Frequency of Important CYP450 Enzyme Gene Polymorphisms in the Iranian Population in Comparison with Other Major Populations: A Comprehensive Review of the Human Data
Abstract
1. Introduction
2. Methods
2.1. Allele Frequency Data
2.2. Allele Nomenclature and Definitions
3. Results and Discussion
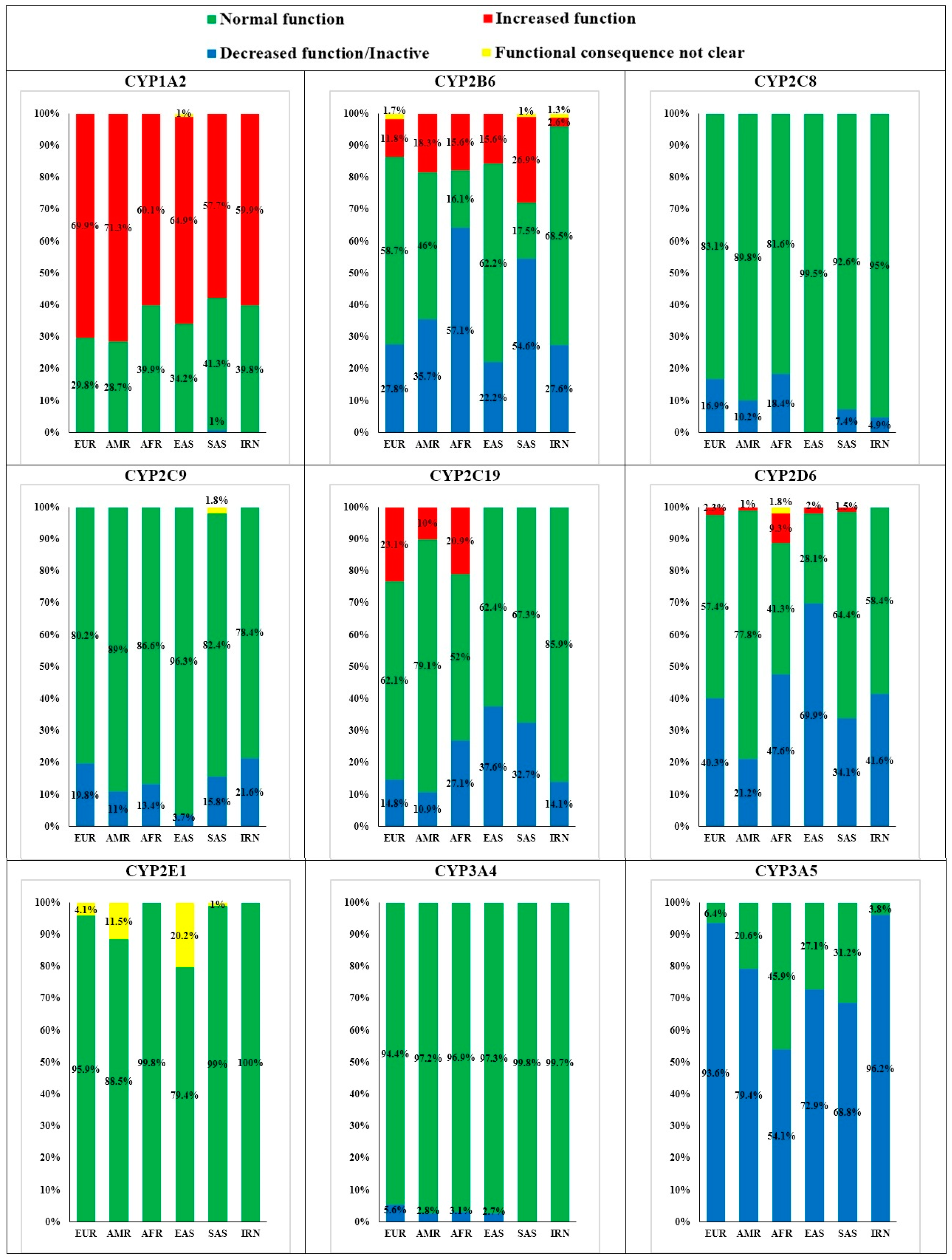
3.1. Cytochrome P450 1A2 (CYP1A2)
3.2. Cytochrome P450 2B6 (CYP2B6)
3.3. Cytochrome P450 2C8 (CYP2C8)
3.4. Cytochrome P450 2C9 (CYP2C9)
3.5. Cytochrome P450 2C19 (CYP2C19)
3.6. Cytochrome P450 2D6 (CYP2D6)
3.7. Cytochrome P450 2E1 (CYP2E1)
3.8. Cytochrome P450 3A4 (CYP3A4)
3.9. Cytochrome P450 3A5 (CYP3A5)
4. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- De Andres, F.; Teran, S.; Hernandez, F.; Teran, E.; LLerena, A. To Genotype or Phenotype for Personalized Medicine? CYP450 Drug Metabolizing Enzyme Genotype-Phenotype Concordance and Discordance in the Ecuadorian Population. OMICS A J. Integr. Biol. 2016, 20, 699–710. [Google Scholar] [CrossRef]
- Chen, Q.; Wei, D. Human cytochrome P450 and personalized medicine. Adv. Exp. Med. Biol. 2015, 827, 341–351. [Google Scholar] [CrossRef]
- Chen, Q.; Zhang, T.; Wang, J.F.; Wei, D.Q. Advances in human cytochrome p450 and personalized medicine. Curr. Drug Metab. 2011, 12, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Testa, B.; Pedretti, A.; Vistoli, G. Reactions and enzymes in the metabolism of drugs and other xenobiotics. Drug Discov. Today 2012, 17, 549–560. [Google Scholar] [CrossRef]
- Nebert, D.W.; Russell, D.W. Clinical importance of the cytochromes P450. Lancet 2002, 360, 1155–1162. [Google Scholar] [CrossRef]
- Rollason, V.; Lloret-Linares, C.; Lorenzini, K.I.; Daali, Y.; Gex-Fabry, M.; Piguet, V.; Besson, M.; Samer, C.; Desmeules, J. Evaluation of Phenotypic and Genotypic Variations of Drug Metabolising Enzymes and Transporters in Chronic Pain Patients Facing Adverse Drug Reactions or Non-Response to Analgesics: A Retrospective Study. J. Pers. Med. 2020, 10, 198. [Google Scholar] [CrossRef]
- Zanger, U.M.; Schwab, M. Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 2013, 138, 103–141. [Google Scholar] [CrossRef]
- He, Z.X.; Chen, X.W.; Zhou, Z.W.; Zhou, S.F. Impact of physiological, pathological and environmental factors on the expression and activity of human cytochrome P450 2D6 and implications in precision medicine. Drug Metab. Rev. 2015, 47, 470–519. [Google Scholar] [CrossRef]
- Neyshaburinezhad, N.; Rouini, M.; Shirzad, N.; Esteghamati, A.; Nakhjavani, M.; Namazi, S.; Ardakani, Y.H. Evaluating the effect of type 2 diabetes mellitus on CYP450 enzymes and P-gp activities, before and after glycemic control: A protocol for a case-control pharmacokinetic study. MethodsX 2020, 7, 100853. [Google Scholar] [CrossRef]
