Radio-lncRNAs: Biological Function and Potential Use as Biomarkers for Personalized Oncology
Abstract
1. lncRNAs Are New Players in Radiogenomics
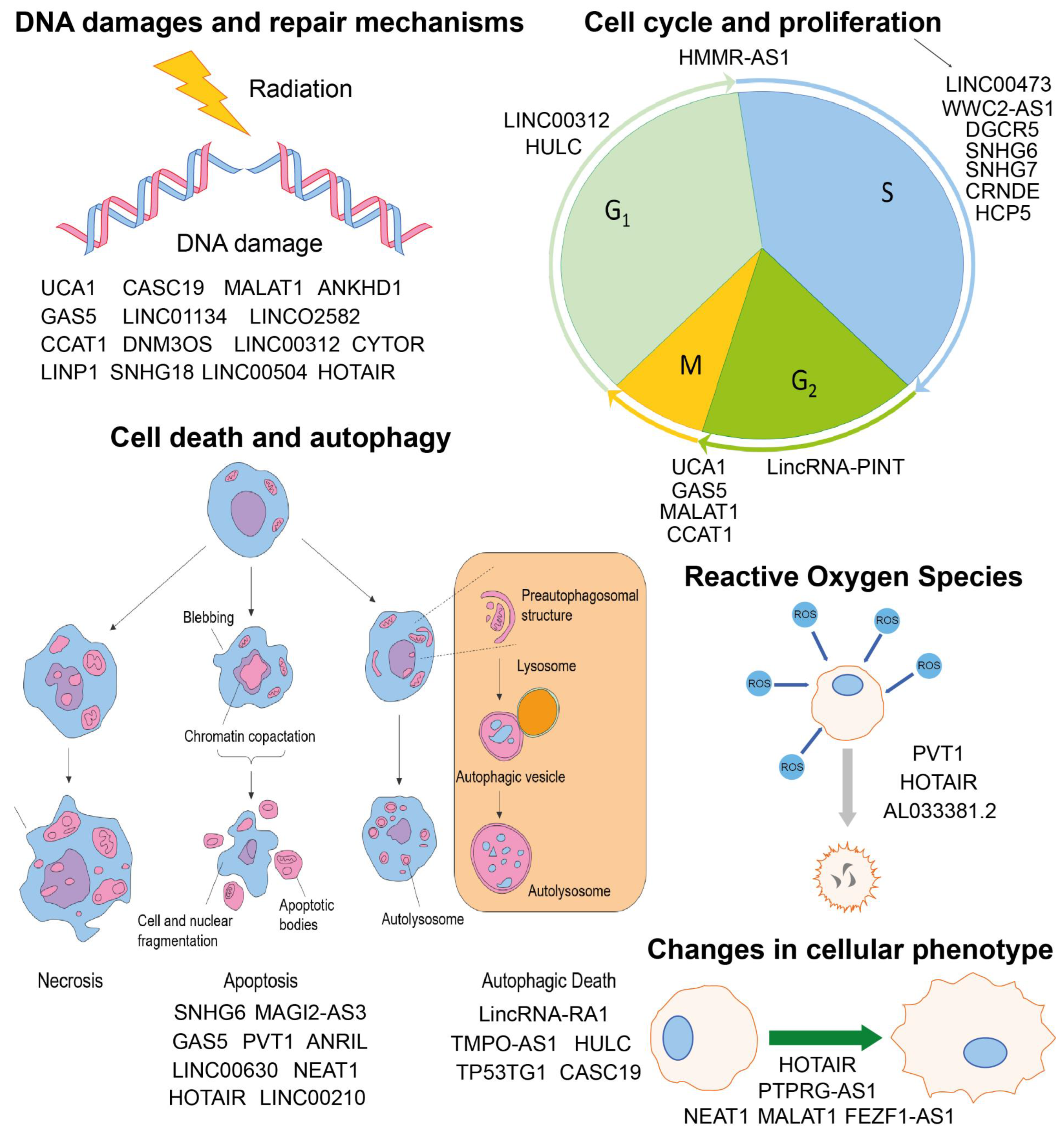
2. Involvement of lncRNAs in Radiobiological Processes
2.1. Cell Cycle and Proliferation
2.2. Cell Death and Autophagy
2.3. Reactive Oxygen Species (ROS)
2.4. DNA Damages and Repair Mechanisms
2.5. Changes in Cellular Phenotype
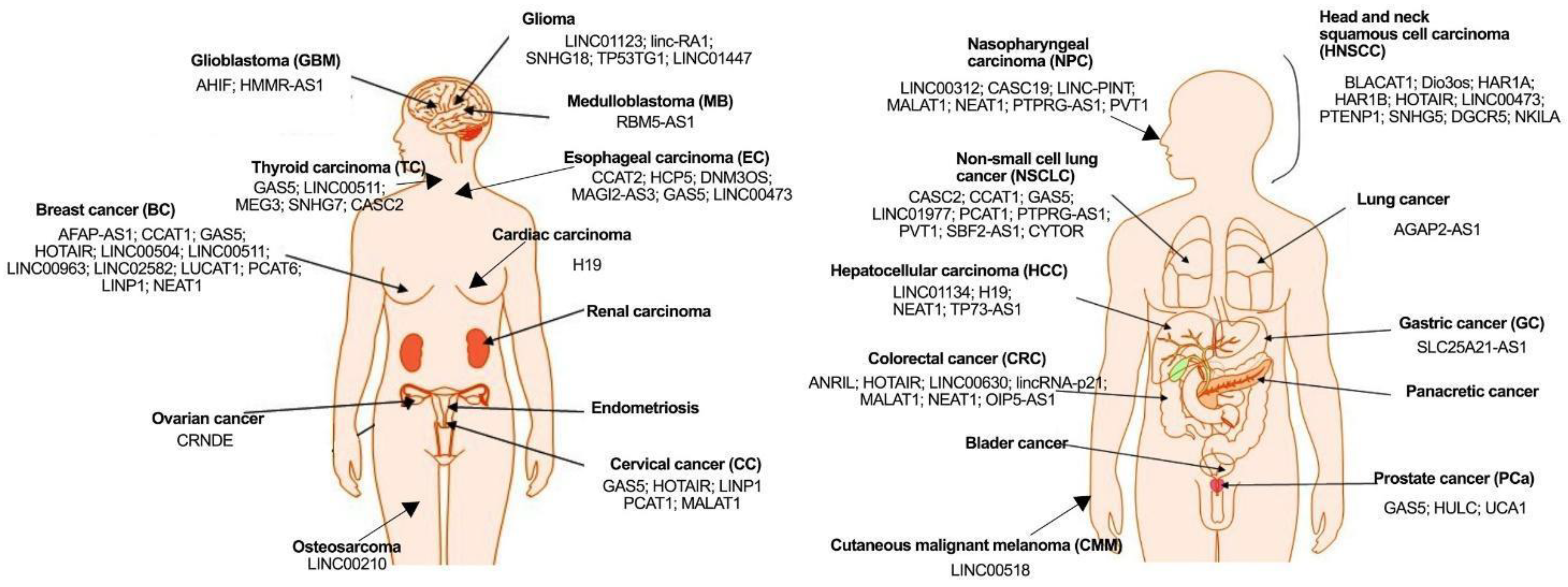
3. Potential Use of lncRNAs as Biomarkers
| Gene Name | Cancer Type | Expression | Impact on Radiotherapy | Targets | Reference |
|---|---|---|---|---|---|
| AFAP-AS1 | Breast cancer (BC) | Upregulated | AFAP-AS1 overexpression enhances radioresistance of BC cells (promoted cell proliferation, invasion, tumor growth, inhibits apoptosis) | Wnt/β-catenin | [92] |
| AGAP2-AS1 | Lung cancer | Upregulated | Promotes the immunologic function after IR | miR-296/NOTCH2 | [93] |
| AHIF | Glioblastoma (GBM) | Upregulated | AHIF knockdown enhances radiosensitivity | Bax/Bcl-2 | [94] |
| ANRIL | Colon cancer | Upregulated | ANRIL suppress radiosensitivity by binding to miR-181a-5p and reversing functions of chitooligosaccharides (COS) | miR-181a-5p/ chitooligosaccharides (COS) | [44] |
| BLACAT1 | Head and neck squamous cell carcinoma (HNSCC) | Upregulated | BLACAT1 knockdown enhances radiosensitivity of HNSCC cells | PSEN1 | [95] |
| CASC19 | Nasopharyngeal carcinoma (NPC) | Upregulated | CASC19 contributes to the radioresistance of NPC cells by promotion of autophagy and inhibition of apoptosis through AMPK/mTOR signaling pathway | PARP1/AMPK/mTOR | [51] |
| CASC2 | Non-small cell lung cancer (NSCLC) | Downregulated | CASC2 overexpression induces radiosensitivity of NSCLC cells | PERK/CHOP | [96] |
| Papillary Thyroid Cancer (PTC) | Downregulated | Low expression of CASC2 causes high IR resistance of PTC cells; overexpression of CASC2 results in higher IR sensitivity of PTC cells (induced cell viability and inhibited post-IR apoptosis); CASC2 enhances radiosensitivity in PTC by sponging miR-155 | miR-155 | [97] | |
| CCAT1 | Breast cancer (BC) | Upregulated | Downregulation of CCAT1 enhances radiosensitivity through miR148b negative regulation (decreased colony formation rate, promoted apoptosis) | miR-148b/miR-218/ZFX | [92,98] |
| Non-small cell lung cancer (NSCLC) | Upregulated | Higher CCAT1 expression correlates with higher radioresistance of NSCLC cells; downregulation of CCAT1 can improve the radiosensitivity of NSCLC cells by mediating cell cycle arrest, DNA damage, and apoptosis | MAPK/MAPK1/ERK/ MEK | [33] | |
| CCAT2 | Esophageal carcinoma (EC) | Upregulated | CCAT2 knockdown results in radiosensitivity enhancement of EC cells (induced apoptosis); overexpressed CCAT2 causes EC cells to gain radioresistant features through inhibiting apoptosis via miR-145/p70S6K1 signaling pathways and by activating the Akt/ERK/p70S6K1 signaling pathways | miR-145/p70S6K1/p53/ c-Myc/Akt signaling pathway | [99] |
| CRNDE | Ovarian cancer | Upregulated | CRNDE silencing resulted in enhanced radiosensitivity, inhibited clone formation and tumor growth in mice | No data | [28] |
| CYTOR | Non-small cell lung cancer (NSCLC) | Upregulated | Silencing CYTOR results in enhanced radiosensitivity of NSCLC cells (weak colony formation, high levels of H2AX); CYTOR binds to miR-206, silencing it and causing upregulation of PTMA, resulting in radioresistance | miR-206/PTMA | [67] |
| Suppresses radiosensitivity through regulating malignant phenotypes | miR-195 | [74] | |||
| DGCR5 | Laryngeal carcinoma (LC) | Upregulated | Knockdown could sensitize tumor cells to radiation through modulating miR-195 | miR-195 | [37] |
| Dio3os | Head and neck squamous cell carcinoma (HNSCC) | Downregulated after IR | No data | MYH7B/SRCAP/ HELZ2/NOS1/CROCC/CEP250/LPP/ABI2/ HERC2/RTEL1/ SMC1A1/HERC1/GAS7/NOTCH2/PKD1/ CFLAR/FAT3/FAT2/ CELSR3/CBL/NCOR2 | [100] |
| DNM3OS | Esophageal squamous cell carcinoma (ESCC) | Upregulated | Decreased DNM3OS cause significantly increased radiosensitivity in vitro and in vivo | Cancer associated fibroblasts (CAFs)/PDGFB/ PDGFRB/FOXO1 | [66] |
| GAS5 | Thyroid carcinoma (TC) | Downregulated | GAS5 overexpression enhances radiosensitivity | miR-362-5p/SMG1 | [101] |
| Cervical cancer (CC) | Downregulated | GAS5 overexpression enhances CC cells radiosensitivity | miR-106b/IER3 | [102] | |
| Breast cancer (BC) | Downregulated after IR further decrease in expression level | Induced overexpression reduces miR-21 expression leading to radiosensitization | miR-21 | [31] | |
| Esophageal squamous cell carcinoma (ESCC) | Downregulated | Simultaneous upregulation with miR-21 knockdown improves radiosensitivity after IR | miR-21/RECK | [41] | |
| Prostate cancer (PCa) | Downregulated | Artificially elevated level enhances α-solanine-induced radiosensitivity | miR18a | [30] | |
| Non-small cell lung cancer (NSCLC) | Downregulated | Enhances radiosensitivity | miR-135b | [103] | |
| H19 | Cardiac carcinoma | Upregulated | H19 knockdown resulted in enhanced radiosensitivity of cardiac carcinoma cells | miR-130a-3p/miR-17-5p | [104] |
| Hepatocellular carcinoma (HCC) | Downregulated | H19 overexpression enhances radiosensitivity of HCC cells through the miR-193a-3p/PSEN1 axis (promoted apoptosis, inhibited DNA double-strand break repair) | miR-193a-3p/PSEN1 | [105] | |
| HAR1A | Head and neck squamous cell carcinoma (HNSCC) | Downregulated after IR | No data | RANBP2/LPP/ABI2/ HELZ/PHC3/HERC1/ MTO5A/FZD3/ CTNNA3/CBL/BMPR2/FAT3/CFLAR | [100] |
| HAR1B | Head and neck squamous cell carcinoma (HNSCC) | Downregulated after IR | No data | LPP/ABI2/RSF1/HERC2/HELZ/FZD3/CFLAR/ FAT3/FRK/FER/PDK1/ FAT1 | [100] |
| HCP5 | Esophageal carcinoma (EC) | Upregulated | Knockdown of HCP5 enhances radiosensitivity trough modulating the Akt signaling pathway | miR-216a-3p/PDK1 | [26] |
| HMMR-AS1 | Glioblastoma (GBM) | Upregulated | Knockdown may suppress and radiosensitize the tumor | may regulate ERK1/2 by altering HMMR expression | [34] |
| HOTAIR | Colorectal cancer (CRC) | Upregulated; increases after IR in dose-dependent manner | Inhibits radiotherapy efficacy through regulating miR-93/ATG12-mediated autophagy | miR-93/ATG | [39] |
