A Novel Approach for the Discovery of Biomarkers of Radiotherapy Response in Breast Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Irradiation of Cells and Development of Radioresistant Cell Lines
2.3. Cell Irradiation and Secretome Sample Preparation
2.4. Liquid Chromatography-Mass Spectrometry and Secretome Analysis
2.5. Lactate Dehydrogenase Assay
2.6. RNA Extraction and Whole-Transcriptome Gene Expression Analysis
2.7. Protein Isolation and Detection
2.8. Murine Xenograft Experiments
2.9. Human Breast Tissue Experiments
2.10. Immunohistochemistry
2.11. Statistical Analysis
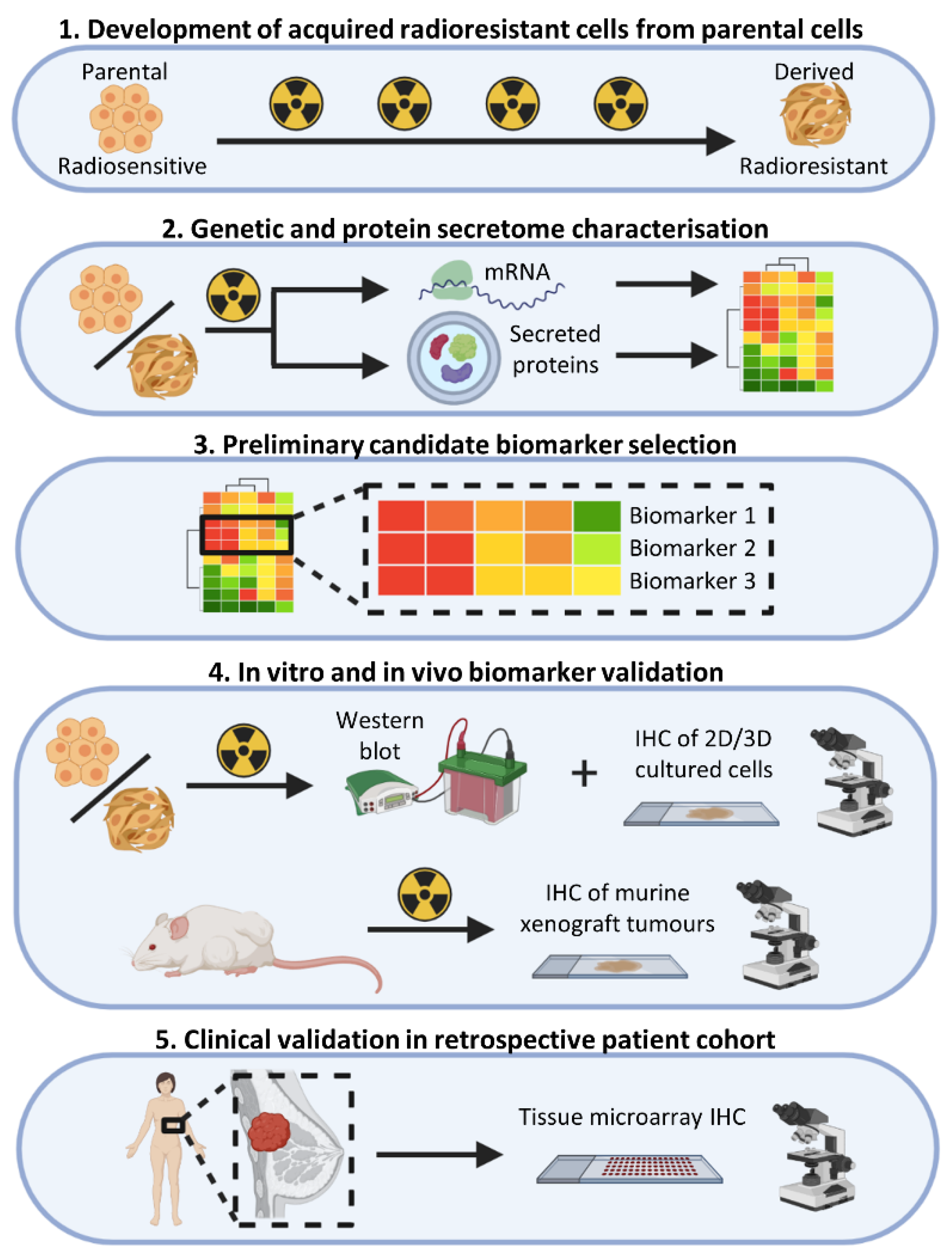
3. Results
