A Systematic Review on the Contribution of Artificial Intelligence in the Development of Medicines for COVID-2019
Abstract
:1. Introduction
2. Materials and Methods
2.1. Research Question
2.2. Searched Period
2.3. Keywords
2.4. Inclusion and Exclusion Criteria
2.4.1. Inclusion Criteria
2.4.2. Exclusion Criteria
2.5. Searched Databases and Period
2.6. Identification of Previous Systematic Reviews on Related Topics
2.6.1. Findings of the Review of Kaushal et al. (2020)
2.6.2. Findings of the Review of Gurung et al. (2021)
3. Results
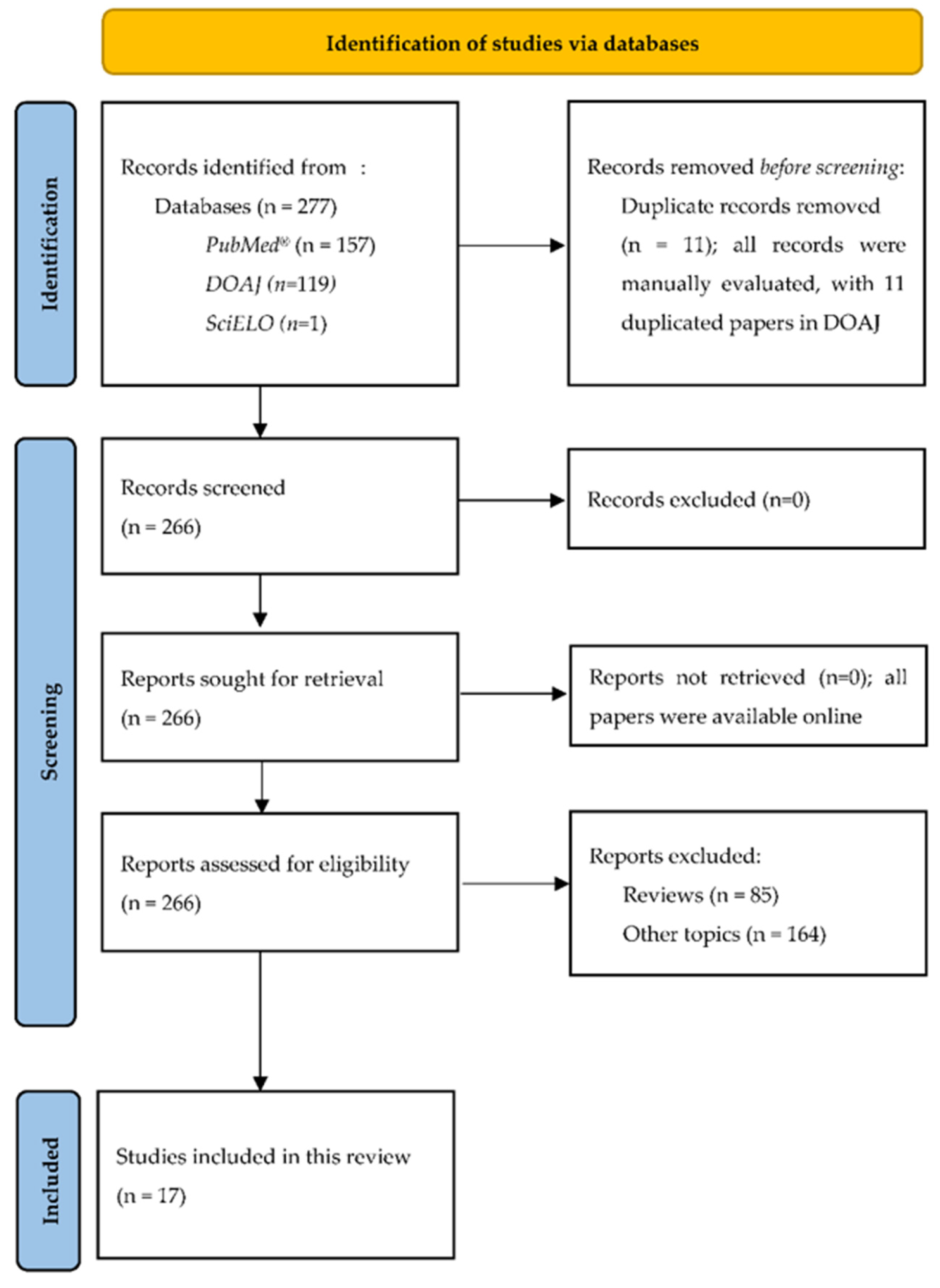
3.1. PRISMA Flow Diagram
3.2. Main Findings of the Selected Studies
3.2.1. Studies with Confirmatory In-Vitro Data
3.2.2. Studies with Confirmatory In-Vitro and/or Clinical Data
3.2.3. Repurposing of Drugs against COVID-2019
- Simeprevir (direct-acting antiviral agent that inhibits HCV NS3/4A protease to treat chronic hepatitis C virus (HCV) infection in adults with HCV genotype 1 or 4; DrugBank),
- Paritaprevir (a direct acting antiviral agent used in combination with other antiviral agents for the treatment of Hepatitis C Virus (HCV) infections; DrugBank),
- Velpatasvir (a NS5A inhibitor used to treat chronic hepatitis C infections in patients without cirrhosis or with compensated cirrhosis; DrugBank),
- Rifapentine (an antibiotic agent used in the treatment of pulmonary tuberculosis; DrugBank),
- Eribulin (a microtubule inhibitor used to treat metastatic breast cancer and metastatic or unresectable liposarcoma; DrugBank),
- Teniposide (a cytotoxic drug used as an adjunct for chemotherapy induction in the treatment of refractory childhood acute lymphoblastic leukemia; DrugBank),
- Trabectedin (an alkylating agent approved for the treatment of unresectable or metastatic soft tissue sarcoma (liposarcoma or leiomyosarcoma); DrugBank),
- Ivermectin (an anti-parasite medication used to treat head lice, onchocerciasis, strongyloidiasis, ascariasis, trichuriasis, and enterobiasis; DrugBank),
- Ledipasvir (a direct-acting antiviral agent used to treat specific hepatitis C virus (HCV) infections in combination with other antiviral agents; DrugBank), and
- Nystatin (a polyene ionophore antifungal used to treat cutaneous, mucocutaneous, and gastrointestinal mycotic infections, particularly those caused by Candida species; DrugBank).