| Colorectal cancer (CRC) | Upregulated | Downregulation of HOTAIR enhanced radiosensitivity via reducing cell proliferation and invasiveness | No data | [75] | |
| Head and neck squamous cell carcinoma (HNSCC) | Upregulated after IR | Higher expression of HOTAIR is correlated with a higher resistance to radiotherapy in colon and breast cancer cell lines; high expression of HOTAIR is connected with the EMT process, maintaining of cancer initiating cells, and aggressive types of HNSCC | LPP/ABI2/NOS1/ CFLAR/REL/FAT3/ PDK1/FZD3/SMAD2/ FRK/SRCAP/WNT2B/ CBL | [100] | |
| Cervical cancer (CC) | Upregulated | Leads to tumor radioresistance via increasing HIF1α expression | HIF1α | [54] | |
| Cervical cancer (CC) | Upregulated | Promotes radioresistance through p21 inhibition | p21 | [76] | |
| Breast cancer (BC) | Upregulated | Downregulation of HOTAIR enhanced radiosensitivity | miR-218 | [106] | |
| Breast cancer (BC) | Upregulated | Overexpression of HOTAIR results in higher radioresistance of BC cells; HOTAIR overexpression enhances the cell proliferation and growth under irradiation stress; HOTAIR knockdown resulted in increased apoptosis and a reduced number of BC cells in the S phase of a cell cycle; its expression is positively correlated with DNA damage repair factors | HSPA1A/ miR-449b-5p/EZH2/PRC2/EED/ SUZ12/Ku70/Ku80/ DNA-Pk/ATM | [61,62] | |
| HULC | Prostate cancer (PCa) | Upregulated after IR | HULC knockdown enhances sensitivity of PCa cell to IR; cell apoptosis and proliferation induced by IR are enhanced by HULC knockdown and decreased by HULC overexpression | Bax/PCNA/cyclinD1/ caspase-3/Beclin-1/ p-4E-BP1 | [50] |
| LINC00210 | Osteosarcoma | Upregulated | Knockdown of LINC00210 results in enhanced radiosensitivity (after IR: decreased cell viability, induced apoptosis, inhibited levels of CyclinD1 and Bcl-2, increased levels of p21 and Bax); regulates radiosensitivity through the miR-342-3p/GFRA1 axis | miR-342-3p/GFRA1/ CyclinD1/Bcl-2/p21/Bax | [45] |
| LINC00312 | Nasopharyngeal carcinoma | Downregulated | Overexpression suppresses radiotherapy resistance | RAD50/MRE11/NBS1/ Ku80 | [57] |
| LINC00473 | Esophageal squamous cell carcinoma (ESCC) | Upregulated | Reduces radiotherapy effectiveness increasing cancer proliferative ability | miR-497-5p/CDC25A, miR-374a-5p/SPIN1 | [35,36] |
| Head and neck squamous cell carcinoma (HNSCC) | Upregulated | LINC00473 knockdown enhances radiosensitivity of HNSCC cells | Bax/Bcl2/Wnt/β-catenin pathway | [107] | |
| LINC00504 | Breast cancer (BC) | Upregulated | Decreases cell radiosensitivity by regulating CPEB2 expression | TAF15/CPEB2 | [59] |
| LINC00511 | Thyroid carcinoma (TC) | Upregulated | Potentiates resistance to radiotherapy by modulating the TAF1/JAK2/STAT3 axis | TAF1/JAK2/STAT3 | [108] |
| Breast cancer (BC) | Upregulated | LINC00511 knockdown enhances radiosensitivity (restricts cell proliferation, promotes apoptosis, and inhibits tumor growth) | STXBP4/miR-185 | [92] | |
| LINC00518 | Cutaneous malignant melanoma (CMM) | Upregulated | Potentiates radioresistance through enhancing glycolytic metabolism | miR-33a-3p/HIF1α/ LDHA | [98] |
| LINC00630 | Colorectal cancer (CRC) | Upregulated | Silencing could increase radiosensitivity by epigenetically repress BEX1 expression | EZH2/BEX1 | [38] |
| LINC00963 | Breast cancer (BC) | Upregulated | Highly expressed LINC00963 causes BC cells to enhance radioresistance; its silencing results in an increase of radiosensitivity (restrains cell proliferation, impairs colony formation and tumor growth, arrest cells at the G0/G1 phase, stimulates apoptosis); LINC00963 induced radiosensitivity through the miR-324-3p/ACK1 axis | miR-324-3p/ACK1/ CDK6/p27/CyclinD1 | [109] |
| LINC01123 | Glioma | Upregulated | Enhances radioresistance by creating the LINC01123/miR-151a/CENPB axis | miR-151a/CENPB | [110] |
| LINC01134 | Hepatocellular carcinoma (HC) | Upregulated | Augments resistance to radiotherapy via modulating the MAPK1 signaling pathway | miR-342-3p/IGF2BP2/ MAPK1 | [55] |
| LINC01447 | Low-grade Glioma | Upregulated | LINC01447 inhibition results in radiosensitivity enhancement in low-grade glioma cells (decreased cell viability, inhibited colony formation, increased apoptosis) | No data | [111] |
| LINC01977 | Non-small cell lung cancer (NSCLC) | Upregulated | LINC01977 inhibition results in enhanced radiosensitivity in NSCLC cells (reduced colony formation, higher expression of H2AX) | No data | [112] |
| LINC02582 | Breast cancer (BC) | Upregulated | Promotes radioresistance via the USP7/CHK1 signaling axis | USP7/CHK1 | [60] |
| LINC-PINT | Nasopharyngeal carcinoma (NPC) | Downregulated | Artificial upregulation potentiates radiosensitivity through an increase in apoptosis rate | ATM/ATR-Chk1/Chk2 pathway and DNA-PKcs | [56] |
| linc-RA1 | Glioma | Upregulated | Strengthens radioresistance via inhibiting autophagy activation | H2Bub1/USP44 | [49] |
| lincRNA-p21 | Colorectal cancer (CRC) | Downregulated IR cause further decrease in expression level | Enhances radiosensitivity after IR through activating pro-apoptotic mechanisms | Wnt/β-catenin/c-myc and cyclin D1 axis, Noxa | [40,113] |
| LINP1 | Cervical cancer (CC) | Upregulated | Augments radioresistance via enhancing dsDNA break repair through the NHEJ pathway | Ku80, DNA-PKcs | [64] |
| Triple negative breast cancer (TNBC) | [63] | ||||
| LUCAT1 | Breast cancer (BC) | Upregulated | LUCAT1 knockdown results in enhanced radiosensitivity of BC cells through the miR-181a-5p/KLF6/KLF15 axis (reduced cell proliferation, migration, viability, and invasion) | miR-181a-5p/KLF6/ KLF15 | [114] |
| MAGI2-AS3 | Esophageal squamous cell carcinoma (ESCC) | Downregulated | MAGI2-AS3 silencing strengthens resistance of ESCC cells to IR in vivo | HOXB7/EZH2/ miR-374b-5p/ CCDC19/miR-15b-5p | [43] |
| MALAT1 | Colorectal cancer (CRC) | Upregulated | Silencing may increase radiosensitivity through the YAP1/AKT axis | ANKHD1/YAP1/AKT | [58] |
| Nasopharyngeal carcinoma (NPC) | Downregulation strengthens IR effects | miR-1/slug | [69] | ||
| High-risk HPV-positive cervical cancer (HR-HPV+ CC) | Increases radioresistance through negatively regulating miR-145 | miR-145 | [32] | ||