- Neyshaburinezhad, N.; Rouini, M.R.; Entezari, H.; Lavasani, H.; Hosseinzadeh Ardakani, Y. Evaluation of changes in cytochrome P450 2C19 activity in type 2 diabetic rats before and after treatment, by using isolated perfused liver model. Iran. J. Basic Med. Sci. 2020, 23, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Neyshaburinezhad, N.; Seidabadi, M.; Rouini, M.; Lavasani, H.; Foroumadi, A.; Ardakani, Y.H. Evaluation of hepatic CYP2D1 activity and hepatic clearance in type I and type II diabetic rat models, before and after treatment with insulin and metformin. DARU J. Pharm. Sci. 2020, 28, 479–487. [Google Scholar] [CrossRef] [PubMed]
- Rezai, S.; Neyshaburinezhad, N.; Rouini, M.; Lavasani, H.; Ardakani, Y.H. Can combination therapy with insulin and metformin improve metabolic function of the liver, in type I diabetic patients? An animal model study on CYP2D1 activity. J. Diabetes Metab. Disord. 2020, 19, 2049–2056. [Google Scholar] [CrossRef] [PubMed]
- Tracy, T.S.; Chaudhry, A.S.; Prasad, B.; Thummel, K.E.; Schuetz, E.G.; Zhong, X.B.; Tien, Y.C.; Jeong, H.; Pan, X.; Shireman, L.M.; et al. Interindividual Variability in Cytochrome P450-Mediated Drug Metabolism. Drug Metab. Dispos. 2016, 44, 343–351. [Google Scholar] [CrossRef]
- Roden, D.M.; Wilke, R.A.; Kroemer, H.K.; Stein, C.M. Pharmacogenomics: The genetics of variable drug responses. Circulation 2011, 123, 1661–1670. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.F. Polymorphism of human cytochrome P450 2D6 and its clinical significance: Part I. Clin. Pharm. 2009, 48, 689–723. [Google Scholar] [CrossRef]
- Zhou, S.F. Polymorphism of human cytochrome P450 2D6 and its clinical significance: Part II. Clin. Pharm. 2009, 48, 761–804. [Google Scholar] [CrossRef]
- Zhou, Y.; Ingelman-Sundberg, M.; Lauschke, V.M. Worldwide Distribution of Cytochrome P450 Alleles: A Meta-analysis of Population-scale Sequencing Projects. Clin. Pharmacol. Ther. 2017, 102, 688–700. [Google Scholar] [CrossRef] [PubMed]
- Lauschke, V.M.; Ingelman-Sundberg, M. The Importance of Patient-Specific Factors for Hepatic Drug Response and Toxicity. Int. J. Mol. Sci. 2016, 17, 1714. [Google Scholar] [CrossRef]
- Helgadottir, A.; Manolescu, A.; Helgason, A.; Thorleifsson, G.; Thorsteinsdottir, U.; Gudbjartsson, D.F.; Gretarsdottir, S.; Magnusson, K.P.; Gudmundsson, G.; Hicks, A.; et al. A variant of the gene encoding leukotriene A4 hydrolase confers ethnicity-specific risk of myocardial infarction. Nat. Genet. 2006, 38, 68–74. [Google Scholar] [CrossRef]
- Food and Drug Administration. Guidance on ethnic factors in the acceptability of foreign clinical data. Fed. Regist. 1998, 63, 31790–31796. [Google Scholar]