- Paritaprevir, trypan blue (a dye used as a visualizing aid to stain the epiretinal membranes during ophthalmic surgical vitrectomy procedures, thereby facilitating removal of the tissue; DrugBank),
- Simeprevir,
- Dihydroergotamine (an ergot alkaloid used in the acute treatment of migraine headache and cluster headache; DrugBank),
- Conivaptan (an antidiuretic hormone inhibitor used to raise serum sodium levels; DrugBank),
- Ergotamine (an alpha-1 selective adrenergic agonist vasoconstrictor used to treat migraines with or without aura and cluster headaches; DrugBank),
- Venetoclax (a BCL-2 inhibitor used to treat chronic lymphocytic leukemia, small lymphocytic lymphoma, or acute myeloid leukemia; DrugBank),
- Idarubicin (an anthracycline antineoplastic agent used to treat acute myeloid leukemia (AML) in adults; DrugBank),
- Ivermectin and
- Nystatin.
- Conivaptan,
- Lifitegrast (a medication used to treat dry eye disease; DrugBank),
- Dihydroergotamine, Ergotamine,
- Eltrombopag (a thrombopoietin receptor agonist used to treat thrombocytopenia or aplastic anemia associated with various etiologies; DrugBank),
- Ponatinib (a kinase inhibitor used to treat patients with various types of chronic myeloid leukemia (CML); DrugBank),
- Lumacaftor (a protein chaperone used in combination with ivacaftor for the treatment of cystic fibrosis in patients who are homozygous for the F508del mutation in the CFTR gene; DrugBank),
- Nilotinib (a kinase inhibitor used for the chronic phase treatment of Chronic Myeloid Leukemia (CML) that is Philadelphia chromosome positive and for the treatment of CML that is resistant to therapy containing imatinib; DrugBank),
- Regorafenib (a kinase inhibitor used to treat patients with metastatic colorectal cancer, unresectable, locally advanced, or metastatic gastrointestinal stromal tumors, and hepatocellular carcinoma; DrugBank), and
- Aprepitant (a substance P/neurokinin 1 receptor antagonist used to treat nausea and vomiting caused by chemotherapy and surger; DrugBank) [41].
3.2.3.1. Repurposing of Drugs against COVID-2019: Association of Drugs
3.2.3.2. Repurposing of Drugs: Alternative Therapies
4. Discussion
4.1. Studies with Confirmatory In-Vitro and/or Clinical Data
4.2. Repurposing of Drugs for COVID-2019, Including Alternative Therapies
- valrubicin,
- aprepitant,
- dihydroergotamine, *
- bivalirudin,
- eltrombopag,
- eribulin,
- fulvestrant,
- idarubicin,
- ivermectin,
- ledipasvir,
- lifitegrast,
- nystatin,
- regorafenib,
- trypan blue,
- vitamin E,
- ruxolitinib,
- nafcillin,
- nabumetone,
- octacosanol,
- cinametic acid,
- trabectedin,
- lauric acid,
- acorbyl palmitrate,
- palmidrol,
- salmeterol,
- simvastatin,
- guaifenesin,
- verteporfin,
- metergoline,
- rescinnamine,
- leuprolide,
- lusutrombopag,
- telotristat,
- fostamatinib,
- tofacitinib,
- etoricoxib,
- ziprasidone,
- interferon-gamma;
- cyclosporine,
- zidovudine,
- methotrexate,
- artemisinin,
- glycyrrhizin acid,
- quinine,
- suramin,
- albuterol,
- ciprofloxacin,
- spirapril,
- lisinopril, and
- captopril.
4.2.1. Drug Repurposing: Combination of Drugs
4.2.2. Drug Repurposing: Natural Therapies
4.3. Future Research
4.4. Study Limitations
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Drug Candidates | DrugBank Summary (Summary and/or Indication) [12,13] | ATC Classification [14] | Drug Candidates | DrugBank Summary (Summary and/or Indication) [12,13] | ATC Classification [14] |
|---|---|---|---|---|---|
| Abacavir | Antiviral nucleoside reverse transcriptase inhibitor used in combination with other antiretrovirals for the treatment of HIV. | J05AF Nucleoside and nucleotide reverse transcriptase inhibitors | Ixazomib | A monoclonal antibody used with other medications to treat multiple myeloma in patients who have received one other therapy already. | L01XG Proteasome inhibitors |
| Adenosine | Medication used in myocardial perfusion scintigraphy and to treat supraventricular tachycardia. | C01EB Other cardiac preparations | Lopinavir | An HIV-1 protease inhibitor used in combination with ritonavir to treat human immunodeficiency virus (HIV) infection. | J05AR Antivirals for treatment of HIV infections, combinations. |
| Afatinib | Antineoplastic agent used for the treatment of locally advanced or metastatic non-small cell lung cancer (NSCLC) with non-resistant EGFR mutations or resistance to platinum-based chemotherapy. | L01EB Epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors | Lumacaftor | A protein chaperone used in combination with ivacaftor for the treatment of cystic fibrosis in patients who are homozygous for the F508del mutation in the CFTR gene. | R07AX Other respiratory system products |
| Almitrine mesylate | Almitrine is a respiratory stimulant that enhances respiration by acting as an agonist of peripheral chemoreceptors located on the carotid bodies. It is used in the treatment of chronic obstructive pulmonary disease. It is also reported to have a potentially beneficial effect in treating the noctural oxygen desaturation without impairing the quality of sleep. | R07AB Respiratory stimulants | Mannitol | A sugar alcohol used to test for asthma, to reduce intracranial and intraocular pressure, to measure glomerular filtration rate, and to manage pulmonary symptoms associated with cystic fibrosis. | A06AD Osmotically acting laxatives |
| Amprenavir | Protease inhibitor used to treat HIV infection. | J05AE Protease inhibitors | Meglumine | An amino sugar found in iodinated contrast media. | P01CB Antimony compounds |
| Asunaprevir | A potent hepatitis C virus (HCV) NS3 protease inhibitor. It has been shown to have a very high efficacy in dual-combination regimens with daclatasvir in patients chronically infected with HCV genotype 1b. | J05AP Antivirals for treatment of HCV infections | Metoprolol tartrate | A beta-blocker used in the treatment of hypertension and angina, and used to reduce mortality due to myocardial infarction. | C07AB Beta blocking agents, selective |
| Atazanavir | Antiviral protease inhibitor used in combination with other antiretrovirals for the treatment of HIV. | J05AE Protease inhibitors | Mitoxantrone | A chemotherapeutic agent used for the treatment of secondary progressive, progressive relapsing, or worsening relapsing-remitting multiple sclerosis. | L01DB Anthracyclines and related substances |