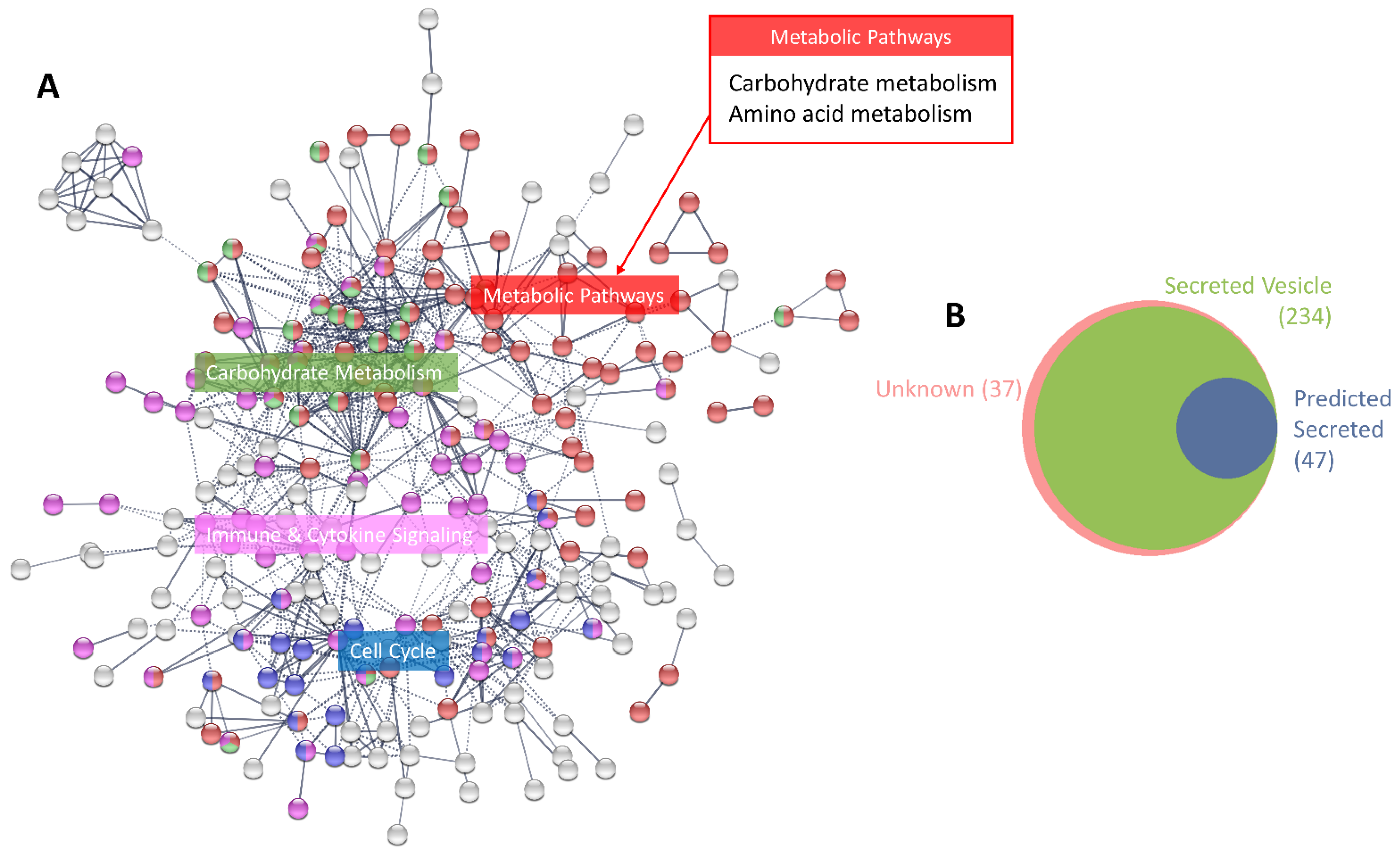
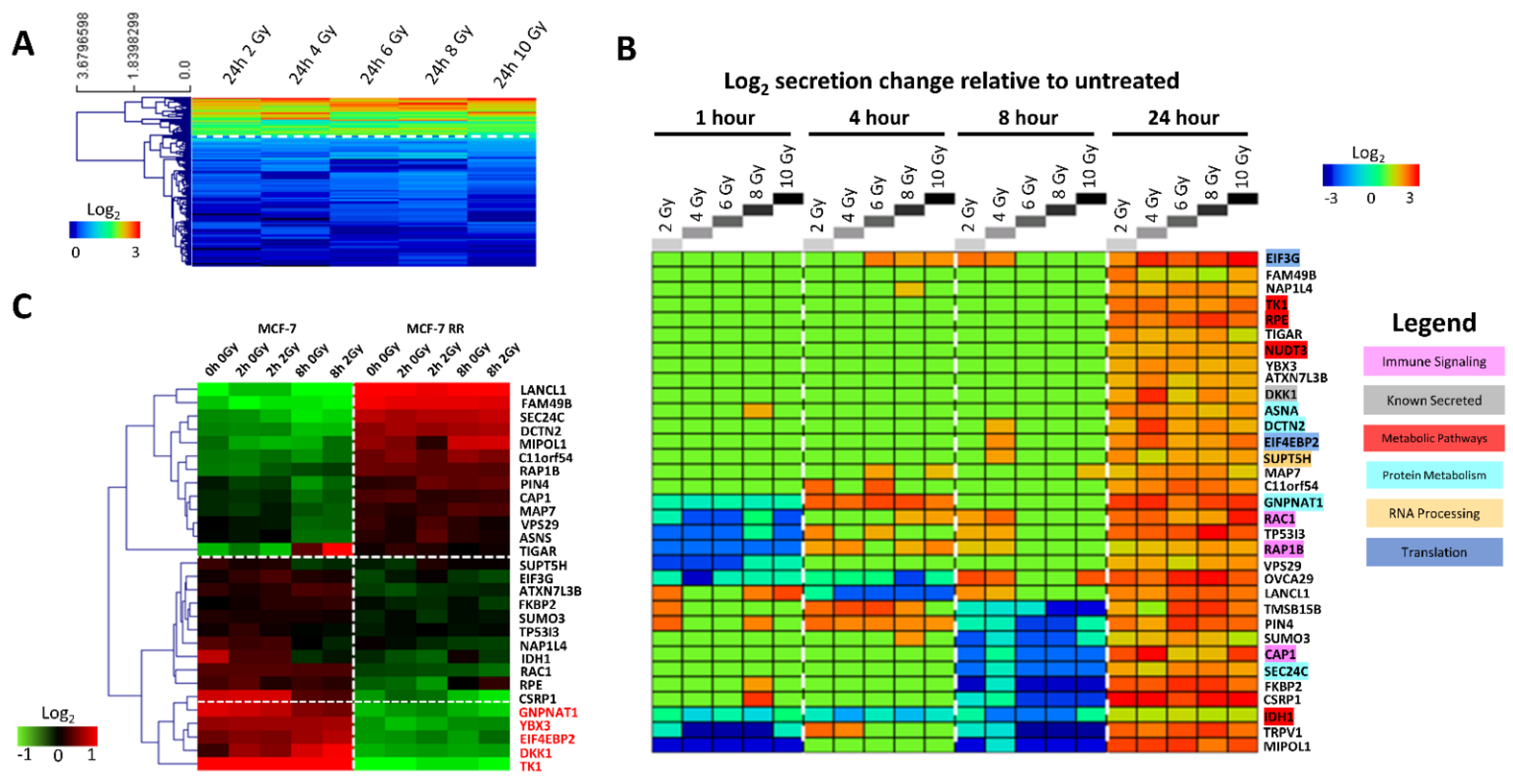
3.1. Characterisation of the MCF-7 Basal Secretome
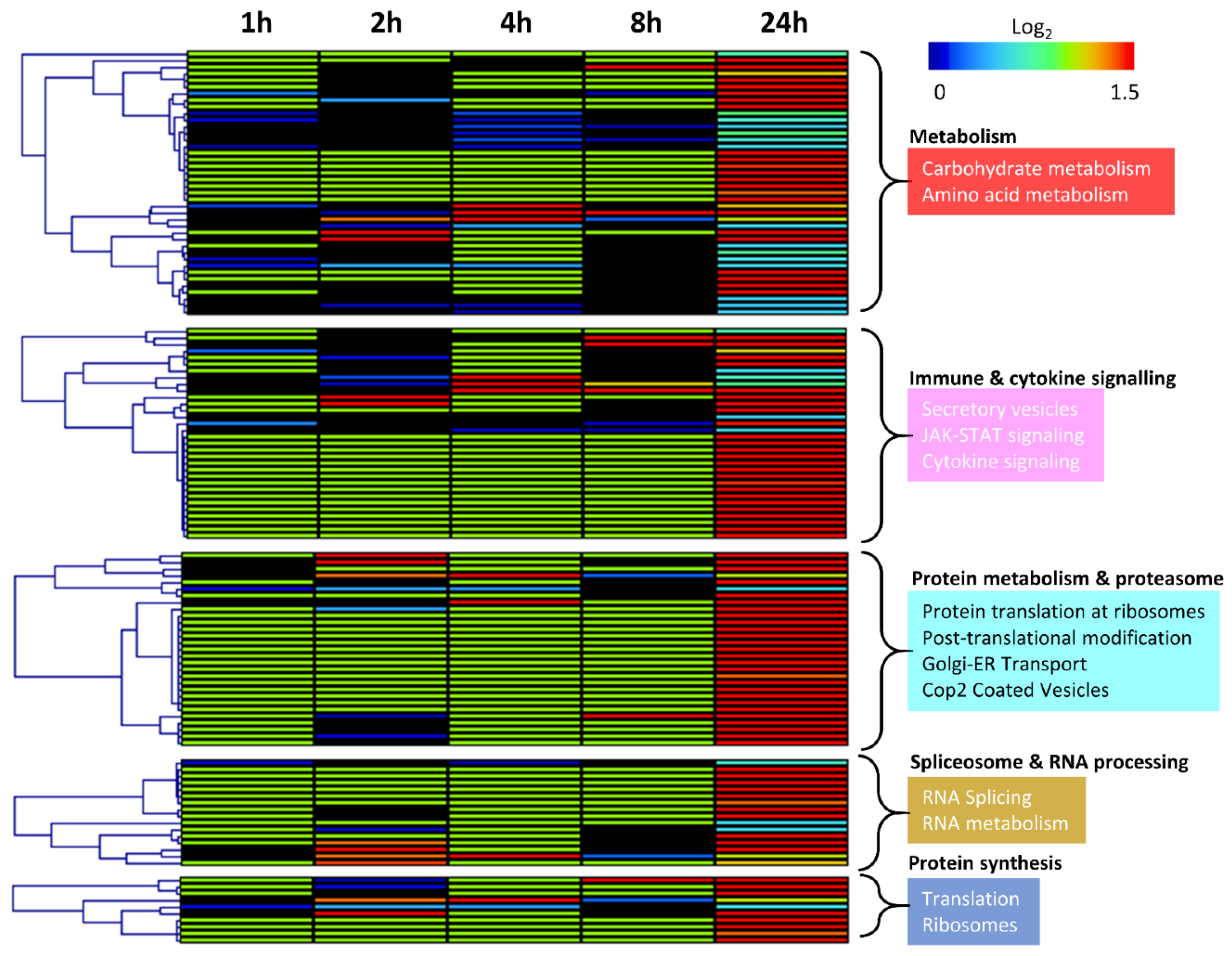
3.2. Characterisation of the MCF-7 Radiation-Induced Secretome
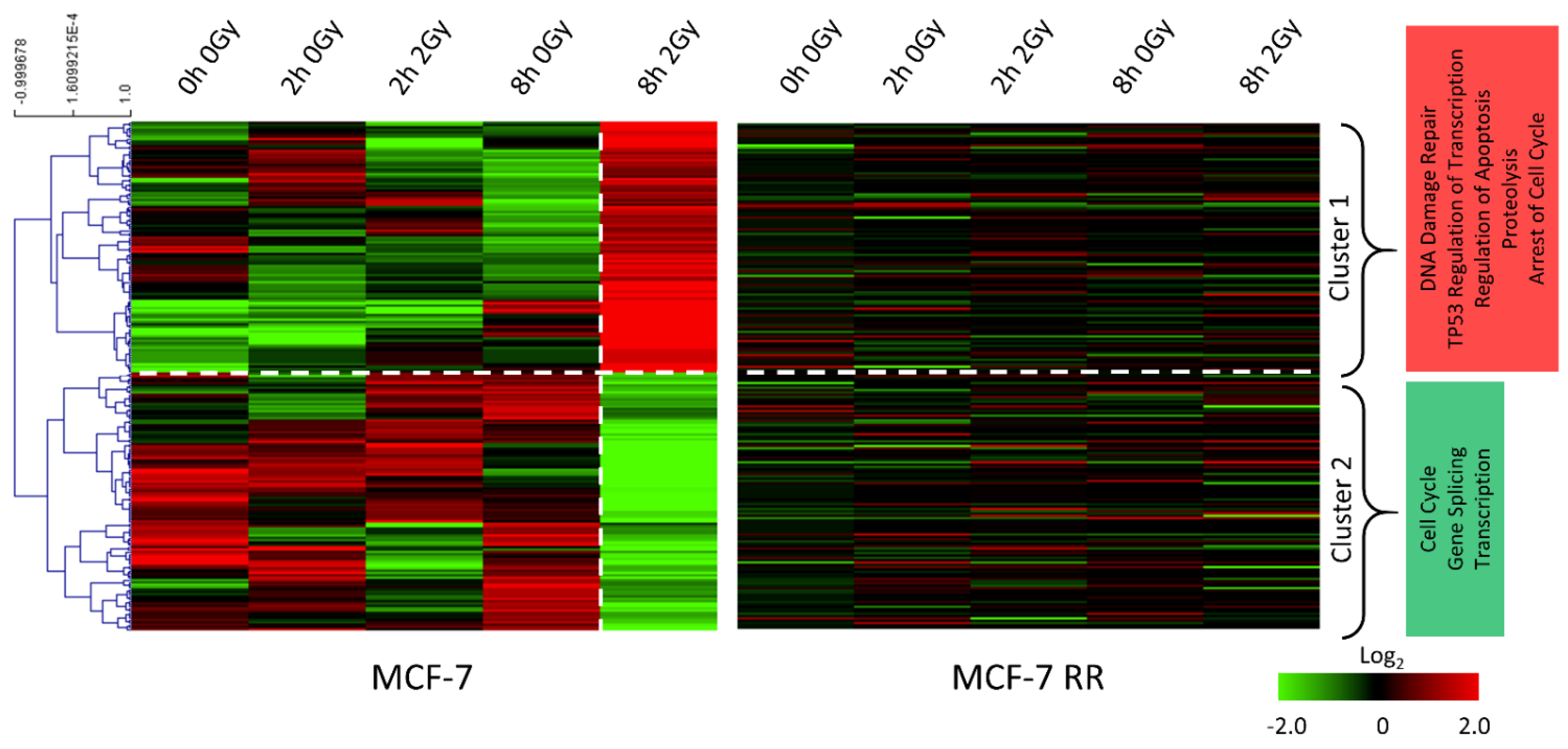
3.3. Gene Expression Changes Associated with Response to Radiation