| MEG3 | Thyroid carcinoma (TC) | Downregulated | MEG3 overexpression results in higher TC cells radiosensitivity (inhibited proliferation, promoted apoptosis, and DNA damage) through miR-182 sponging | miR-182 | [115] |
| NEAT1 | Colorectal cancer (CRC) | Upregulated; increases after IR in time-dependent manner | Augments radioresistance by promoting IR-induced pyroptosis | miR-448/GSDME | [46] |
| Triple negative breast cancer (TNBC) | Upregulated | Knockdown improves cell sensitivity to radiation via positive regulation of NQO1 | NQO1/miR-218 | [70,92] | |
| Nasopharyngeal carcinoma (NPC) | Upregulated | NEAT1 downregulation sensitizes NPC cells to radiation | miR-204/ZEB1 | [116] | |
| Hepatocellular carcinoma (HCC) | Upregulated | NEAT1_2 down-regulation enhances radiosensitivity of HCC cells | miR-101-3p/WEE1 | [117] | |
| NKILA | Laryngeal carcinoma (LC) | Downregulated | Overexpression of NKILA reduces radioresistance of LC cells by inhibiting p65 nuclear translocation (suppresses cell viability, DNA synthesis capability, and migration ability) | p65 | [118] |
| OIP5-AS1 | Colorectal cancer (CRC) | Downregulated | Overexpressed OIP5-AS1 impedes cell viability, promotes radio-induced apoptosis, and enhances radiosensitivity of CRC cells through the miR-369-3p/DYRK1A axis | miR-369-3p/DYRK1A | [119] |
| PCAT1 | Non-small cell lung cancer (NSCLC) | Upregulated | Inhibition of PCAT1/SOX2 together with radiation promotes IR-induced anti-tumor immune responses | SOX2/cGAS/STING | [120] |
| Cervical cancer (CC) | Upregulated | PCAT1 knockdown enhances radiosensitivity of CC cells through regulating the miR-128/GOLM1 axis (inhibited cell proliferation, migration, and invasion) | miR-128/GOLM1 | [121] | |
| PCAT6 | Breast cancer (BC) | Upregulated | PCAT6 knockdown results in enhanced radiosensitivity (reduced cell proliferation, promoted apoptosis) | TPD52/miR-185-5p | [92] |
| PTPRG-AS1 | Non-small cell lung cancer (NSCLC) | Upregulated | Diminishes radiotherapy efficacy by modulating miR-200c-3p/TCF4 | miR-200c-3p/TCF4 | [122] |
| Nasopharyngeal carcinoma (NPC) | Silencing leads to significant improvement of radiosensitivity | miR-194-3p/PRC1 | [73] | ||
| PTENP1 | Head and neck squamous cell carcinoma (HNSCC) | Upregulated after IR | Inhibition of miR-21 causes cell radiosensitivity by increasing the PTEN protein expression in HNSCC | miR-21 | [100] |
| PVT1 | Non-small cell lung cancer (NSCLC) | Upregulated; the highest levels reached under hypoxia | Induced downregulation could weaken radioresistance by reducing hypoxia | HIF1α/miR-199a-5p | [53] |
| Nasopharyngeal carcinoma (NPC) | Upregulated | Induces radioresistance; knockdown of PVT1 enhances the radiosensitivity of NPC cell lines | No data | [42] | |
| RBM5-AS1 | Medulloblastoma (MB) | Upregulated; the highest levels in radioresistant medulloblastoma cells | Promotes radioresistance and cancer stemness | SIRT6 | [68] |
| SBF2-AS1 | Non-small cell lung cancer (NSCLC) | Upregulated | Downregulation of SBF2-AS1 enhances NSCLC cells’ radiosensitivity through the miR-302a/MBNL3 axis (inhibited cell proliferation, enhanced apoptosis, reduced tumor growth in mice) | miR-302a/MBNL3 | [123] |
| SLC25A21-AS1 | Gastric cancer (GC) | Downregulated | Overexpression of SLC25A21-AS1 enhances the radiosensitivity and inhibits the malignant behaviors of GC cells by upregulating the miR-15a-5p/SNCG axis | miR-15a-5p/SNCG | [124] |
| SNHG5 | Head and neck squamous cell carcinoma (HNSCC) | Downregulated after IR | No data | LPP/ABI2/HELZ/ RANBP2/CEP250/ CDK6/HERC1/PHC3/ MYO5A/FZD3/CFLAR/FAT3/FAT1/FRK/ SMAD2/BMPR2/PTEN/ZMAT3/MDM4/FAT2/APC/FER | [100] |
| SNHG7 | Thyroid carcinoma (TC) | Upregulated | Induced depletion may diminish radioactive iodine resistance through the PI3K/Akt pathway | miR-9-5p/DPP4/PI3K/ Akt | [25] |
| SNHG18 | Glioma | Upregulated | Potentiates radioresistance via modulating levels of DNA damage response proteins | Sema5A | [65] |
| TP53TG1 | Glioma | Upregulated | Induced downregulation could lead to radiation-mediated cancer growth inhibition | miR-524-5p/RAB5A | [48] |
| TP73-AS1 | Hepatocellular carcinoma (HCC) | Upregulated | TP73-AS1 knockdown results in radiosensitivity enhancement of HCC cells through the PTEN/Akt signaling pathway (reduced proliferation, reduced colony formation ability, and induced apoptosis) | PTEN | [125] |
| TTN-AS1 | Large intestine cancer | Upregulated | TTN-AS1 knockdown resulted in radiosensitivity enhancement in large intestine cancer cells | miR-134-5p/PAK3 | [126] |
| UCA1 | Prostate cancer (PCa) | Upregulated | UCA1 knockdown enhances radiosensitivity of PCa cells | miR-331-3p/EIF4G1 | [127] |
| XIST | Neuroblastoma (NB) | Upregulated | Higher XIST expression is correlated with higher NB cells’ radioresistance; its silencing results in inhibition of cell cycle progression, cell proliferation, colony formation, and enhanced post-IR apoptosis rate; it regulates IT through the miR-375/L1CAM axis | miR-375/L1CAM | [128] |
4. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cabrera-Licona, A.; Pérez-Añorve, I.X.; Flores-Fortis, M.; Moral-Hernández, O.D.; González-de la Rosa, C.H.; Suárez-Sánchez, R.; Chávez-Saldaña, M.; Aréchaga-Ocampo, E. Deciphering the epigenetic network in cancer radioresistance. Radiother. Oncol. 2021, 159, 48–59. [Google Scholar] [CrossRef]
- Smits, K.M.; Melotte, V.; Niessen, H.E.; Dubois, L.; Oberije, C.; Troost, E.G.; Starmans, M.H.; Boutros, P.C.; Vooijs, M.; van Engeland, M.; et al. Epigenetics in radiotherapy: Where are we heading? Radiother. Oncol. 2014, 111, 168–177. [Google Scholar] [CrossRef]
- Story, M.D.; Durante, M. Radiogenomics. Med. Phys. 2018, 45, e1111–e1122. [Google Scholar] [CrossRef] [PubMed]
- Ouellette, M.M.; Zhou, S.; Yan, Y. Cell Signaling Pathways That Promote Radioresistance of Cancer Cells. Diagnostics 2022, 12, 656. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Han, Y.; Jin, Y.; He, Q.; Wang, Z. The Advances in Epigenetics for Cancer Radiotherapy. Int. J. Mol. Sci. 2022, 23, 5654. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.J.; Tay, Y. Noncoding RNA:RNA Regulatory Networks in Cancer. Int. J. Mol. Sci. 2018, 19, 1310. [Google Scholar] [CrossRef]