| Baricitinib | A Janus kinase inhibitor used to treat moderate to severe rheumatoid arthritis that has responded poorly to at least one TNF antagonist. | L04AA Selective immunosuppressants | Niclosamide | An anthelmintic indicated in the treatment of beef, pork, fish, and dwarf tapeworm infections in adults and children. | P02DA Salicylic acid derivatives |
| Bedaquiline | Diarylquinoline antimycobacterial used in combination with other antibacterials to treat pulmonary multidrug resistant tuberculosis (MDR-TB). | J04AK Other drugs for treatment of tuberculosis | Nilotinib | A kinase inhibitor used for the chronic phase treatment of Chronic Myeloid Leukemia (CML) that is Philadelphia chromosome positive and for the treatment of CML that is resistant to therapy containing imatinib. | L01EA BCR-ABL tyrosine kinase inhibitors |
| Bictegravir | Bictegravir is an integrase inhibitor used to treat HIV infections. | J05AR Antivirals for treatment of HIV infections, combinations | Nitazoxanide | A thiazolide anti-infective used to treat infections by protozoa, helminths, anaerobic bacteria, microaerophilic bacteria, and viruses. | P01AX Other agents against amoebiasis and other protozoal diseases |
| Bortezomib | A proteasome inhibitor used to treat multiple myeloma in patients who have not been successfully treated with at least two previous therapies. | L01XG Proteasome inhibitors | Nitrofurantoin | An antibiotic used to treat urinary tract infections. | J01XE Nitrofuran derivatives |
| Brefeldin A | A metabolite from Penicillium brefeldianum that exhibits a wide range of antibiotic activity. | n.a. | Ombitasvir | A direct acting antiviral agent used in combination with other antiviral agents for the treatment of Hepatitis C Virus (HCV) infections. | J05AP Antivirals for treatment of HCV infections |
| Brequinar | Synthetic quinolinecarboxylic acid analogue with antineoplastic properties. | n.a. | Oritavancin | Antibacterial agent used to treat acute bacterial skin and skin structure infections caused by susceptible Gram-positive bacteria. | J01XA Glycopeptide antibacterials |
| Brigatinib | Anaplastic lymphoma kinase inhibitor used to treat anaplastic lymphoma kinase positive metastatic non-small cell lung cancer. | L01ED Anaplastic lymphoma kinase (ALK) inhibitors | Paritaprevir | A direct acting antiviral agent used in combination with other antiviral agents for the treatment of Hepatitis C Virus (HCV) infections. | J05AP Antivirals for treatment of HCV infections |
| Capecitabine | Nucleoside metabolic inhibitor indicated to treat colon, colorectal and breast cancer. | L01BC Pyrimidine analogues | Pemirolast | A medication used to treat allergies such as hay fever and allergic conjunctivitis. | n.a. |
| Carbocisteine | A expectorant mucolytic used in the relief of respiratory of COPD and other conditions associated with increased mucus viscosity. | R05CB Mucolytics | Pibrentasvir | Hepatitis C NS5A inhibitor used to treat Hepatitis C. | J05AP Antivirals for treatment of HCV infections |
| Carfilzomib | Proteasome inhibitor used either alone or in conjunction with a chemotherapy regimen to treat patients with relapsed or refractory multiple myeloma. | L01XG Proteasome inhibitors | Piperacillin | A penicillin antibiotic combined with tazobactam to treat piperacillin-resistant, piperacillin/tazobactam susceptible, β-lactamase generating strains of several bacteria. | J01CA Penicillins with extended spectrum |
| Celecoxib | An NSAID used to treat osteoarthritis, rheumatoid arthritis, acute pain, menstrual symptoms, and to reduce polyps is familial adenomatous polyposis. | M01AH Coxibs | Ponatinib | Kinase inhibitor used to treat patients with various types of chronic myeloid leukemia (CML). | L01EA BCR-ABL tyrosine kinase inhibitors |
| Cefotiam | A cephalosporin antibiotic used to treat a variety of bacterial infections. | J01DC Second-generation cephalosporins | Posaconazole | A triazole antifungal drug that is used to treat invasive infections by Candida species and Aspergillus species in severely immunocompromised patients. | J02AC Triazole derivatives |
| Chloroquine | An antimalarial drug used to treat susceptible infections with P. vivax, P. malariae, P. ovale, and P. falciparum. It is also used for second line treatment for rheumatoid arthritis. | P01BA Aminoquinolines | Procaterol | A beta-2 adrenergic receptor agonist and bronchodilator used for the treatment of asthma and chronic obstructive pulmonary disease (COPD). | R03AC Selective beta-2-adrenoreceptor agonists |
| Clofazimine | Riminophenazine antimycobacterial used to treat leprosy. | J04BA Drugs for treatment of lepra | Remdesivir | A nucleoside analog used to treat RNA virus infections including COVID-19. | n.a. |
| Conivaptan | Antidiuretic hormone inhibitor used to raise serum sodium levels. | C03XA Vasopressin antagonists | Reserpine | Antihypertensive and antipsychotic. | C02AA Rauwolfia alkaloids |
| Darunavir | A HIV protease inhibitor used in the treatment of human immunodeficiency virus (HIV) infection in patients with history of prior antiretroviral therapies. | J05AE Protease inhibitors | Rifabutin | An antibiotic used to treat mycobacterium avium complex disease in patients with HIV. | J04AB Antibiotics |
| D-Mannitol | Mannitol is a sugar alcohol used to test for asthma, to reduce intracranial and intraocular pressure, to measure glomerular filtration rate, and to manage pulmonary symptoms associated with cystic fibrosis. | V04CX Other diagnostic agents | Rifamycin | An antibacterial used to treat traveler’s diarrhea. | A07AA Antibiotics |
| D-Sorbitol | Laxative to relieve constipation, urologic irrigating fluid, and pharmaceutical sweetener. | A06AD Osmotically acting laxatives | Rifapentine | Antibiotic agent used in the treatment of pulmonary tuberculosis. | J04AB Antibiotics |
| Daclatasvir | Direct-acting antiviral agent used to treat specific hepatitis C virus (HCV) infections in combination with other antiviral agents. | J05AP Antivirals for treatment of HCV infections | Ritonavir | An HIV protease inhibitor used in combination with other antivirals in the treatment of HIV infection. | J05AE Protease inhibitors |