in Parental Radiosensitive and Derived Radioresistant MCF-7 Cells
3.4. MCF-7 Candidate Biomarker Selection
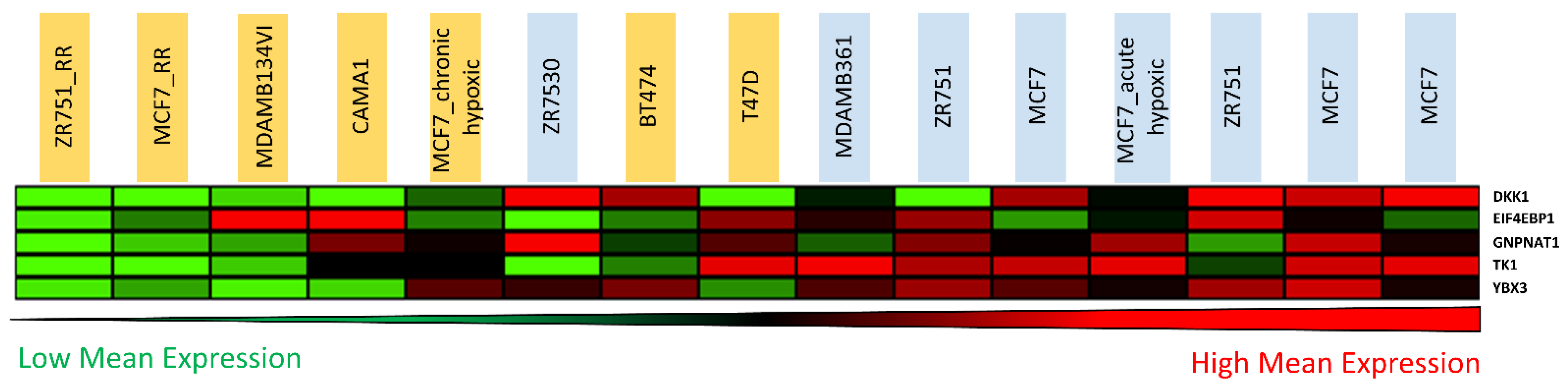
3.5. Candidate Biomarker Expression and Intrinsic Sensitivity to Radiation
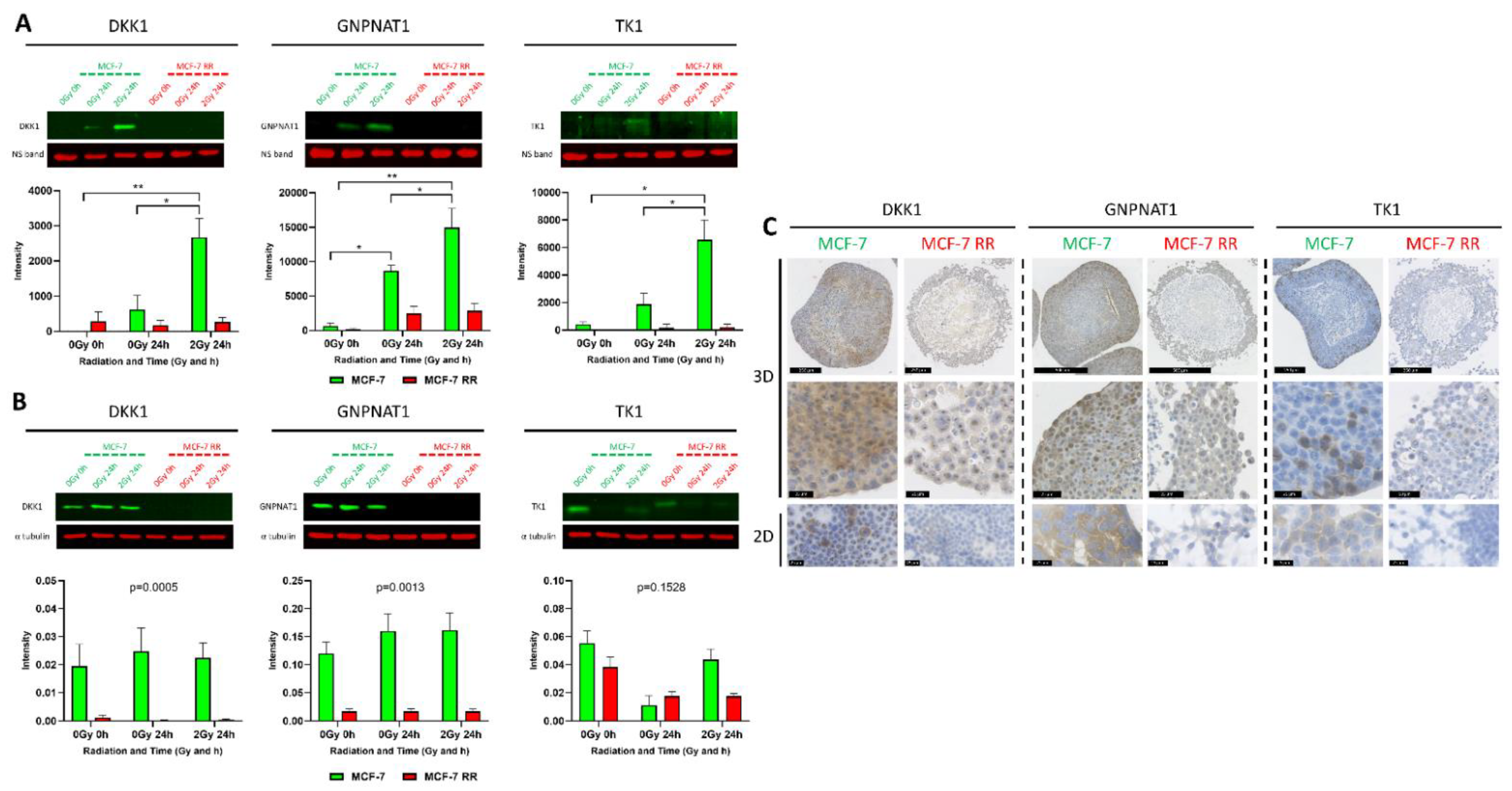
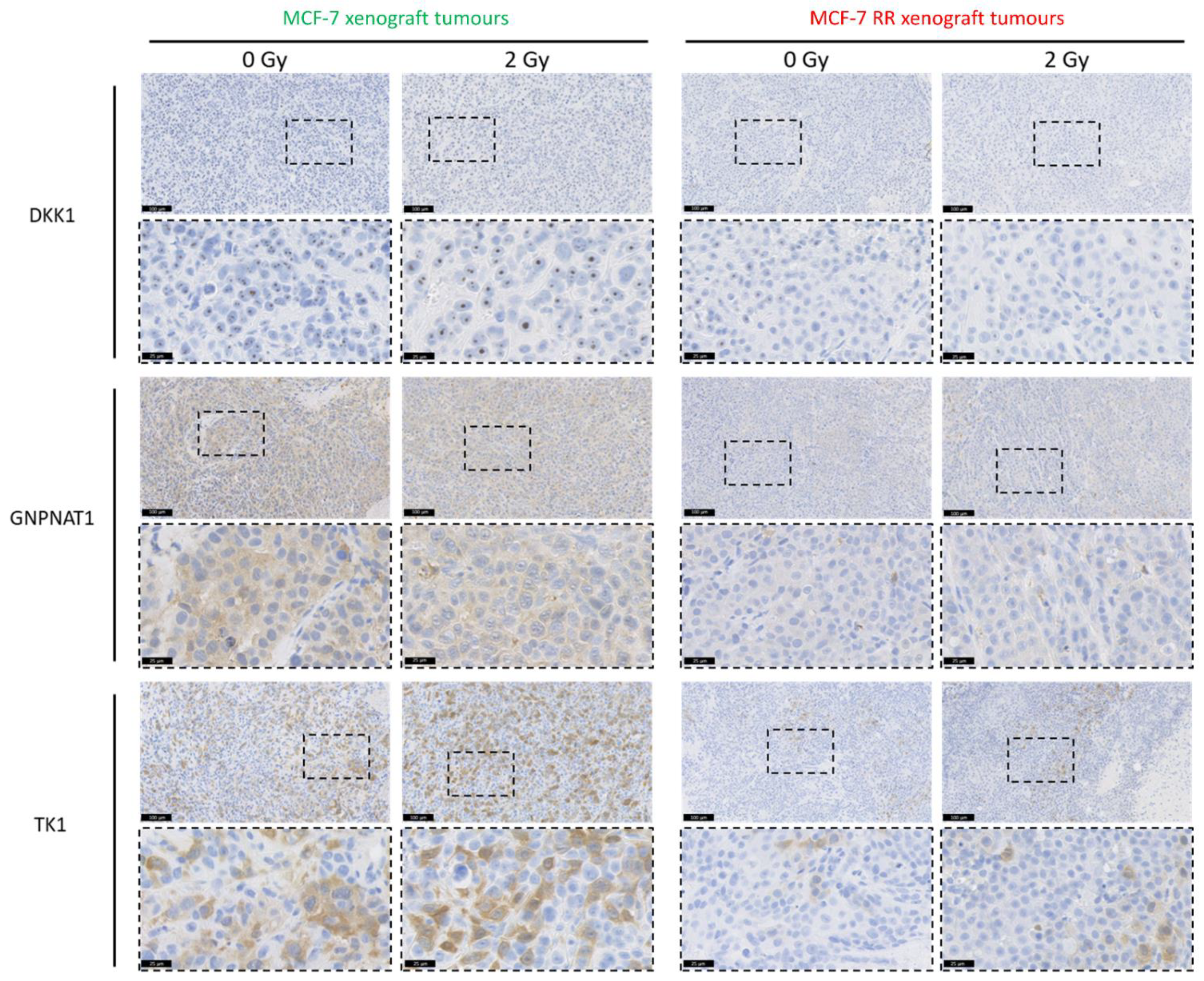