- Johansson, I.; Ingelman-Sundberg, M. Genetic polymorphism and toxicology—with emphasis on cytochrome p450. Toxicol. Sci. 2011, 120, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Pikuleva, I.A.; Waterman, M.R. Cytochromes p450: Roles in diseases. J. Biol. Chem. 2013, 288, 17091–17098. [Google Scholar] [CrossRef]
- Preissner, S.C.; Hoffmann, M.F.; Preissner, R.; Dunkel, M.; Gewiess, A.; Preissner, S. Polymorphic cytochrome P450 enzymes (CYPs) and their role in personalized therapy. PLoS ONE 2013, 8, e82562. [Google Scholar] [CrossRef] [PubMed]
- McGraw, J.; Waller, D. Cytochrome P450 variations in different ethnic populations. Expert Opin. Drug Metab. Toxicol. 2012, 8, 371–382. [Google Scholar] [CrossRef]
- Roco, A.; Quinones, L.; Agundez, J.A.; Garcia-Martin, E.; Squicciarini, V.; Miranda, C.; Garay, J.; Farfan, N.; Saavedra, I.; Caceres, D.; et al. Frequencies of 23 functionally significant variant alleles related with metabolism of antineoplastic drugs in the chilean population: Comparison with caucasian and asian populations. Front. Genet. 2012, 3, 229. [Google Scholar] [CrossRef] [PubMed]
- Azarpira, N.; Ashraf, M.J.; Khademi, B.; Darai, M.; Hakimzadeh, A.; Abedi, E. Study the polymorphism of CYP3A5 and CYP3A4 loci in Iranian population with laryngeal squamous cell carcinoma. Mol. Biol. Rep. 2011, 38, 5443–5448. [Google Scholar] [CrossRef]
- Bagheri, A.; Kamalidehghan, B.; Haghshenas, M.; Azadfar, P.; Akbari, L.; Sangtarash, M.H.; Vejdandoust, F.; Ahmadipour, F.; Meng, G.Y.; Houshmand, M. Prevalence of the CYP2D6*10 (C100T), *4 (G1846A), and *14 (G1758A) alleles among Iranians of different ethnicities. Drug Des. Devel. Ther. 2015, 9, 2627–2634. [Google Scholar] [CrossRef]
- Tabari, M.G.; Naseri, F.; Ataby, M.A.; Marjani, A. Genetic Polymorphism of Cytochrome p450 (2C9) Enzyme in Iranian Baluch Ethnic Group. Open Biochem. J. 2015, 9, 37–41. [Google Scholar] [CrossRef][Green Version]
- Tabari, R.G.; Marjani, A.; Ataby, O.A.; Mansourian, A.R.; Samai, N.M. Genetic Polymorphism of Cytochrome p450 (2C19) Enzyme in Iranian Turkman Ethnic Group. Oman. Med. J. 2013, 28, 237–244. [Google Scholar] [CrossRef]
- Fricke-Galindo, I.; Cespedes-Garro, C.; Rodrigues-Soares, F.; Naranjo, M.E.; Delgado, A.; de Andres, F.; Lopez-Lopez, M.; Penas-Lledo, E.; LLerena, A. Interethnic variation of CYP2C19 alleles, ‘predicted’ phenotypes and ‘measured’ metabolic phenotypes across world populations. Pharm. J. 2016, 16, 113–123. [Google Scholar] [CrossRef]
- Gaedigk, A.; Sangkuhl, K.; Whirl-Carrillo, M.; Klein, T.; Leeder, J.S. Prediction of CYP2D6 phenotype from genotype across world populations. Genet. Med. 2017, 19, 69–76. [Google Scholar] [CrossRef]