| Darunavir | HIV protease inhibitor used in the treatment of human immunodeficiency virus (HIV) infection in patients with history of prior antiretroviral therapies. | J05AE Protease inhibitors | Roflumilast | A selective phosphodiesterase 4 inhibitor indicated to decrease the risk of exacerbations in patients with severe chronic obstructive pulmonary disease (COPD) associated with a history of exacerbations and chronic bronchitis. | R03DX Other systemic drugs for obstructive airway diseases |
| Dolutegravir | Antiviral agent used for the treatment of HIV-1 infections in combination with other antiretroviral agents. | J05AJ Integrase inhibitors | Saquinavir | HIV protease inhibitor used in combination with other antiretroviral agents for the treatment of HIV-1 with advanced immunodeficiency. | J05AE Protease inhibitors |
| Efavirenz | Non-nucleoside reverse transcriptase inhibitor used to treat HIV infection or prevent the spread of HIV. | J05AG Non-nucleoside reverse transcriptase inhibitors | Simeprevir | Antiviral agent that inhibits HCV NS3/4A protease to treat chronic hepatitis C virus (HCV) infection in adults with HCV genotype 1 or 4. | J05AP Antivirals for treatment of HCV infections |
| Elbasvir | Antiviral and NS5A inhibitor used to treat hepatitis C infections. | J05AP Antivirals for treatment of HCV infections | Sodium gluconate | Gluconic acid or gluconate is used to maintain the cation-anion balance on electrolyte solutions e.g., fluid loss. | n.a. |
| Emricasan | A caspase inhibitor that protects liver cells from excessive apoptosis. | n.a. | Sulfamethoxazole | An oral sulfonamide antibiotic, given in combination with trimethoprim, used to treat a variety of infections of the urinary tract, respiratory system, and gastrointestinal tract. | J01EC Intermediate-acting sulfonamides |
| Ergotamine | A alpha-1 selective adrenergic agonist vasoconstrictor used to treat migraines with or without aura and cluster headaches. | N02CA Ergot alkaloids | Sulfanilamide | A sulfonamide anti-infective used to treat vulvovaginal candidiasis caused by Candida albicans. | D06BA Sulfonamides |
| Flutamide | Antiandrogen used for locally confined stage B2-C and D-2 metastatic prostate carcinoma. | L02BB Anti-androgens | Telithromycin | A ketolide used to treat community acquired pneumonia of mild to moderate severity. | J01FA Macrolides |
| Fiboflapon sodium | Investigated for use/treatment in inflammatory disorders (unspecified). | n.a. | Teniposide | A cytotoxic drug used as an adjunct for chemotherapy induction in the treatment of refractory childhood acute lymphoblastic leukemia. | L01CB Podophyllotoxin derivatives |
| Fosamprenavir | Antiretroviral agent used for the treatment and postexposure prophylaxis of human immunodeficiency virus (HIV-1) infection. | J05AE Protease inhibitors | Tiotropium-bromide | A long-acting bronchodilator used in the management of chronic obstructive pulmonary disease (COPD). | R03BB Anticholinergics |
| Ganciclovir | A DNA polymerase inhibitor used to treat cytomegalovirus and herpetic keratitis of the eye. | J05AB Nucleosides and nucleotides excl. reverse transcriptase inhibitors | Tirofiban Hydrochloride | A platelet aggregation inhibitor used to prevent thrombotic events in non-ST elevated acute coronary syndrome. | B01AC Platelet aggregation inhibitors excl. heparin |
| Gemcitabine | A nucleoside metabolic inhibitor used as adjunct therapy in the treatment of certain types of ovarian cancer, non-small cell lung carcinoma, metastatic breast cancer, and as a single agent for pancreatic cancer. | L01BC Pyrimidine analogues | Tipranavir | A protease inhibitor used to treat HIV-1 resistant to more than 1 protease inhibitor. | J05AE Protease inhibitors |
| Glecaprevir | A Hepatitis C NS3/4A protease inhibitor used to treat Hepatitis C. | J05AP Antivirals for treatment of HCV infections | Tolcapone | A catechol-O-methyltransferase (COMT) inhibitor used as adjunct therapy in the symptomatic management of idiopathic Parkinson’s disease. | N04BX Other dopaminergic agents |
| Glutamine | An amino acid commonly found as a component in total parenteral nutrition. | A16AA Amino acids and derivatives | Valaciclovir | An guanine nucleoside antiviral used to treat herpes exacerbations. | J05AB Nucleosides and nucleotides excl. reverse transcriptase inhibitors |
| Glutathione | For nutritional supplementation, also for treating dietary shortage or imbalance. | V03AB Antidotes | Vancomycin | A glycopeptide antibiotic used to treat severe but susceptible bacterial infections such as MRSA (methicillin-resistant Staphylococcus aureus) infections. | J01XA Glycopeptide antibacterials |
| Grazoprevir | An antiviral and NS3/4A protease inhibitor used to treat hepatitis C infections. | J05AP Antivirals for treatment of HCV infections | Velpatasvir | A NS5A inhibitor used to treat chronic hepatitis C infections in patients without cirrhosis or with compensated cirrhosis. | J05AP Antivirals for treatment of HCV infections |
| Hydroxychloroquine | An antimalarial medication used to treat uncomplicated cases of malaria and for chemoprophylaxis in specific regions. Also a disease modifying anti-rheumatic drug (DMARD) indicated for treatment of rheumatoid arthritis and lupus erythematosus. | P01BA Aminoquinolines | Venetoclax | A BCL-2 inhibitor used to treat chronic lymphocytic leukemia, small lymphocytic lymphoma, or acute myeloid leukemia. | L01XX Other antineoplastic agents |
| Indinavir | A protease inhibitor used to treat HIV infection. | J05AE Protease inhibitors | Vidarabine | An antiviral agent used to treat various viral infections. | J05AB Nucleosides and nucleotides excl. reverse transcriptase inhibitors |
| Itraconazole | An antifungal agent used for the treatment of various fungal infections in immunocompromised and non-immunocompromised patients. | J02AC Triazole derivatives | Vismodegib | A hedgehog pathway inhibitor used to treat patients with locally advanced or metastatic basal cell carcinoma. | L01XJ Hedgehog pathway inhibitors |
References
- Dhar Chowdhury, S.; Oommen, A.M. Epidemiology of COVID-19. J. Dig. Endosc. 2020, 11, 3–7. [Google Scholar]
- WHO. Archived: WHO Timeline—COVID-19. 2020. Available online: https://www.who.int/news/item/27-04-2020-who-timeline---covid-19 (accessed on 18 July 2021).
- WHO. WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 18 July 2021).