- Podralska, M.; Ciesielska, S.; Kluiver, J.; van den Berg, A.; Dzikiewicz-Krawczyk, A.; Slezak-Prochazka, I. Non-Coding RNAs in Cancer Radiosensitivity: MicroRNAs and lncRNAs as Regulators of Radiation-Induced Signaling Pathways. Cancers 2020, 12, 1662. [Google Scholar] [CrossRef]
- Schofield, P.N.; Kulka, U.; Tapio, S.; Grosche, B. Big data in radiation biology and epidemiology; an overview of the historical and contemporary landscape of data and biomaterial archives. Int. J. Radiat. Biol. 2019, 95, 861–878. [Google Scholar] [CrossRef]
- De Mey, S.; Dufait, I.; De Ridder, M. Radioresistance of Human Cancers: Clinical Implications of Genetic Expression Signatures. Front. Oncol. 2021, 11, 761901. [Google Scholar] [CrossRef]
- Coates, J.; Souhami, L.; El Naqa, I. Big Data Analytics for Prostate Radiotherapy. Front. Oncol. 2016, 6, 149. [Google Scholar] [CrossRef][Green Version]
- Kozłowska, J.; Kolenda, T.; Poter, P.; Sobocińska, J.; Guglas, K.; Stasiak, M.; Bliźniak, R.; Teresiak, A.; Lamperska, K. Long Intergenic Non-Coding RNAs in HNSCC: From “Junk DNA” to Important Prognostic Factor. Cancers 2021, 13, 2949. [Google Scholar] [CrossRef] [PubMed]
- Stasiak, M.; Kolenda, T.; Kozłowska-Masłoń, J.; Sobocińska, J.; Poter, P.; Guglas, K.; Paszkowska, A.; Bliźniak, R.; Teresiak, A.; Kazimierczak, U.; et al. The World of Pseudogenes: New Diagnostic and Therapeutic Targets in Cancers or Still Mystery Molecules? Life 2021, 11, 1354. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.L. Linking Long Noncoding RNA Localization and Function. Trends Biochem. Sci. 2016, 41, 761–772. [Google Scholar] [CrossRef]
- Ransohoff, J.D.; Wei, Y.; Khavari, P.A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2018, 19, 143–157. [Google Scholar] [CrossRef]
- Guglas, K.; Bogaczyńska, M.; Kolenda, T.; Ryś, M.; Teresiak, A.; Bliźniak, R.; Łasińska, I.; Mackiewicz, J.; Lamperska, K. lncRNA in HNSCC: Challenges and potential. Contemp. Oncol. 2017, 21, 259–266. [Google Scholar] [CrossRef]
- Kelley, D.; Rinn, J. Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biol. 2012, 13, R107. [Google Scholar] [CrossRef]
- Hoffmann, M.J.; Dehn, J.; Droop, J.; Niegisch, G.; Niedworok, C.; Szarvas, T.; Schulz, W.A. Truncated Isoforms of lncRNA ANRIL Are Overexpressed in Bladder Cancer, But Do Not Contribute to Repression of INK4 Tumor Suppressors. Noncoding RNA 2015, 1, 266–284. [Google Scholar] [CrossRef]
- Smith, K.N.; Miller, S.C.; Varani, G.; Calabrese, J.M.; Magnuson, T. Multimodal Long Noncoding RNA Interaction Networks: Control Panels for Cell Fate Specification. Genetics 2019, 213, 1093–1110. [Google Scholar] [CrossRef]
- Li, Y.; Ge, C.; Feng, G.; Xiao, H.; Dong, J.; Zhu, C.; Jiang, M.; Cui, M.; Fan, S. Low dose irradiation facilitates hepatocellular carcinoma genesis involving HULC. Mol. Carcinog. 2018, 57, 926–935. [Google Scholar] [CrossRef]
- Aryankalayil, M.J.; Chopra, S.; Levin, J.; Eke, I.; Makinde, A.; Das, S.; Shankavaram, U.; Vanpouille-Box, C.; Demaria, S.; Coleman, C.N. Radiation-Induced Long Noncoding RNAs in a Mouse Model after Whole-Body Irradiation. Radiat. Res. 2018, 189, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Wang, B.; Wang, H.; Meng, L.; Zhao, Q.; Li, X.; Xin, Y.; Jiang, X. Radiation-Induced Normal Tissue Damage: Oxidative Stress and Epigenetic Mechanisms. Oxid. Med. Cell. Longev. 2019, 2019, 3010342. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Chen, S.; Yang, B.; Mao, W.; Yang, X.; Cai, J. Molecular mechanisms of lncRNAs in regulating cancer cell radiosensitivity. Biosci. Rep. 2019, 39, BSR20190590. [Google Scholar] [CrossRef]
- Zhou, J.M.; Liang, R.; Zhu, S.Y.; Wang, H.; Zou, M.; Zou, W.J.; Nie, S.L. LncRNA WWC2-AS1 functions AS a novel competing endogenous RNA in the regulation of FGF2 expression by sponging miR-16 in radiation-induced intestinal fibrosis. BMC Cancer 2019, 19, 647. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Yu, J.; Xie, R.; Zhou, T.; Xiong, C.; Zhang, S.; Zhong, M. Roles of the SNHG7/microRNA-9-5p/DPP4 ceRNA network in the growth and 131I resistance of thyroid carcinoma cells through PI3K/Akt activation. Oncol. Rep. 2021, 45, 3. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Wang, L.; Yang, H.; Ding, N. Knockdown long non-coding RNA HCP5 enhances the radiosensitivity of esophageal carcinoma by modulating AKT signaling activation. Bioengineered 2022, 13, 884–893. [Google Scholar] [CrossRef]
- Wang, Y.Q.; Huang, G.; Chen, J.; Cao, H.; Xu, W.T. LncRNA SNHG6 promotes breast cancer progression and epithelial-mesenchymal transition via miR-543/LAMC1 axis. Breast Cancer Res. Treat. 2021, 188, 1–14. [Google Scholar] [CrossRef]
- Yang, W.; Li, X.; Zhao, L.; Zhao, F. Reversal of Radiotherapy Resistance of Ovarian Cancer Cell Strain CAOV3/R by Targeting lncRNA CRNDE. J. Healthc. Eng. 2021, 2021, 8556965. [Google Scholar] [CrossRef]
- Fotouhi Ghiam, A.; Taeb, S.; Huang, X.; Huang, V.; Ray, J.; Scarcello, S.; Hoey, C.; Jahangiri, S.; Fokas, E.; Loblaw, A.; et al. Long non-coding RNA urothelial carcinoma associated 1 (UCA1) mediates radiation response in prostate cancer. Oncotarget 2017, 8, 4668–4689. [Google Scholar] [CrossRef]
- Yang, J.; Hao, T.; Sun, J.; Wei, P.; Zhang, H. Long noncoding RNA GAS5 modulates α-Solanine-induced radiosensitivity by negatively regulating miR-18a in human prostate cancer cells. Biomed. Pharmacother. 2019, 112, 108656. [Google Scholar] [CrossRef]
- Ma, Y.; Yu, L.; Yan, W.; Qiu, L.; Zhang, J.; Jia, X. lncRNA GAS5 Sensitizes Breast Cancer Cells to Ionizing Radiation by Inhibiting DNA Repair. Biomed. Res. Int. 2022, 2022, 1987519. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; He, Y.; Lin, L.; Qi, Z.; Ma, L.; Li, L.; Su, Y. Long non-coding RNA MALAT1 modulates radiosensitivity of HR-HPV+ cervical cancer via sponging miR-145. Tumour Biol. 2016, 37, 1683–1691. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z. LncRNA CCAT1 downregulation increases the radiosensitivity of non-small cell lung cancer cells. Kaohsiung J. Med. Sci. 2021, 37, 654–663. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ji, X.; Wang, H. Targeting Long Noncoding RNA HMMR-AS1 Suppresses and Radiosensitizes Glioblastoma. Neoplasia 2018, 20, 456–466. [Google Scholar] [CrossRef]