- Kalman, L.V.; Agundez, J.; Appell, M.L.; Black, J.L.; Bell, G.C.; Boukouvala, S.; Bruckner, C.; Bruford, E.; Caudle, K.; Coulthard, S.A.; et al. Pharmacogenetic allele nomenclature: International workgroup recommendations for test result reporting. Clin. Pharmacol. Ther. 2016, 99, 172–185. [Google Scholar] [CrossRef]
- Williams, J.A.; Ring, B.J.; Cantrell, V.E.; Jones, D.R.; Eckstein, J.; Ruterbories, K.; Hamman, M.A.; Hall, S.D.; Wrighton, S.A. Comparative metabolic capabilities of CYP3A4, CYP3A5, and CYP3A7. Drug Metab. Dispos. 2002, 30, 883–891. [Google Scholar] [CrossRef]
- Payan, M.; Tajik, N.; Rouini, M.R.; Ghahremani, M.H. Genotype and allele frequency of CYP2C19*17 in a healthy Iranian population. Med. J. Islam. Repub. Iran. 2015, 29, 269. [Google Scholar]
- Saghafi, F.; Salehifar, E.; Janbabai, G.; Zaboli, E.; Hedayatizadeh-Omran, A.; Amjadi, O.; Moradi, S. CYP2D6*3 (A2549del), *4 (G1846A), *10 (C100T) and *17 (C1023T) genetic polymorphisms in Iranian breast cancer patients treated with adjuvant tamoxifen. Biomed. Rep. 2018, 9, 446–452. [Google Scholar] [CrossRef] [PubMed]
- Zakeri, S.; Amiri, N.; Pirahmadi, S.; Dinparast, N.D. Genetic variability of CYP2B6 polymorphisms in southeast Iranian population: Implications for malaria and HIV/AIDS treatment. Arch. Iran. Med. 2014, 17, 685–691. [Google Scholar] [PubMed]
- Zhou, S.F.; Yang, L.P.; Zhou, Z.W.; Liu, Y.H.; Chan, E. Insights into the substrate specificity, inhibitors, regulation, and polymorphisms and the clinical impact of human cytochrome P450 1A2. AAPS J. 2009, 11, 481–494. [Google Scholar] [CrossRef] [PubMed]
- Sim, S.C. CYP1A2*1E [corrected] contains the -163C>A substitution and is highly inducible. Pharm. Genom. 2013, 23, 104–105. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.F.; Wang, B.; Yang, L.P.; Liu, J.P. Structure, function, regulation and polymorphism and the clinical significance of human cytochrome P450 1A2. Drug Metab. Rev. 2010, 42, 268–354. [Google Scholar] [CrossRef]
- Myrand, S.P.; Sekiguchi, K.; Man, M.Z.; Lin, X.; Tzeng, R.Y.; Teng, C.H.; Hee, B.; Garrett, M.; Kikkawa, H.; Lin, C.Y.; et al. Pharmacokinetics/genotype associations for major cytochrome P450 enzymes in native and first- and third-generation Japanese populations: Comparison with Korean, Chinese, and Caucasian populations. Clin. Pharmacol. Ther. 2008, 84, 347–361. [Google Scholar] [CrossRef] [PubMed]
- Lang, T.; Klein, K.; Fischer, J.; Nussler, A.K.; Neuhaus, P.; Hofmann, U.; Eichelbaum, M.; Schwab, M.; Zanger, U.M. Extensive genetic polymorphism in the human CYP2B6 gene with impact on expression and function in human liver. Pharmacogenetics 2001, 11, 399–415. [Google Scholar] [CrossRef]