- Yang, L.; Xie, X.; Tu, Z.; Fu, J.; Xu, D.; Zhou, Y. The signal pathways and treatment of cytokine storm in COVID-19. Signal Transduct. Target. Ther. 2021, 6, 255. [Google Scholar] [CrossRef]
- Gurung, A.B.; Ali, M.A.; Lee, J.; Farah, M.A.; Al-Anazi, K.M. An Updated Review of Computer-Aided Drug Design and Its Application to COVID-19. BioMed Res. Int. 2021, 2021, 8853056. [Google Scholar] [CrossRef] [PubMed]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.M.; Martins, T.B.; Peterson, L.K.; Hill, H.R. Clinical significance of measuring serum cytokine levels as inflammatory biomarkers in adult and pediatric COVID-19 cases: A review. Cytokine 2021, 142, 155478. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.M.; Hill, H.R. Role of Host Immune and Inflammatory Responses in COVID-19 Cases with Underlying Primary Immunodeficiency: A Review. J. Interferon Cytokine Res. 2020, 40, 549–554. [Google Scholar] [CrossRef] [PubMed]
- Aleem, A.; Akbar Samad, A.B.; Slenker, A.K. Emerging Variants of SARS-CoV-2 And Novel Therapeutics Against Coronavirus (COVID-19); StatPearls Publishing: Treasure Island, FL, USA, 2021. Available online: https://www.ncbi.nlm.nih.gov/books/NBK570580/#:~:text=Available%20from%3A%20https%3A//www.ncbi.nlm.nih.gov/books/NBK570580/ (accessed on 31 July 2021).
- Centers for Disease Control and Prevention. SARS-CoV-2 Variant Classifications and Definitions. Available online: https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html (accessed on 18 July 2021).
- EMA. Treatments and Vaccines for COVID-19. Available online: https://www.ema.europa.eu/en/human-regulatory/overview/public-health-threats/coronavirus-disease-covid-19/treatments-vaccines-covid-19 (accessed on 18 July 2021).
- Law, V.; Knox, C.; Djoumbou, Y.; Jewison, T.; Guo, A.C.; Liu, Y.; Maciejewski, A.; Arndt, D.; Wilson, M.; Neveu, V.; et al. DrugBank 4.0: Shedding new light on drug metabolism. Nucleic Acids Res. 2014, 42, D1091–D1097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- WHO. WHO Collaborating Centre for Drug Statistics Methodology: ATC/DDD Index 2021. Available online: https://www.whocc.no/atc_ddd_index/ (accessed on 18 July 2021).
- Gatti, M.; Turrini, E.; Raschi, E.; Sestili, P.; Fimognari, C. Janus Kinase Inhibitors and Coronavirus Disease (COVID)-19: Rationale, Clinical Evidence and Safety Issues. Pharmaceuticals 2021, 14, 738. [Google Scholar] [CrossRef]
- WHO. WHO COVID-19 Solidarity Therapeutics Trial. 2021. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/global-research-on-novel-coronavirus-2019-ncov/solidarity-clinical-trial-for-covid-19-treatments (accessed on 15 August 2021).
- Naqvi, A.A.T.; Fatima, K.; Mohammad, T.; Fatima, U.; Singh, I.K.; Singh, A.; Atif, S.M.; Hariprasad, G.; Hasan, G.M.; Hassan, I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta-Mol. Basis Dis. 2020, 1866, 165878. [Google Scholar] [CrossRef]
- Zumla, A.; Chan, J.F.; Azhar, E.I.; Hui, D.S.; Yuen, K.Y. Coronaviruses—Drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016, 15, 327–347. [Google Scholar] [CrossRef] [Green Version]
- Kaushal, K.; Sarma, P.; Rana, S.V.; Medhi, B.; Naithani, M. Emerging role of artificial intelligence in therapeutics for COVID-19: A systematic review. J. Biomol. Struct. Dyn. 2020, 10, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Torres, P.; Sodero, A.; Jofily, P.; Silva-Junior, F.P. Key Topics in Molecular Docking for Drug Design. Int. J. Mol. Sci. 2019, 20, 4574. [Google Scholar] [CrossRef] [Green Version]
- Srinivasan, S.; Batra, R.; Chan, H.; Kamath, G.; Cherukara, M.J.; Sankaranarayanan, S. Artificial Intelligence-Guided De Novo Molecular Design Targeting COVID-19. ACS Omega 2021, 6, 12557–12566. [Google Scholar] [CrossRef] [PubMed]
- Bera, I.; Payghan, P.V. Use of Molecular Dynamics Simulations in Structure-Based Drug Discovery. Curr. Pharm. Des. 2019, 25, 3339–3349. [Google Scholar] [CrossRef]
- Hospital, A.; Goñi, J.R.; Orozco, M.; Gelpí, J.L. Molecular dynamics simulations: Advances and applications. Adv. Appl. Bioinform. Chem. 2015, 8, 37–47. [Google Scholar] [PubMed] [Green Version]
- Temml, V.; Kutil, Z. Structure-based molecular modeling in SAR analysis and lead optimization. Comput. Struct. Biotechnol. J. 2021, 19, 1431–1444. [Google Scholar] [CrossRef]
- Voet, A.; Zhang, K.Y. Pharmacophore modelling as a virtual screening tool for the discovery of small molecule protein-protein interaction inhibitors. Curr. Pharm. Des. 2012, 18, 4586–4598. [Google Scholar] [CrossRef]
- Bai, Q.; Tan, S.; Xu, T.; Liu, H.; Huang, J.; Yao, X. MolAICal: A soft tool for 3D drug design of protein targets by artificial intelligence and classical algorithm. Brief. Bioinform. 2021, 22, bbaa161. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Ke, Y.-Y.; Peng, T.-T.; Yeh, T.-K.; Huang, W.-Z.; Chang, S.-E.; Wu, S.-H.; Hung, H.-C.; Hsu, T.-A.; Lee, S.-J.; Song, J.-S.; et al. Artificial intelligence approach fighting COVID-19 with repurposing drugs. Biomed. J. 2020, 43, 355–362. [Google Scholar] [CrossRef]
- Gysi, D.M.; Do Valle, Í.; Zitnik, M.; Ameli, A.; Gan, X.; Varol, O.; Ghiassian, S.D.; Patten, J.J.; Davey, R.A.; Loscalzo, J.; et al. Network medicine framework for identifying drug-repurposing opportunities for COVID-19. Proc. Natl. Acad. Sci. USA 2021, 118, e2025581118. [Google Scholar] [CrossRef] [PubMed]
- Abdulla, A.; Wang, B.; Qian, F.; Kee, T.; Blasiak, A.; Ong, Y.H.; Hooi, L.; Parekh, F.; Soriano, R.; Olinger, G.G.; et al. Project IDentif.AI: Harnessing Artificial Intelligence to Rapidly Optimize Combination Therapy Development for Infectious Disease Intervention. Adv. Ther. 2020, 3, 2000034. [Google Scholar] [CrossRef]