- Liu, W.H.; Qiao, H.Y.; Xu, J.; Wang, W.Q.; Wu, Y.L.; Wu, X. LINC00473 contributes to the radioresistance of esophageal squamous cell carcinoma by regulating microRNA-497-5p and cell division cycle 25A. Int. J. Mol. Med. 2020, 46, 571–582. [Google Scholar] [CrossRef]
- Chen, W.; Zhang, Y.; Wang, H.; Pan, T.; Zhang, Y.; Li, C. LINC00473/miR-374a-5p regulates esophageal squamous cell carcinoma via targeting SPIN1 to weaken the effect of radiotherapy. J. Cell. Biochem. 2019, 120, 14562–14572. [Google Scholar] [CrossRef]
- Tang, T.; Shan, G.; Zeng, F. Knockdown of DGCR5 enhances the radiosensitivity of human laryngeal carcinoma cells via inducing miR-195. J. Cell. Physiol. 2019, 234, 12918–12925. [Google Scholar] [CrossRef]
- Liu, F.; Huang, W.; Hong, J.; Cai, C.; Zhang, W.; Zhang, J.; Kang, Z. Long noncoding RNA LINC00630 promotes radio-resistance by regulating BEX1 gene methylation in colorectal cancer cells. IUBMB Life 2020, 72, 1404–1414. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, X.; Chen, X.; Liu, J.; Gu, H.; Fan, R.; Ge, H. Long non-coding RNA HOTAIR knockdown enhances radiosensitivity through regulating microRNA-93/ATG12 axis in colorectal cancer. Cell Death Dis. 2020, 11, 175. [Google Scholar] [CrossRef]
- Chen, L.; Yuan, D.; Yang, Y.; Ren, M. LincRNA-p21 enhances the sensitivity of radiotherapy for gastric cancer by targeting the β-catenin signaling pathway. J. Cell. Biochem. 2019, 120, 6178–6187. [Google Scholar] [CrossRef]
- Lin, J.; Liu, Z.; Liao, S.; Li, E.; Wu, X.; Zeng, W. Elevation of long non-coding RNA GAS5 and knockdown of microRNA-21 up-regulate RECK expression to enhance esophageal squamous cell carcinoma cell radio-sensitivity after radiotherapy. Genomics 2020, 112, 2173–2185. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Jing, Y.; Wei, F.; Tang, Y.; Yang, L.; Luo, J.; Yang, P.; Ni, Q.; Pang, J.; Liao, Q.; et al. Long non-coding RNA PVT1 predicts poor prognosis and induces radioresistance by regulating DNA repair and cell apoptosis in nasopharyngeal carcinoma. Cell Death Dis. 2018, 9, 235. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.; Shi, X.; Lin, M.; Yao, Q.; Ma, J.; Li, J. LncRNA MAGI2-AS3 Overexpression Sensitizes Esophageal Cancer Cells to Irradiation Through Down-Regulation of HOXB7 via EZH2. Front. Cell Dev. Biol. 2020, 8, 552822. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Shen, C.; Zhang, Y.; Hu, C. LncRNA ANRIL negatively regulated chitooligosaccharide-induced radiosensitivity in colon cancer cells by sponging miR-181a-5p. Adv. Clin. Exp. Med. 2021, 30, 55–65. [Google Scholar] [CrossRef]
- He, P.; Xu, Y.Q.; Wang, Z.J.; Sheng, B. LncRNA LINC00210 regulated radiosensitivity of osteosarcoma cells via miR-342-3p/GFRA1 axis. J. Clin. Lab. Anal. 2020, 34, e23540. [Google Scholar] [CrossRef]
- Su, F.; Duan, J.; Zhu, J.; Fu, H.; Zheng, X.; Ge, C. Long non-coding RNA nuclear paraspeckle assembly transcript 1 regulates ionizing radiation-induced pyroptosis via microRNA-448/gasdermin E in colorectal cancer cells. Int. J. Oncol. 2021, 59, 79. [Google Scholar] [CrossRef]
- Jiang, A.; Liu, N.; Bai, S.; Wang, J.; Gao, H.; Zheng, X.; Fu, X.; Ren, M.; Zhang, X.; Tian, T.; et al. Identification and validation of an autophagy-related long non-coding RNA signature as a prognostic biomarker for patients with lung adenocarcinoma. J. Thorac. Dis. 2021, 13, 720–734. [Google Scholar] [CrossRef]
- Gao, W.; Qiao, M.; Luo, K. Long Noncoding RNA TP53TG1 Contributes to Radioresistance of Glioma Cells Via miR-524-5p/RAB5A Axis. Cancer Biother. Radiopharm. 2021, 36, 600–612. [Google Scholar] [CrossRef]
- Zheng, J.; Wang, B.; Zheng, R.; Zhang, J.; Huang, C.; Zheng, R.; Huang, Z.; Qiu, W.; Liu, M.; Yang, K.; et al. Linc-RA1 inhibits autophagy and promotes radioresistance by preventing H2Bub1/USP44 combination in glioma cells. Cell Death Dis. 2020, 11, 758. [Google Scholar] [CrossRef]
- Chen, C.; Wang, K.; Wang, Q.; Wang, X. LncRNA HULC mediates radioresistance via autophagy in prostate cancer cells. Braz. J. Med. Biol. Res. 2018, 51, e7080. [Google Scholar] [CrossRef]
- Liu, H.; Zheng, W.; Chen, Q.; Zhou, Y.; Pan, Y.; Zhang, J.; Bai, Y.; Shao, C. lncRNA CASC19 Contributes to Radioresistance of Nasopharyngeal Carcinoma by Promoting Autophagy via AMPK-mTOR Pathway. Int. J. Mol. Sci. 2021, 22, 1407. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zhu, L.; Xue, Q.; Tang, C.; Tang, W.; Zhang, N.; Dai, C.; Chen, Z. Novel lncRNA AL033381.2 Promotes Hepatocellular Carcinoma Progression by Upregulating PRKRA Expression. Oxid. Med. Cell. Longev. 2022, 2022, 1125932. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Han, C.; Zhang, Y.; Liu, F. LncRNA PVT1 regulate expression of HIF1α via functioning as ceRNA for miR-199a-5p in non-small cell lung cancer under hypoxia. Mol. Med. Rep. 2018, 17, 1105–1110. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Meng, D.D.; Gao, L.; Xu, Y.; Liu, P.J.; Tian, Y.W.; Yi, Z.Y.; Zhang, Y.; Tie, X.J.; Xu, Z.Q. Overexpression of HOTAIR leads to radioresistance of human cervical cancer via promoting HIF-1α expression. Radiat. Oncol. 2018, 13, 210. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, X.; Rong, Z.; Dai, L.; Qin, C.; Wang, S.; Geng, W. LncRNA LINC01134 Contributes to Radioresistance in Hepatocellular Carcinoma by Regulating DNA Damage Response viaMAPK Signaling Pathway. Front. Pharmacol. 2022, 12, 791889. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.H.; Guo, Z.; An, L.; Zhou, Y.; Xu, H.; Xiong, J.; Liu, Z.Q.; Chen, X.P.; Zhou, H.H.; Li, X.; et al. LINC-PINT impedes DNA repair and enhances radiotherapeutic response by targeting DNA-PKcs in nasopharyngeal cancer. Cell Death Dis. 2021, 12, 454. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Wang, Y.H.; Xu, H.; Yuan, C.S.; Zhou, H.H.; Huang, W.H.; Wang, H.; Zhang, W. LncRNA linc00312 suppresses radiotherapy resistance by targeting DNA-PKcs and impairing DNA damage repair in nasopharyngeal carcinoma. Cell Death Dis. 2021, 12, 69. [Google Scholar] [CrossRef]
- Yao, P.A.; Wu, Y.; Zhao, K.; Li, Y.; Cao, J.; Xing, C. The feedback loop of ANKHD1/lncRNA MALAT1/YAP1 strengthens the radioresistance of CRC by activating YAP1/AKT signaling. Cell Death Dis. 2022, 13, 103. [Google Scholar] [CrossRef]