- Zanger, U.M.; Klein, K. Pharmacogenetics of cytochrome P450 2B6 (CYP2B6): Advances on polymorphisms, mechanisms, and clinical relevance. Front. Genet. 2013, 4, 24. [Google Scholar] [CrossRef]
- Bahadur, N.; Leathart, J.B.S.; Mutch, E.; Steimel-Crespi, D.; Dunn, S.A.; Gilissen, R.; Houdt, J.V.; Hendrickx, J.; Mannens, G.; Bohets, H.; et al. CYP2C8 polymorphisms in Caucasians and their relationship with paclitaxel 6 alpha-hydroxylase activity in human liver microsomes. Biochem. Pharmacol. 2002, 64, 1579–1589. [Google Scholar] [CrossRef]
- Kudzi, W.; Dodoo, A.N.; Mills, J.J. Characterisation of CYP2C8, CYP2C9 and CYP2C19 polymorphisms in a Ghanaian population. BMC Med. Genet. 2009, 10, 124. [Google Scholar] [CrossRef]
- Del Tredici, A.L.; Malhotra, A.; Dedek, M.; Espin, F.; Roach, D.; Zhu, G.D.; Voland, J.; Moreno, T.A. Frequency of CYP2D6 Alleles Including Structural Variants in the United States. Front. Pharmacol. 2018, 9, 305. [Google Scholar] [CrossRef]
- Zhang, H.; Sridar, C.; Kenaan, C.; Amunugama, H.; Ballou, D.P.; Hollenberg, P.F. Polymorphic variants of cytochrome P450 2B6 (CYP2B6.4-CYP2B6.9) exhibit altered rates of metabolism for bupropion and efavirenz: A charge-reversal mutation in the K139E variant (CYP2B6.8) impairs formation of a functional cytochrome p450-reductase complex. J. Pharmacol. Exp. Ther. 2011, 338, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Forrester, L.M.; Henderson, C.J.; Glancey, M.J.; Back, D.J.; Park, B.K.; Ball, S.E.; Kitteringham, N.R.; McLaren, A.W.; Miles, J.S.; Skett, P.; et al. Relative expression of cytochrome P450 isoenzymes in human liver and association with the metabolism of drugs and xenobiotics. Biochem. J. 1992, 281, 359–368. [Google Scholar] [CrossRef]
- Rahman, A.; Korzekwa, K.R.; Grogan, J.; Gonzalez, F.J.; Harris, J.W. Selective biotransformation of taxol to 6 alpha-hydroxytaxol by human cytochrome P450 2C8. Cancer Res. 1994, 54, 5543–5546. [Google Scholar] [PubMed]
- Fleming, I.; Michaelis, U.R.; Bredenkotter, D.; Fisslthaler, B.; Dehghani, F.; Brandes, R.P.; Busse, R. Endothelium-derived hyperpolarizing factor synthase (Cytochrome P450 2C9) is a functionally significant source of reactive oxygen species in coronary arteries. Circ. Res. 2001, 88, 44–51. [Google Scholar] [CrossRef]
- Mutch, E.; Blain, P.G.; Williams, F.M. The role of metabolism in determining susceptibility to parathion toxicity in man. Toxicol. Lett. 1999, 107, 177–187. [Google Scholar] [CrossRef]
- Agundez, J.A.; Garcia-Martin, E.; Martinez, C. Genetically based impairment in CYP2C8- and CYP2C9-dependent NSAID metabolism as a risk factor for gastrointestinal bleeding: Is a combination of pharmacogenomics and metabolomics required to improve personalized medicine? Expert. Opin. Drug Metab. Toxicol. 2009, 5, 607–620. [Google Scholar] [CrossRef]