- Blasiak, A.; Lim, J.J.; Seah, S.G.K.; Kee, T.; Remus, A.; Chye, D.H.; Wong, P.S.; Hooi, L.; Truong, A.T.L.; Le, N.; et al. IDentif.AI: Rapidly optimizing combination therapy design against severe Acute Respiratory Syndrome Coronavirus 2 (SARS-Cov-2) with digital drug development. Bioeng. Transl. Med. 2020, 6, e10196. [Google Scholar] [CrossRef] [PubMed]
- Schultz, M.B.; Vera, D.; Sinclair, D.A. Can artificial intelligence identify effective COVID-19 therapies? EMBO Mol. Med. 2020, 12, e12817. [Google Scholar] [CrossRef] [PubMed]
- Stebbing, J.; Krishnan, V.; De Bono, S.; Ottaviani, S.; Casalini, G.; Richardson, P.J.; Monteil, V.; Lauschke, V.M.; Mirazimi, A.; Youhanna, S.; et al. Mechanism of baricitinib supports artificial intelligence-predicted testing in COVID-19 patients. EMBO Mol. Med. 2020, 12, e12697. [Google Scholar] [CrossRef] [PubMed]
- Nayarisseri, A.; Khandelwal, R.; Madhavi, M.; Selvaraj, C.; Panwar, U.; Sharma, K.; Hussain, T.; Singh, S.K. Shape-based Machine Learning Models for the Potential Novel COVID-19 Protease Inhibitors Assisted by Molecular Dynamics Simulation. Curr. Top. Med. Chem. 2020, 20, 2146–2167. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Zhang, J.; Cha, Y.; Kolitz, S.; Funt, J.; Chong, R.E.; Barrett, S.; Kusko, R.; Zeskind, B.; Kaufman, H. Advanced bioinformatics rapidly identifies existing therapeutics for patients with coronavirus disease-2019 (COVID-19). J. Transl. Med. 2020, 18, 257. [Google Scholar] [CrossRef]
- Das, G.; Das, T.; Chowdhury, N.; Chatterjee, D.; Bagchi, A.; Ghosh, Z. Repurposed drugs and nutraceuticals targeting envelope protein: A possible therapeutic strategy against COVID-19. Genomics 2021, 113 Pt 2, 1129–1140. [Google Scholar] [CrossRef]
- Rajput, A.; Thakur, A.; Mukhopadhyay, A.; Kamboj, S.; Rastogi, A.; Gautam, S.; Jassal, H.; Kumar, M. Prediction of repurposed drugs for Coronaviruses using artificial intelligence and machine learning. Comput. Struct. Biotechnol. J. 2021, 19, 3133–3148. [Google Scholar] [CrossRef]
- Li, Z.; Yao, Y.; Cheng, X.; Chen, Q.; Zhao, W.; Ma, S.; Li, Z.; Zhou, H.; Li, W.; Fei, T. A computational framework of host-based drug repositioning for broad-spectrum antivirals against RNA viruses. iScience 2021, 24, 102148. [Google Scholar] [CrossRef] [PubMed]
- McCoy, K.; Gudapati, S.; He, L.; Horlander, E.; Kartchner, D.; Kulkarni, S.; Mehra, N.; Prakash, J.; Thenot, H.; Vanga, S.; et al. Biomedical Text Link Prediction for Drug Discovery: A Case Study with COVID-19. Pharmaceutics 2021, 13, 794. [Google Scholar] [CrossRef] [PubMed]
- Chakravarty, K.; Antontsev, V.G.; Khotimchenko, M.; Gupta, N.; Jagarapu, A.; Bundey, Y.; Hou, H.; Maharao, N.; Varshney, J. Accelerated Repurposing and Drug Development of Pulmonary Hypertension Therapies for COVID-19 Treatment Using an AI-Integrated Biosimulation Platform. Molecules 2021, 26, 1912. [Google Scholar] [CrossRef] [PubMed]
- Kadioglu, O.; Saeed, M.; Greten, H.J.; Efferth, T. Identification of novel compounds against three targets of SARS CoV-2 coronavirus by combined virtual screening and supervised machine learning. Comput. Biol. Med. 2021, 133, 104359. [Google Scholar] [CrossRef] [PubMed]
- Artigas, L.; Coma, M.; Matos-Filipe, P.; Aguirre-Plans, J.; Farrés, J.; Valls, R.; Fernandez-Fuentes, N.; De La Haba-Rodriguez, J.; Olvera, A.; Barbera, J.; et al. In-silico drug repurposing study predicts the combination of pirfenidone and melatonin as a promising candidate therapy to reduce SARS-CoV-2 infection progression and respiratory distress caused by cytokine storm. PLoS ONE 2020, 15, e0240149. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, L.; Song, M.; Yan, J.; Shi, J.; Yao, Y. Evaluating the Traditional Chinese Medicine (TCM) Officially Recommended in China for COVID-19 Using Ontology-Based Side-Effect Prediction Framework (OSPF) and Deep Learning. J. Ethnopharmacol. 2021, 272, 113957. [Google Scholar] [CrossRef]
- Acharya, A.; Agarwal, R.; Baker, M.B.; Baudry, J.; Bhowmik, D.; Boehm, S.; Byler, K.G.; Chen, S.Y.; Coates, L.; Cooper, C.J.; et al. Supercomputer-Based Ensemble Docking Drug Discovery Pipeline with Application to Covid-19. J. Chem. Inf. Model. 2020, 60, 5832–5852. [Google Scholar] [CrossRef]
- Fontelo, P.; Liu, F. A review of recent publication trends from top publishing coutries. BMC Syst. Rev. 2018, 7, 147. [Google Scholar]
- Pan, H.; Peto, R.; Henao-Restrepo, A.M.; Preziosi, M.P.; Sathiyamoorthy, V.; Karim, Q.A.; Alejandria, M.M.; Hernández García, C.; Kieny, M.P.; Malekzadeh, R.; et al. Repurposed Antiviral Drugs for Covid-19—Interim WHO Solidarity Trial Results. New Engl. J. Med. 2021, 384, 497–511. [Google Scholar]
- Rehman, S.U.; Rehman, S.U.; Yoo, H.H. COVID-19 challenges and its therapeutics. Biomed. Pharmacother. 2021, 142, 112015. [Google Scholar] [CrossRef]
- Silva, V.; Rosa, M.N.; Miranda-Gonçalves, V.; Costa, A.M.; Tansini, A.; Evangelista, A.F. Euphol, a tetracyclic triterpene, from Euphorbia tirucalli induces autophagy and sensitizes temozolomide cytotoxicity on glioblastoma cells. Investig. New Drugs 2019, 37, 223–237. [Google Scholar] [CrossRef] [PubMed]
- Rastelli, G.; Pellati, F.; Pinzi, L.; Gamberini, M. Repositioning Natural Products in Drug Discovery. Molecules 2020, 25, 1154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, Y.; Han, Y.; Liu, J.; Qiu, Y.; Tan, Q.; Zhou, Z.; Yu, Y.-Z.; Chen, J.; Giger, M.L.; Lure, F.Y.M.; et al. Tailoring steroids in the treatment of COVID-19 pneumonia assisted by CT scans: Three case reports. J. X-ray Sci. Technol. 2020, 28, 885–892. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Mehran, M.T.; Haq, Z.U.; Ullah, Z.; Naqvi, S.R.; Ihsan, M.; Abbass, H. Applications of artificial intelligence in COVID-19 pandemic: A comprehensive review. Expert Syst. Appl. 2021, 185, 115695. [Google Scholar] [CrossRef]