- Feng, J.; Li, Y.; Zhu, L.; Zhao, Q.; Li, D.; Li, Y.; Wu, T. STAT1 mediated long non-coding RNA LINC00504 influences radio-sensitivity of breast cancer via binding to TAF15 and stabilizing CPEB2 expression. Cancer Biol. Ther. 2021, 22, 630–639. [Google Scholar] [CrossRef]
- Wang, B.; Zheng, J.; Li, R.; Tian, Y.; Lin, J.; Liang, Y.; Sun, Q.; Xu, A.; Zheng, R.; Liu, M.; et al. Long noncoding RNA LINC02582 acts downstream of miR-200c to promote radioresistance through CHK1 in breast cancer cells. Cell Death Dis. 2019, 10, 764. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, B.; Xiao, H.; Dong, J.; Li, Y.; Zhu, C.; Jin, Y.; Li, H.; Cui, M.; Fan, S. LncRNA HOTAIR enhances breast cancer radioresistance through facilitating HSPA1A expression via sequestering miR-449b-5p. Thorac. Cancer 2020, 11, 1801–1816. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Fei, Q.; Zhang, H.; Qiu, M.; Zhang, B.; Wang, Q.; Yu, Y.; Guo, C.; Ren, Y.; Mei, M.; et al. lncRNA HOTAIR Promotes DNA Repair and Radioresistance of Breast Cancer via EZH2. DNA Cell Biol. 2020, 39, 2166–2173. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; He, Q.; Hu, Z.; Feng, Y.; Fan, L.; Tang, Z.; Yuan, J.; Shan, W.; Li, C.; Hu, X.; et al. Long noncoding RNA LINP1 regulates repair of DNA double-strand breaks in triple-negative breast cancer. Nat. Struct Mol. Biol. 2016, 23, 522–530. [Google Scholar] [CrossRef]
- Wang, X.; Liu, H.; Shi, L.; Yu, X.; Gu, Y.; Sun, X. LINP1 facilitates DNA damage repair through non-homologous end joining (NHEJ) pathway and subsequently decreases the sensitivity of cervical cancer cells to ionizing radiation. Cell Cycle 2018, 17, 439–447. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.; Yao, Q.; Ren, C.; Liu, Y.; Yang, H.; Xie, G.; Du, S.; Yang, K.; Yuan, Y. Upregulation of Long Noncoding RNA Small Nucleolar RNA Host Gene 18 Promotes Radioresistance of Glioma by Repressing Semaphorin 5A. Int. J. Radiat. Oncol. Biol. Phys. 2016, 96, 877–887. [Google Scholar] [CrossRef]
- Zhang, H.; Hua, Y.; Jiang, Z.; Yue, J.; Shi, M.; Zhen, X.; Zhang, X.; Yang, L.; Zhou, R.; Wu, S. Cancer-associated Fibroblast-promoted LncRNA DNM3OS Confers Radioresistance by Regulating DNA Damage Response in Esophageal Squamous Cell Carcinoma. Clin. Cancer Res. 2019, 25, 1989–2000. [Google Scholar] [CrossRef]
- Jiang, G.; Yu, H.; Li, Z.; Zhang, F. lncRNA cytoskeleton regulator reduces non-small cell lung cancer radiosensitivity by downregulating miRNA-206 and activating prothymosin α. Int. J. Oncol. 2021, 59, 88. [Google Scholar] [CrossRef]
- Zhu, C.; Li, K.; Jiang, M.; Chen, S. RBM5-AS1 promotes radioresistance in medulloblastoma through stabilization of SIRT6 protein. Acta Neuropathol. Commun. 2021, 9, 123. [Google Scholar] [CrossRef]
- Jin, C.; Yan, B.; Lu, Q.; Lin, Y.; Ma, L. The role of MALAT1/miR-1/slug axis on radioresistance in nasopharyngeal carcinoma. Tumour Biol. 2016, 37, 4025–4033. [Google Scholar] [CrossRef]
- Lin, L.C.; Lee, H.T.; Chien, P.J.; Huang, Y.H.; Chang, M.Y.; Lee, Y.C.; Chang, W.W. NAD(P)H:quinone oxidoreductase 1 determines radiosensitivity of triple negative breast cancer cells and is controlled by long non-coding RNA NEAT1. Int. J. Med. Sci. 2020, 17, 2214–2224. [Google Scholar] [CrossRef]
- Ye, H.; Zhou, Q.; Zheng, S.; Li, G.; Lin, Q.; Ye, L.; Wang, Y.; Wei, L.; Zhao, X.; Li, W.; et al. FEZF1-AS1/miR-107/ZNF312B axis facilitates progression and Warburg effect in pancreatic ductal adenocarcinoma. Cell Death Dis. 2018, 9, 34. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; He, D.; Xiao, M.; Zhu, Y.; Zhou, J.; Cao, K. Long noncoding RNA LINC00518 induces radioresistance by regulating glycolysis through an miR-33a-3p/HIF-1α negative feedback loop in melanoma. Cell Death Dis. 2021, 12, 245. [Google Scholar] [CrossRef] [PubMed]
- Yi, L.; Ouyang, L.; Wang, S.; Li, S.S.; Yang, X.M. Long noncoding RNA PTPRG-AS1 acts as a microRNA-194-3p sponge to regulate radiosensitivity and metastasis of nasopharyngeal carcinoma cells via PRC1. J Cell Physiol. 2019, 234, 19088–19102. [Google Scholar] [CrossRef]
- Zhang, J.; Li, W. Long noncoding RNA CYTOR sponges miR-195 to modulate proliferation, migration, invasion and radiosensitivity in nonsmall cell lung cancer cells. Biosci. Rep. 2018, 38, BSR20181599. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.D.; Xu, H.T.; Xu, X.H.; Ru, G.; Liu, W.; Zhu, J.J.; Wu, Y.Y.; Zhao, K.; Wu, Y.; Xing, C.G.; et al. Knockdown of long non-coding RNA HOTAIR inhibits proliferation and invasiveness and improves radiosensitivity in colorectal cancer. Oncol. Rep. 2016, 35, 479–487. [Google Scholar] [CrossRef]
- Jing, L.; Yuan, W.; Ruofan, D.; Jinjin, Y.; Haifeng, Q. HOTAIR enhanced aggressive biological behaviors and induced radio-resistance via inhibiting p21 in cervical cancer. Tumour Biol. 2015, 36, 3611–3619. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, Z.; Zhang, G.; Xue, L.; Yang, H.; Luo, Y.; Zheng, X.; Zhang, Y.; Yuan, Y.; Lei, R.; et al. A three-lncRNA signature of pretreatment biopsies predicts pathological response and outcome in esophageal squamous cell carcinoma with neoadjuvant chemoradiotherapy. Clin. Transl. Med. 2020, 10, e156. [Google Scholar] [CrossRef]
- Li, Z.Y.; Li, H.F.; Zhang, Y.Y.; Zhang, X.L.; Wang, B.; Liu, J.T. Value of long non-coding RNA Rpph1 in esophageal cancer and its effect on cancer cell sensitivity to radiotherapy. World J. Gastroenterol. 2020, 26, 1775–1791. [Google Scholar] [CrossRef]
- Eissa, S.; Matboli, M.; Essawy, N.O.; Shehta, M.; Kotb, Y.M. Rapid detection of urinary long non-coding RNA urothelial carcinoma associated one using a PCR-free nanoparticle-based assay. Biomarkers 2015, 20, 212–217. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, L.; Wang, Y.X.; Xi, M.; Liu, S.L.; Luo, L.L. Long non-coding RNA HOTTIP is correlated with progression and prognosis in tongue squamous cell carcinoma. Tumour Biol. 2015, 36, 8805–8809. [Google Scholar] [CrossRef]
- Gezer, U.; Özgür, E.; Cetinkaya, M.; Isin, M.; Dalay, N. Long non-coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell Biol. Int. 2014, 38, 1076–1079. [Google Scholar] [CrossRef] [PubMed]