- Hirota, T.; Eguchi, S.; Ieiri, I. Impact of genetic polymorphisms in CYP2C9 and CYP2C19 on the pharmacokinetics of clinically used drugs. Drug Metab. Pharm. 2013, 28, 28–37. [Google Scholar] [CrossRef]
- Haining, R.L.; Hunter, A.P.; Veronese, M.E.; Trager, W.F.; Rettie, A.E. Allelic variants of human cytochrome P450 2C9: Baculovirus-mediated expression, purification, structural characterization, substrate stereoselectivity, and prochiral selectivity of the wild-type and I359L mutant forms. Arch. Biochem. Biophys. 1996, 333, 447–458. [Google Scholar] [CrossRef] [PubMed]
- Rettie, A.E.; Wienkers, L.C.; Gonzalez, F.J.; Trager, W.F.; Korzekwa, K.R. Impaired (S)-warfarin metabolism catalysed by the R144C allelic variant of CYP2C9. Pharmacogenetics 1994, 4, 39–42. [Google Scholar] [CrossRef] [PubMed]
- Scott, S.A.; Sangkuhl, K.; Shuldiner, A.R.; Hulot, J.S.; Thorn, C.F.; Altman, R.B.; Klein, T.E. PharmGKB summary: Very important pharmacogene information for cytochrome P450, family 2, subfamily C, polypeptide 19. Pharm. Genom. 2012, 22, 159–165. [Google Scholar] [CrossRef]
- Hulot, J.S.; Bura, A.; Villard, E.; Azizi, M.; Remones, V.; Goyenvalle, C.; Aiach, M.; Lechat, P.; Gaussem, P. Cytochrome P450 2C19 loss-of-function polymorphism is a major determinant of clopidogrel responsiveness in healthy subjects. Blood 2006, 108, 2244–2247. [Google Scholar] [CrossRef] [PubMed]
- Mega, J.L.; Simon, T.; Collet, J.P.; Anderson, J.L.; Antman, E.M.; Bliden, K.; Cannon, C.P.; Danchin, N.; Giusti, B.; Gurbel, P.; et al. Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: A meta-analysis. JAMA 2010, 304, 1821–1830. [Google Scholar] [CrossRef]
- Manolopoulos, V.G. Pharmacogenomics and adverse drug reactions in diagnostic and clinical practice. Clin. Chem. Lab. Med. 2007, 45, 801–814. [Google Scholar] [CrossRef] [PubMed]
- Guengerich, F.P.; Kim, D.H.; Iwasaki, M. Role of human cytochrome P-450 IIE1 in the oxidation of many low molecular weight cancer suspects. Chem. Res. Toxicol. 1991, 4, 168–179. [Google Scholar] [CrossRef]
- Peter, R.; Bocker, R.; Beaune, P.H.; Iwasaki, M.; Guengerich, F.P.; Yang, C.S. Hydroxylation of chlorzoxazone as a specific probe for human liver cytochrome P-450IIE1. Chem. Res. Toxicol. 1990, 3, 566–573. [Google Scholar] [CrossRef]
- Bolt, H.M.; Roos, P.H.; Thier, R. The cytochrome P-450 isoenzyme CYP2E1 in the biological processing of industrial chemicals: Consequences for occupational and environmental medicine. Int. Arch. Occup. Environ. Health 2003, 76, 174–185. [Google Scholar] [CrossRef]