| Author(s); Year Reference | Country(ies) of Origin | Key Repurposed Drug(s) Potentially Relevant against the Treatment of COVID-2019 | Key Used AI Methodologies |
|---|---|---|---|
| (Ke, Peng, Yeh, et al., 2020) [28] | Taiwan | Bedaquiline Brequinar Celecoxib Clofazimine Conivaptan Gemcitabine Tolcapone Vismodegib | First, an an AI platform was defined to identify potential old/repurposed drugs with anti-coronavirus activities (or potential anti-coronavirus activity). Second, AI predicted drugs were tested for their activity against a feline coronavirus in vitro. Third, results of assays were introduced in the AI system. A Deep Neural Network algorithm was used to identify the most relevant descriptors, with the generation of different weightings to generate AI prediction models. |
| (Morselli, do Valle, Zitnik, et al., 2021) [29] | USA, Turkey, Hungary | Auranofin Azelastine Fluvastatin Methotrexate Vinblastine | AI based on algorithms, network diffusion, and network proximity, tasking to rank 6,340 drugs, regarding their potential efficacy against SARS-CoV-2. Multimodal technology was required to fuse the prediction of all algorithms, since the predictive algorithm did not offer consistently reliable outcomes. The top-ranked drugs were screened in human cells. |
| (Abdulla, Wang, Qian, et al., 2020) [30] * | China, Singapore | Amantadine Azithromycin Chloroquine Omeprazole Sodium (optional) Ribavirin (optional) - These repurposing drugs were evaluated in combination. | An AI-based platform was used to interrogate 12 drug/dose parameters space. Combination therapies that optimally inhibit A549 lung cell infection by vesicular stomatitis virus within 3 days of project start were identified. This AI project utilized a quadratic relationship between drug/dose inputs and efficacy/safety outputs to successfully identify the drug-dose parameter space. |
| (Blasiak, Lim, Seah, et al., 2020) [31] * | China, Singapore, USA | Lopinavir Remdesivir Ritonavir - These repurposing drugs were evaluated in combination. | A platform (IDentif.AI) that pairs experimental validation with AI and digital drug development was used. Workflow of the project IDentif.AI: 1) clinically relevant concentrations based on and dose–response curves and maximal plasma concentration; 2) in vitro testing of combination therapies; combination therapies were determined through an orthogonal array composite (OACD) design; 3) IDentif.AI analysis: drug–drug interactions and clinically relevant drug-dosage combinations; and 4) biological validation. |
| Studies with confirmatory in-vitro and/or clinical data (Section 3.2.2) | |||
| (Schultz, Vera, Sinclair et al., 2020) [32] | USA | Baricitinib | |
| (Stebbing, Krishnan, de Bono et al., 2020) [33] | UK, USA, Italy, Sweden, Singapore | Baricitinib | BenevolentAI (an artificial AI platform) identified baricitinib as a potential COVID-19 drugs. Details/information on BenevolentAI works were limited. |
| Repurposing of drugs against COVID-2019 (Section 3.2.3) | |||
| (Nayarisseri, Khandelwal, Madhavi et al., 2020) [34] | India, Saudi Arabia | Aprepitant Fulvestrant Remdesivir Valrubicin | A machine learning approach was employed. Particularly, repurposed drugs were selected based on their capacity of targeting the main coronavirus protease (6LU7) using ligand-receptor Docking (optimized potential for liquid simulations algorithms to identify high affinity compounds). Additionally, candidates were subjected to Molecular Dynamic Simulations followed by ADMET (absorption, distribution, metabolism, excretion, and toxicity) studies. |
| (Kim, Zhang, Cha et al., 2020) [35] | USA | Emricasan Fosamprenavir Glutamine Glutathione Piperacillin sodium Ruxolitinib Vitamin E | Two computational approaches were applied. Fist, a high-throughput AI-based binding affinity prediction platform was used to identify FDA approved drugs with potential capacity to block coronaviruses from entering cells by binding to ACE2 (angiotensin-converting enzyme) or TMPRSS2 (Transmembrane Serine Protease 2). Second, the Disease Cancelling Technology (DCT) platform was used to identify FDA approved drugs, which may attenuate the gene expression patterns induced by coronaviruses. |
| (Das G., Das, T., Chowdhury et al., 2021) [36] | India | Ascorbyl palmitrate Cinametic acid Guaifenesin Lauric acid Nabumetone Nafcillin Octacosanol Palmidrol Salmeterol | AI deep learning techniques, in silico methodologies) and pattern recognition techniques were used to screen FDA approved pharmaceuticals and nutraceuticals to target CoV envelope (E) protein. A protein involved in the assembly and release of the virus inside the host. Multiple opensource drug databases were considered, such as ChEMBL v.26, Enamine Bio reference Compounds (https://www.enaminestore.com/products/bioreference-compounds, accessed on 17 September 2021) and Chemoinformatic tools and database (https://chemoinfo.ipmc.cnrs.fr/TMP/tmp.32396/e-Drug3D_1930_v3.sdf, accessed on 17 September 2021). |
| (Rajput, Thakur, Mukhopadhyay et al., 2021) [37] | India | Alatrofloxacin Metergoline Rescinnamine Rescinnamine Telotristat ethyl Verteporfin | Robust computational methods using machine learning techniques, such as Support Vector Machine, Random Forest, k-Nearest Neighbour, Artificial Neural Network, and Deep Learning were developed by the authors to predict the repurposed drugs. |
