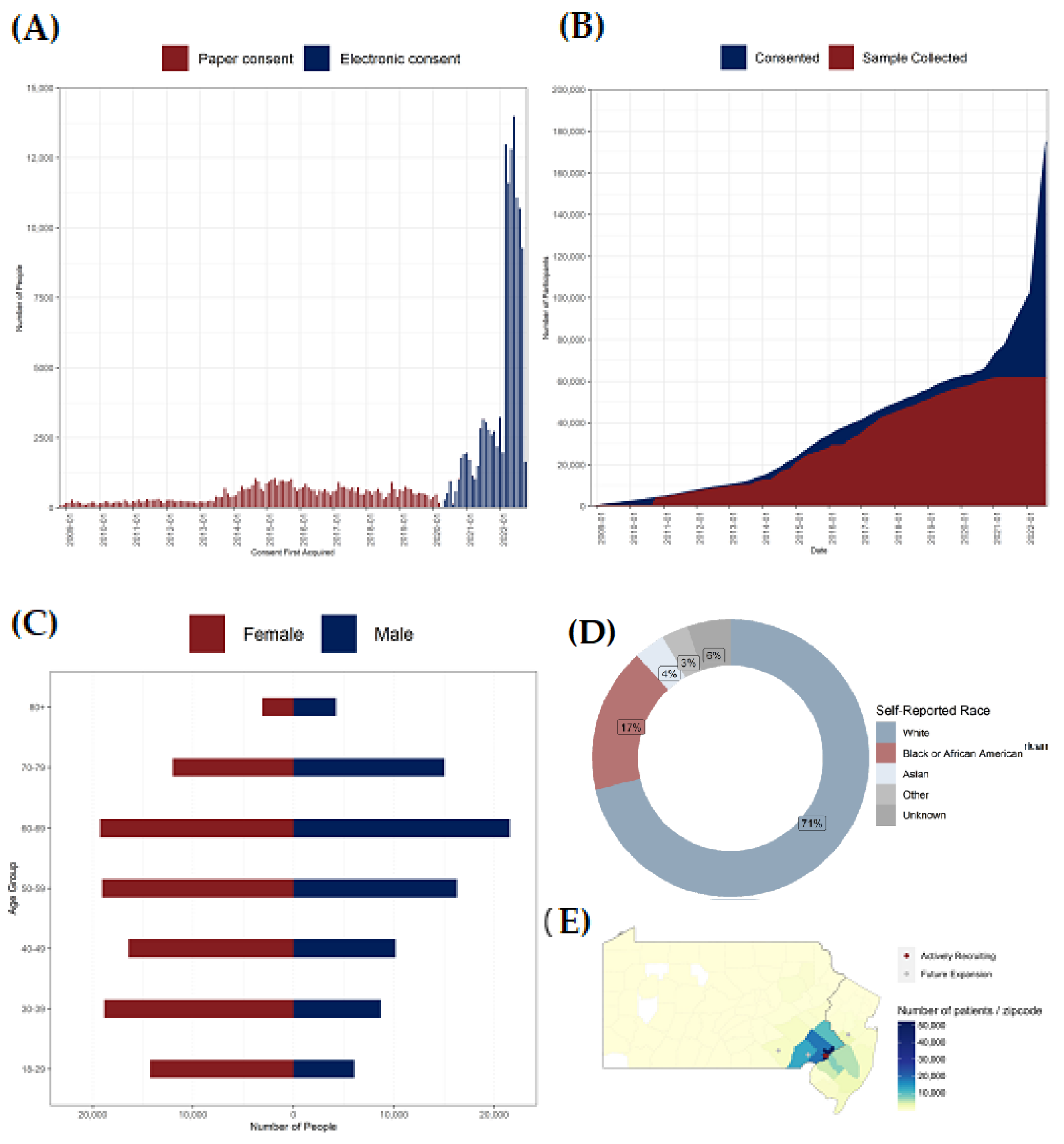
| PMBB Participants n (%) | Genotyped Participants n (%) | UPHS Patients n (%) † | |
|---|---|---|---|
| Total | 174,712 | 43,884 | 3,688,610 |
| Gender | |||
| Female | 97,674 (55.9%) | 21,965 (50.1%) | 2,042,868 (55.4%) |
| Male | 77,055 (44.1%) | 21,918 (49.9%) | 1,604,210 (43.5%) |
| Other | 17 (<1%) | 1 (<1%) | 107 (<1%) |
| Age Range (Years) | 18–103 | 18–103 | 0–121 |
| Age Groups | |||
| 18–29 | 17,815 (10.2%) | 4302 (9.8%) | 392,532 (10.5%) |
| 30–39 | 27,355 (15.7%) | 5406 (12.3%) | 518,679 (14.1%) |
| 40–49 | 25,819 (14.8%) | 5688 (13.0%) | 430,489 (11.7%) |
| 50–59 | 33,827 (19.4%) | 9519 (21.7%) | 458,952 (12.4%) |
| 60–69 | 40,268 (23.0%) | 10,839 (24.7%) | 507,397 (13.8%) |
| 70–79 | 28,582 (16.4%) | 5941 (13.5%) | 400,016 (10.8%) |
| 80+ | 8811 (5.0%) | 2189 (5.0%) | 333,781 (9.0%) |
| Self-reported Race | |||
| African American | 29,372 (16.8%) | 10,815 (24.6%) | 672,461 (18.2%) |
| White | 124,406 (71.2%) | 29,329 (66.8%) | 2,029,684 (55.0%) |
| Asian | 7156 (4.1%) | 979 (2.2%) | 152,615 (4.1%) |
| Other | 9386 (5.4%) | 1372 (3.1%) | 370,313 (10.0%) |
| Unknown | 7499 (4.3%) | 1761 (4.0%) | 463,537 (12.6%) |
| Self-reported Ethnicity | |||
| Hispanic | 5715 (3.3%) | 1112 (2.5%) | 174,179 (4.7%) |
| Non-Hispanic | 165,713 (94.8%) | 42,425 (96.7%) | 3,290,018 (89.2%) |
| Unknown | 3284 (1.9%) | 347 (0.8%) | 183,723 (5.0%) |
| Genetically-Inferred Ancestry | |||
| African | N/A | 11,300 (25.7%) | N/A |
| European | N/A | 30,360 (69.2%) | N/A |
| East Asian | N/A | 680 (1.5%) | N/A |
| South Asian | N/A | 573 (1.3%) | N/A |
| Admixed American | N/A | 711 (1.6%) | N/A |
| Other | N/A | 301 (0.7%) | N/A |
| Median period of EHR follow-up since enrollment | 7 years | 5.7 years | N/A |
| Event Type | Total Number of Events | Mean Number of Events (SD) * |
|---|---|---|
| Encounter | 46,738,773 | 268 (321) |