- Elens, L.; van Gelder, T.; Hesselink, D.A.; Haufroid, V.; van Schaik, R.H. CYP3A4*22: Promising newly identified CYP3A4 variant allele for personalizing pharmacotherapy. Pharmacogenomics 2013, 14, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Okubo, M.; Murayama, N.; Shimizu, M.; Shimada, T.; Guengerich, F.P.; Yamazaki, H. CYP3A4 intron 6 C>T polymorphism (CYP3A4*22) is associated with reduced CYP3A4 protein level and function in human liver microsomes. J. Toxicol. Sci. 2013, 38, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Guo, Y.; Wrighton, S.A.; Cooke, G.E.; Sadee, W. Intronic polymorphism in CYP3A4 affects hepatic expression and response to statin drugs. Pharm. J. 2011, 11, 274–286. [Google Scholar] [CrossRef] [PubMed]
- Lamba, J.; Hebert, J.M.; Schuetz, E.G.; Klein, T.E.; Altman, R.B. PharmGKB summary: Very important pharmacogene information for CYP3A5. Pharm. Genom. 2012, 22, 555–558. [Google Scholar] [CrossRef]
- Lee, S.J.; Goldstein, J.A. Functionally defective or altered CYP3A4 and CYP3A5 single nucleotide polymorphisms and their detection with genotyping tests. Pharmacogenomics 2005, 6, 357–371. [Google Scholar] [CrossRef]

| Population | Description | Genomes |
|---|---|---|
| IRN | Iranian Based on Iranome database. Available online: http://www.iranome.ir (accessed on 14 August 2021) | 800 |
| AMR | Latino/Admixed American Based on Genome database. Available online: https://gnomad.broadinstitute.org (accessed on 14 August 2021) | 6835 |
| EUR | Non-Finnish European Based on Genome database. Available online: https://gnomad.broadinstitute.org (accessed on 14 August 2021) | 32,299 |
| AFR | Based on Genome database. Available online: https://gnomad.broadinstitute.org (accessed on 14 August 2021) | 21,042 |
| EAS | East Asian Based on Genome database. Available online: https://gnomad.broadinstitute.org (accessed on 14 August 2021) | 1567 |
| SAS | South Asian Based on Genome database. Available online: https://gnomad.broadinstitute.org (accessed on 14 August 2021) | 1526 |
| CAUC | Caucasian (based on literature review) | ~100–250 |
| CYP450 Enzyme | Allele | SNP ID | Frequency in IRNs | Similarity with Five Major Populations (Freq.) |
|---|---|---|---|---|
| CYP1A2 | *1F | (rs762551) | 59.9% | AFR (60.1%), SAS (57.7%) |
| CYP2B6 | *9 | (rs3745274) | 26.6% | EUR (24%), Cauc. (28.6%) |
| *5 | (rs3211371) | 8.5% | SAS (8%) | |
| *2 | (rs8192709) | 5.3% | EUR (5.6%), Cauc. (5.3%) | |
| *22 | (rs34223104) | 2.6% | AFR (2.8%), Cauc. (2.4%) | |
| *3 | (rs45482602) | 1.3% | No similarity | |
| CYP2C8 | *4 | (rs1058930) | 2.6% | AMR (2.7%) |
| *2 | (rs11572103) | 2.3% | No similarity | |
| CYP2C9 | *2 | (rs1799853) | 10.5% | No similarity |
| *3 | (rs1057910) | 10.2% | SAS (11%) | |
| CYP2C19 | *2 | (rs4244285) | 13.1% | EUR (14.6%), Cauc. (13.6%) |