- Fayda, M.; Isin, M.; Tambas, M.; Guveli, M.; Meral, R.; Altun, M.; Sahin, D.; Ozkan, G.; Sanli, Y.; Isin, H.; et al. Do circulating long non-coding RNAs (lncRNAs) (LincRNA-p21, GAS 5, HOTAIR) predict the treatment response in patients with head and neck cancer treated with chemoradiotherapy? Tumour Biol. 2016, 37, 3969–3978. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Wu, Z.; Zhang, J.; Su, B. Salivary lncRNA as a potential marker for oral squamous cell carcinoma diagnosis. Mol. Med. Rep. 2013, 7, 761–766. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Yin, C.; Dang, Y.; Ye, F.; Zhang, G. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci. Rep. 2015, 5, 11516. [Google Scholar] [CrossRef]
- Fang, Z.; Zhang, S.; Wang, Y.; Shen, S.; Wang, F.; Hao, Y.; Li, Y.; Zhang, B.; Zhou, Y.; Yang, H.; et al. Long non-coding RNA MALAT-1 modulates metastatic potential of tongue squamous cell carcinomas partially through the regulation of small proline rich proteins. BMC Cancer 2016, 16, 706. [Google Scholar] [CrossRef]
- Feng, Y.; Hu, X.; Zhang, Y.; Zhang, D.; Li, C.; Zhang, L. Methods for the study of long noncoding RNA in cancer cell signaling. Methods Mol. Biol. 2014, 1165, 115–143. [Google Scholar]
- Wojdacz, T.K.; Dobrovic, A.; Algar, E.M. Rapid detection of methylation change at H19 in human imprinting disorders using methylation-sensitive high-resolution melting. Hum. Mutat. 2008, 29, 1255–1260. [Google Scholar] [CrossRef]
- Dodd, D.W.; Gagnon, K.T.; Corey, D.R. Digital quantitation of potential therapeutic target RNAs. Nucleic Acid Ther. 2013, 23, 188–194. [Google Scholar] [CrossRef]
- Chen, H.H.W.; Kuo, M.T. Improving radiotherapy in cancer treatment: Promises and challenges. Oncotarget 2017, 8, 62742–62758. [Google Scholar] [CrossRef]
- Ree, A.H.; Redalen, K.R. Personalized radiotherapy: Concepts, biomarkers and trial design. Br. J. Radiol. 2015, 88, 20150009. [Google Scholar] [CrossRef]
- Niemantsverdriet, M.; van Goethem, M.J.; Bron, R.; Hogewerf, W.; Brandenburg, S.; Langendijk, J.A.; van Luijk, P.; Coppes, R.P. High and low LET radiation differentially induce normal tissue damage signals. Int. J. Radiat. Oncol. Biol. Phys. 2012, 83, 1291–1297. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, Z.; Wang, F.; Zhu, Y.; Guo, T.; Lin, M. Long Noncoding RNAs Regulate the Radioresistance of Breast Cancer. Anal. Cell. Pathol. 2021, 2021, 9005073. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Sang, Y.; Chen, D.; Wu, X.; Wang, X.; Yang, W.; Chen, Y. M2 macrophage-derived exosomal long non-coding RNA AGAP2-AS1 enhances radiotherapy immunity in lung cancer by reducing microRNA-296 and elevating NOTCH2. Cell Death Dis. 2021, 12, 467. [Google Scholar] [CrossRef] [PubMed]
- Liao, K.; Ma, X.; Chen, B.; Lu, X.; Hu, Y.; Lin, Y.; Huang, R.; Qiu, Y. Upregulated AHIF-mediated radioresistance in glioblastoma. Biochem. Biophys. Res. Commun. 2019, 509, 617–623. [Google Scholar] [CrossRef] [PubMed]
- Gou, C.; Han, P.; Li, J.; Gao, L.; Ji, X.; Dong, F.; Su, Q.; Zhang, Y.; Liu, X. Knockdown of lncRNA BLACAT1 enhances radiosensitivity of head and neck squamous cell carcinoma cells by regulating PSEN1. Br. J. Radiol. 2020, 93, 20190154. [Google Scholar] [CrossRef]
- Ding, Z.; Kang, J.; Yang, Y. Long non-coding RNA CASC2 enhances irradiation-induced endoplasmic reticulum stress in NSCLC cells through PERK signaling. 3 Biotech 2020, 10, 449. [Google Scholar] [CrossRef]
- Tao, L.; Tian, P.; Yang, L.; Guo, X. lncRNA CASC2 Enhances Sensitivity in Papillary Thyroid Cancer by Sponging miR-155. Biomed. Res. Int. 2020, 2020, 7183629. [Google Scholar] [CrossRef]
- Lai, Y.; Chen, Y.; Lin, Y.; Ye, L. Down-regulation of LncRNA CCAT1 enhances radiosensitivity via regulating miR-148b in breast cancer. Cell Biol. Int. 2018, 42, 227–236. [Google Scholar] [CrossRef]
- Wang, M.; Wang, L.; He, X.; Zhang, J.; Zhu, Z.; Zhang, M.; Li, X. lncRNA CCAT2 promotes radiotherapy resistance for human esophageal carcinoma cells via the miR-145/p70S6K1 and p53 pathway. Int. J. Oncol. 2020, 56, 327–336. [Google Scholar] [CrossRef]
- Guglas, K.; Kolenda, T.; Teresiak, A.; Kopczyńska, M.; Łasińska, I.; Mackiewicz, J.; Mackiewicz, A.; Lamperska, K. lncRNA Expression after Irradiation and Chemoexposure of HNSCC Cell Lines. Noncoding RNA 2018, 4, 33. [Google Scholar] [CrossRef]
- Li, L.; Lin, X.; Xu, P.; Jiao, Y.; Fu, P. LncRNA GAS5 sponges miR-362-5p to promote sensitivity of thyroid cancer cells to 131 I by upregulating SMG1. IUBMB Life 2020, 72, 2420–2431. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Liu, L.; Li, G.; Cai, M.; Tan, C.; Han, X.; Han, L. LncRNA GAS5 confers the radio sensitivity of cervical cancer cells via regulating miR-106b/IER3 axis. Int. J. Biol. Macromol. 2019, 126, 994–1001. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Ni, T.; Jiang, Y.; Li, Y. Long Noncoding RNA GAS5 Inhibits Tumorigenesis and Enhances Radiosensitivity by Suppressing miR-135b Expression in Non-Small Cell Lung Cancer. Oncol. Res. 2017, 25, 1305–1316. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Zhang, X.; Zhan, D.; Li, J.; Li, Z.; Li, H.; Qian, J. LncRNA H19 interacted with miR-130a-3p and miR-17-5p to modify radio-resistance and chemo-sensitivity of cardiac carcinoma cells. Cancer Med. 2019, 8, 1604–1618. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Yuan, L.; Li, W.; Xu, K.; Yang, L. The LncRNA H19/miR-193a-3p axis modifies the radio-resistance and chemotherapeutic tolerance of hepatocellular carcinoma cells by targeting PSEN1. J. Cell. Biochem. 2018, 119, 8325–8335. [Google Scholar] [CrossRef]
- Hu, X.; Ding, D.; Zhang, J.; Cui, J. Knockdown of lncRNA HOTAIR sensitizes breast cancer cells to ionizing radiation through activating miR-218. Biosci Rep. 2019, 39, BSR20181038. [Google Scholar] [CrossRef]
- Han, P.B.; Ji, X.J.; Zhang, M.; Gao, L.Y. Upregulation of lncRNA LINC00473 promotes radioresistance of HNSCC cells through activating Wnt/β-catenin signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 7305–7313. [Google Scholar]
- Chen, Y.; Bao, C.; Zhang, X.; Lin, X.; Fu, Y. Knockdown of LINC00511 promotes radiosensitivity of thyroid carcinoma cells via suppressing JAK2/STAT3 signaling pathway. Cancer Biol. Ther. 2019, 20, 1249–1257. [Google Scholar] [CrossRef]
- Zhang, N.; Zeng, X.; Sun, C.; Guo, H.; Wang, T.; Wei, L.; Zhang, Y.; Zhao, J.; Ma, X. LncRNA LINC00963 Promotes Tumorigenesis and Radioresistance in Breast Cancer by Sponging miR-324-3p and Inducing ACK1 Expression. Mol. Ther. Nucleic Acids 2019, 18, 871–881. [Google Scholar] [CrossRef]