| Diagnosis Code (number of condition-related visits) | 10,023,922 | 57.4 (58.7) |
| Procedure Code | 3,621,056 | 20.7 (26.7) |
| Medication Order | 27,914,486 | 159.8 (271.8) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Verma, A.; Damrauer, S.M.; Naseer, N.; Weaver, J.; Kripke, C.M.; Guare, L.; Sirugo, G.; Kember, R.L.; Drivas, T.G.; Dudek, S.M.; et al. The Penn Medicine BioBank: Towards a Genomics-Enabled Learning Healthcare System to Accelerate Precision Medicine in a Diverse Population. J. Pers. Med. 2022, 12, 1974. https://doi.org/10.3390/jpm12121974
Verma A, Damrauer SM, Naseer N, Weaver J, Kripke CM, Guare L, Sirugo G, Kember RL, Drivas TG, Dudek SM, et al. The Penn Medicine BioBank: Towards a Genomics-Enabled Learning Healthcare System to Accelerate Precision Medicine in a Diverse Population. Journal of Personalized Medicine. 2022; 12(12):1974. https://doi.org/10.3390/jpm12121974
Chicago/Turabian StyleVerma, Anurag, Scott M. Damrauer, Nawar Naseer, JoEllen Weaver, Colleen M. Kripke, Lindsay Guare, Giorgio Sirugo, Rachel L. Kember, Theodore G. Drivas, Scott M. Dudek, and et al. 2022. "The Penn Medicine BioBank: Towards a Genomics-Enabled Learning Healthcare System to Accelerate Precision Medicine in a Diverse Population" Journal of Personalized Medicine 12, no. 12: 1974. https://doi.org/10.3390/jpm12121974
APA StyleVerma, A., Damrauer, S. M., Naseer, N., Weaver, J., Kripke, C. M., Guare, L., Sirugo, G., Kember, R. L., Drivas, T. G., Dudek, S. M., Bradford, Y., Lucas, A., Judy, R., Verma, S. S., Meagher, E., Nathanson, K. L., Feldman, M., Ritchie, M. D., Rader, D. J., & For The Penn Medicine BioBank. (2022). The Penn Medicine BioBank: Towards a Genomics-Enabled Learning Healthcare System to Accelerate Precision Medicine in a Diverse Population. Journal of Personalized Medicine, 12(12), 1974. https://doi.org/10.3390/jpm12121974