| (Li, Yao, Cheng et al., 2021) [38] | China, USA | Baricitinib Bivalirudin Fostamatinib Lusutrombopag Simvastatin | 1) Public genetic screening data were successively interrogated to identify human-specific host dependency genes, i.e., indispensable genes for effective viral infections. 2) Extensive drug-target interactions were interrogated through diverse methodologies, such as database retrieval, literature mining and de novo prediction using AI-based algorithms. |
| (McCoy, Gudapati, He, Horlander, 2021) [39] | USA | Amprenavir Albuterol Artemisinin Chloroquine Ciprofloxacin Cyclosporine Fluoroquinolones Hydroxymethylglutaryl-CoA reductase inhibitors Methotrexate Quinolone antibacterial agents Suramin Zidovudine | A link prediction model was developed (an AI text mining model). The biomedical knowledge graph, SemNe was used to predict missing links in biomedical literature, regarding drug repurposing. TransE, CompleX, and RotatE based methods were used to visualize knowledge graph embeddings and link prediction results using in a web application. |
| (Chakravarty, Antontsev, Khotimchenko et al., 2021) [40] | USA | Captopril Lisinopril Spirapril | The plataform BIOiSIM (an AI-integrated mechanistic modeling platform) was used to simulate systemic therapy of Calcium Channel Blockers (CCBs) and ACE compounds in tissues related to the COVID-19 pathogenesis, namely the disposition and site-of-action penetration (in silico modeling). BIOiSIM is a dynamic, biology-driven platform that provides a scalable computational prediction of in vivo pharmacokinetic-pharmacodynamic (PK-PD) phenomena. |
| (Kadioglu, Saeed & Efferth, 2021) [41] | Germany | Conivaptan Dihydroergotamine Eltrombopag Ergotamine Eribulin Idarubicin Ivermectin Ledipasvir Lifitegrast Lumacaftor Nilotinib Nystatin Paritaprevir Ponatinib Regorafenib Rifapentine Simeprevir Teniposide Trabectedin Trypan blue Velpatasvir Venetoclax | Diverse combined in silico methods (virtual drug screening, molecular docking, and supervised machine learning algorithms) were used in a workflow to identify repurposed drug against COVID-19. |
| Repurposing of drugs against COVID-2019: association of drugs (Section 3.2.3.1) | |||
| (Artigas, Coma, Matos-Filipe et al., 2020) [42] | Spain | Pirfenidone plus melatonin | The mechanism of action of pirfenidone and melatonin was simulated by using the previously described Therapeutic Performance Mapping System (TPMS) technology (an AI-based approach). GUILDify v2.0 web server was used to confirm the effect of pirfenidone and melatonin against SARS-CoV-2 infection. This web server is able to calculate the neighbourhoods of the human biological network related to the host-key points (e.g., for SARS-CoV infection) and simultaneously affected by specific drugs. |
| Note: References [30,31] are also related to the combination of repurposing medicines. | |||
| Repurposing of drugs: alternative therapies (Section 3.2.3.2) | |||
| (Wang, Li; Song, et al., 2021) [43] | China, Australia | GCT-CJ Hanshiyufen fang (HSYF-F) Huashi Baidu Formula (HSBD-F) Qingfei Paidu Decoction (QFPD-T) Shenfu zhusheye (SF-ZSY) Zhongqifangzi (PMSP) Recommended as supplementary treatment against COVID-2019. | An ontology-based side-effect prediction framework (OSPF) was developed based on a previous work and Artificial Neural Network (ANN)-based deep learning. The Traditional Chinese Medicine prescriptions for the treatment of COVID-19 (officially recommended in China) were evaluated. |
| (Li, Yao, Cheng et al., 2021) [38] | China, USA | Atropine (Lycii Cortex, Hyoscyami Semen) Dehydroeffusal (Junci Medulla) Lysergol (Pharbitidis Semen) Solanocapsine (Solanum Nigrum) Solanocapsine (Solanum Nigrum) Vitexifolin C (Viticis Fructus) | 1) Public genetic screening data were successively interrogated to identify human-specific host dependency genes, i.e., indispensable genes for effective viral infections. 2) Extensive drug-target interactions were interrogated through diverse methodologies, such as database retrieval, literature mining and de novo prediction using AI-based algorithms. |
| (Kadioglu, Saeed & Efferth, 2021) [41] | Germany | Amyrin Baicalin Crinine Euphol Forsythiaside Friedelin Hoslunddiol IlexsaponinB1 IlexsaponinB2 IlexsaponinB3 Loniflavone Procyanidin Punicalagin Quercetin Quercetin-3-o-rutinoside Rutin Strictinin TirucallinA TingeninB Wogonoside ZINC252515584 ZINC27215482 ZINC15675938 | Diverse combined in silico methods (virtual drug screening, molecular docking, and supervised machine learning algorithms) were used in a workflow to identify repurposed drug against COVID-19. |
| (Acharya, Agarwal, Baker et al., 2020) [44] | USA, Italy | Hypericin Quercetin | An enhanced sampling molecular dynamics (MD) and ensemble docking was used supercomputer-driven pipeline for in silico drug discovery. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pires, C. A Systematic Review on the Contribution of Artificial Intelligence in the Development of Medicines for COVID-2019. J. Pers. Med. 2021, 11, 926. https://doi.org/10.3390/jpm11090926
Pires C. A Systematic Review on the Contribution of Artificial Intelligence in the Development of Medicines for COVID-2019. Journal of Personalized Medicine. 2021; 11(9):926. https://doi.org/10.3390/jpm11090926
Chicago/Turabian StylePires, Carla. 2021. "A Systematic Review on the Contribution of Artificial Intelligence in the Development of Medicines for COVID-2019" Journal of Personalized Medicine 11, no. 9: 926. https://doi.org/10.3390/jpm11090926
APA StylePires, C. (2021). A Systematic Review on the Contribution of Artificial Intelligence in the Development of Medicines for COVID-2019. Journal of Personalized Medicine, 11(9), 926. https://doi.org/10.3390/jpm11090926