- Tian, W.; Zhang, Y.; Liu, H.; Jin, H.; Sun, T. LINC01123 potentially correlates with radioresistance in glioma through the miR-151a/CENPB axis. Neuropathology 2022, 42, 3–15. [Google Scholar] [CrossRef]
- Lin, W.; Huang, Z.; Xu, Y.; Chen, X.; Chen, T.; Ye, Y.; Ding, J.; Chen, Z.; Chen, L.; Qiu, X.; et al. A three-lncRNA signature predicts clinical outcomes in low-grade glioma patients after radiotherapy. Aging 2020, 12, 9188–9204. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Zhang, S.; Sun, Y.; Gu, J.; Ye, Z.; Sun, X.; Tang, Q. A Radioresponse-Related lncRNA Biomarker Signature for Risk Classification and Prognosis Prediction in Non-Small-Cell Lung Cancer. J. Oncol. 2021, 2021, 4338838. [Google Scholar] [CrossRef] [PubMed]
- Ogunwobi, O.O.; Mahmood, F.; Akingboye, A. Biomarkers in Colorectal Cancer: Current Research and Future Prospects. Int. J. Mol. Sci. 2020, 21, 5311. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Cheng, T.; Du, Y.; Hu, X.; Xia, W. LncRNA LUCAT1/miR-181a-5p axis promotes proliferation and invasion of breast cancer via targeting KLF6 and KLF15. BMC Mol. Cell Biol. 2020, 21, 69. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yue, P.; Zhou, T.; Zhang, F.; Wang, H.; Chen, X. LncRNA MEG3 enhances 131I sensitivity in thyroid carcinoma via sponging miR-182. Biomed. Pharmacother. 2018, 105, 1232–1239. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Li, T.; Wei, G.; Liu, L.; Chen, Q.; Xu, L.; Zhang, K.; Zeng, D.; Liao, R. The long non-coding RNA NEAT1 regulates epithelial to mesenchymal transition and radioresistance in through miR-204/ZEB1 axis in nasopharyngeal carcinoma. Tumour Biol. 2016, 37, 11733–11741. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, N. Downregulation of lncRNA NEAT1_2 radiosensitizes hepatocellular carcinoma cells through regulation of miR-101-3p/WEE1 axis. Cell Biol. Int. 2019, 43, 44–55. [Google Scholar] [CrossRef]
- Yang, T.; Li, S.; Liu, J.; Yin, D.; Yang, X.; Tang, Q. lncRNA-NKILA/NF-κB feedback loop modulates laryngeal cancer cell proliferation, invasion, and radioresistance. Cancer Med. 2018, 7, 2048–2063. [Google Scholar] [CrossRef]
- Zou, Y.; Yao, S.; Chen, X.; Liu, D.; Wang, J.; Yuan, X.; Rao, J.; Xiong, H.; Yu, S.; Yuan, X.; et al. LncRNA OIP5-AS1 regulates radioresistance by targeting DYRK1A through miR-369-3p in colorectal cancer cells. Eur. J. Cell Biol. 2018, 97, 369–378. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, N.; Zeng, Z.; Wu, Q.; Jiang, X.; Li, S.; Sun, W.; Zhang, J.; Li, Y.; Li, J.; et al. LncRNA PCAT1 activates SOX2 and suppresses radioimmune responses via regulating cGAS/STING signalling in non-small cell lung cancer. Clin. Transl. Med. 2022, 12, e792. [Google Scholar] [CrossRef]
- Ge, X.; Gu, Y.; Li, D.; Jiang, M.; Zhao, S.; Li, Z.; Liu, S. Knockdown of lncRNA PCAT1 Enhances Radiosensitivity of Cervical Cancer by Regulating miR-128/GOLM1 Axis. OncoTargets Ther. 2020, 13, 10373–10385. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Niu, R.; Huang, W.; Da, L.; Tang, Y.; Jiang, D.; Xi, Y.; Zhang, C. Long Noncoding RNA PTPRG Antisense RNA 1 Reduces Radiosensitivity of Nonsmall Cell Lung Cancer Cells Via Regulating MiR-200c-3p/TCF4. Technol Cancer Res. Treat. 2020, 19, 1533033820942615. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Wang, G.; Zhang, C.; Liu, Y.; Chen, W.; Wang, H.; Liu, H. LncRNA SBF2-AS1 affects the radiosensitivity of non-small cell lung cancer via modulating microRNA-302a/MBNL3 axis. Cell Cycle 2020, 19, 300–316. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Z.; Wang, L. Long Noncoding RNA Solute Carrier Family 25 Member 21 Antisense RNA 1 Inhibits Cell Malignant Behaviors and Enhances Radiosensitivity of Gastric Cancer Cells by Upregulating Synuclein Gamma Expression. Tohoku J. Exp. Med. 2022, 257, 225–239. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Zhang, J.; Xia, Q.; Sun, M. Down-regulated lncRNA TP73-AS1 reduces radioresistance in hepatocellular carcinoma via the PTEN/Akt signaling pathway. Cell Cycle 2019, 18, 3177–3188. [Google Scholar] [CrossRef]
- Zuo, Z.; Ji, S.; He, L.; Zhang, Y.; Peng, Z.; Han, J. LncRNA TTN-AS1/miR-134-5p/PAK3 axis regulates the radiosensitivity of human large intestine cancer cells through the P21 pathway and AKT/GSK-3β/β-catenin pathway. Cell Biol. Int. 2020, 44, 2284–2292. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Yang, J. Down-regulation of lncRNA UCA1 enhances radiosensitivity in prostate cancer by suppressing EIF4G1 expression via sponging miR-331-3p. Cancer Cell Int. 2020, 20, 449. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, X.; Zhao, Y.; Sun, G.; Zhang, J.; Gao, Y.; Liu, Q.; Zhang, W.; Zhu, H. Downregulation of lncRNA XIST Represses Tumor Growth and Boosts Radiosensitivity of Neuroblastoma via Modulation of the miR-375/L1CAM Axis. Neurochem. Res. 2020, 45, 2679–2690. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kozłowska-Masłoń, J.; Guglas, K.; Paszkowska, A.; Kolenda, T.; Podralska, M.; Teresiak, A.; Bliźniak, R.; Lamperska, K. Radio-lncRNAs: Biological Function and Potential Use as Biomarkers for Personalized Oncology. J. Pers. Med. 2022, 12, 1605. https://doi.org/10.3390/jpm12101605
Kozłowska-Masłoń J, Guglas K, Paszkowska A, Kolenda T, Podralska M, Teresiak A, Bliźniak R, Lamperska K. Radio-lncRNAs: Biological Function and Potential Use as Biomarkers for Personalized Oncology. Journal of Personalized Medicine. 2022; 12(10):1605. https://doi.org/10.3390/jpm12101605
Chicago/Turabian StyleKozłowska-Masłoń, Joanna, Kacper Guglas, Anna Paszkowska, Tomasz Kolenda, Marta Podralska, Anna Teresiak, Renata Bliźniak, and Katarzyna Lamperska. 2022. "Radio-lncRNAs: Biological Function and Potential Use as Biomarkers for Personalized Oncology" Journal of Personalized Medicine 12, no. 10: 1605. https://doi.org/10.3390/jpm12101605
APA StyleKozłowska-Masłoń, J., Guglas, K., Paszkowska, A., Kolenda, T., Podralska, M., Teresiak, A., Bliźniak, R., & Lamperska, K. (2022). Radio-lncRNAs: Biological Function and Potential Use as Biomarkers for Personalized Oncology. Journal of Personalized Medicine, 12(10), 1605. https://doi.org/10.3390/jpm12101605