| CYP2D6 | *2 | (rs16947, rs1135840) | 47% | No similarity |
| *10 | (rs1065852, rs1135840) | 15.1% | No similarity | |
| *41 | (rs28371725) | 14% | SAS (13.5%) | |
| *4 | (rs3892097) | 11.2% | AMR (11.9%) | |
| CYP2E1 | *4 | (rs6413419) | 5.6% | No similarity |
| CYP3A4 | - | - | - | - |
| CYP3A5 | *3 | (rs776746) | 96.1% | EUR (93%), Cauc. (95.5%) |
| CYP450 Enzyme | Allele | SNP ID | Frequency in IRNs | Frequency in CAUC |
|---|---|---|---|---|
| CYP1A2 | *1F | (rs762551) | 59.9% | 73.7% [40] |
| CYP2B6 | *9 | (rs3745274) | 26.6% | 28.6% [41] |
| *5 | (rs3211371) | 8.5% | 10.9% [41] | |
| *2 | (rs8192709) | 5.3% | 5.3% [41] | |
| *22 | (rs34223104) | 2.6% | 2.4% [42] | |
| *3 | (rs45482602) | 1.3% | <1% [41] | |
| CYP2C8 | *4 | (rs1058930) | 2.6% | 7.5% [43] |
| *2 | (rs11572103) | 2.3% | <1% [44] | |
| CYP2C9 | *2 | (rs1799853) | 10.5% | 13.3% [40] |
| *3 | (rs1057910) | 10.2% | 5.6% [40] | |
| CYP2C19 | *2 | (rs4244285) | 13.1% | 13.6% [40] |
| CYP2D6 | *2 | (rs16947, rs1135840) | 47% | 32.8–52.5% [40] |
| *10 | (rs1065852, rs1135840) | 15.1% | 19.6% [40] | |
| *41 | (rs28371725) | 14% | 9.6% [45] | |
| *4 | (rs3892097) | 11.2% | 18.2% [40] | |
| CYP2E1 | *4 | (rs6413419) | 5.6% | NA |
| CYP3A4 | - | - | - | - |
| CYP3A5 | *3 | (rs776746) | 96.1% | 95.5% [40] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neyshaburinezhad, N.; Ghasim, H.; Rouini, M.; Daali, Y.; Ardakani, Y.H. Frequency of Important CYP450 Enzyme Gene Polymorphisms in the Iranian Population in Comparison with Other Major Populations: A Comprehensive Review of the Human Data. J. Pers. Med. 2021, 11, 804. https://doi.org/10.3390/jpm11080804
Neyshaburinezhad N, Ghasim H, Rouini M, Daali Y, Ardakani YH. Frequency of Important CYP450 Enzyme Gene Polymorphisms in the Iranian Population in Comparison with Other Major Populations: A Comprehensive Review of the Human Data. Journal of Personalized Medicine. 2021; 11(8):804. https://doi.org/10.3390/jpm11080804
Chicago/Turabian StyleNeyshaburinezhad, Navid, Hengameh Ghasim, Mohammadreza Rouini, Youssef Daali, and Yalda H. Ardakani. 2021. "Frequency of Important CYP450 Enzyme Gene Polymorphisms in the Iranian Population in Comparison with Other Major Populations: A Comprehensive Review of the Human Data" Journal of Personalized Medicine 11, no. 8: 804. https://doi.org/10.3390/jpm11080804
APA StyleNeyshaburinezhad, N., Ghasim, H., Rouini, M., Daali, Y., & Ardakani, Y. H. (2021). Frequency of Important CYP450 Enzyme Gene Polymorphisms in the Iranian Population in Comparison with Other Major Populations: A Comprehensive Review of the Human Data. Journal of Personalized Medicine, 11(8), 804. https://doi.org/10.3390/jpm11080804