3.6. In Vitro and In Vivo Validation of Candidate Biomarkers
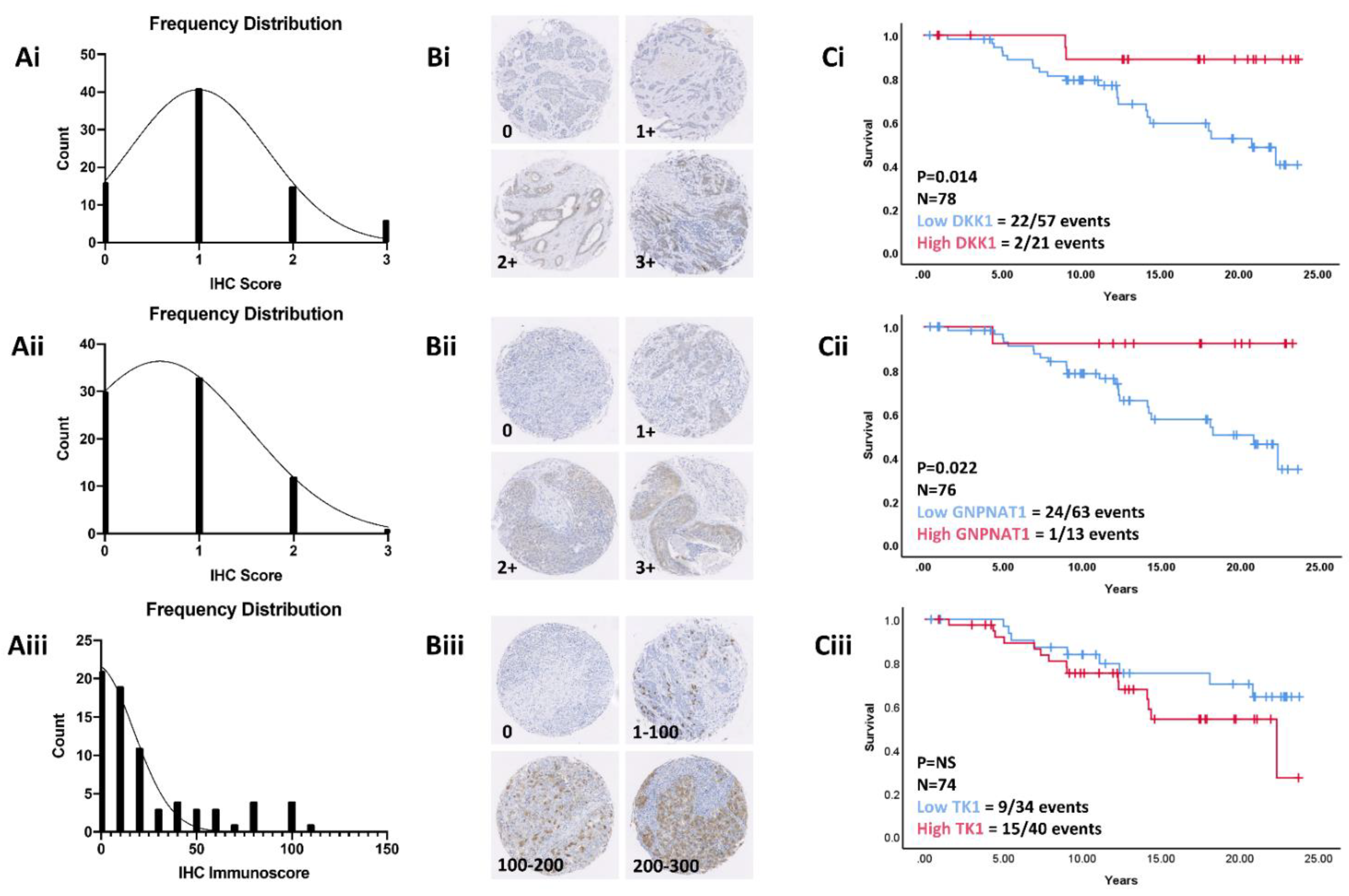
3.7. Validation in a Retrospective Patient Cohort
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Connell, P.P.; Hellman, S. Advances in radiotherapy and implications for the next century: A historical perspective. Cancer Res. 2009, 69, 383–392. [Google Scholar] [CrossRef]
- Delaney, G.; Jacob, S.; Featherstone, C.; Barton, M. The role of radiotherapy in cancer treatment: Estimating optimal utilization from a review of evidence-based clinical guidelines. Cancer Interdiscip. Int. J. Am. Cancer Soc. 2005, 104, 1129–1137. [Google Scholar] [CrossRef]
- Onitilo, A.A.; Engel, J.M.; Stankowski, R.V.; Doi, S.A. Survival comparisons for breast conserving surgery and mastectomy revisited: Community experience and the role of radiation therapy. Clin. Med. Res. 2015, 13, 65–73. [Google Scholar] [CrossRef]
- Cao, J.; Olson, R.; Tyldesley, S. Comparison of recurrence and survival rates after breast-conserving therapy and mastectomy in young women with breast cancer. Curr. Oncol. 2013, 20, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Poortmans, P. Evidence based radiation oncology: Breast cancer. Radiother. Oncol. 2007, 84, 84–101. [Google Scholar] [CrossRef] [PubMed]
- Allemani, C.; Sant, M.; Weir, H.K.; Richardson, L.C.; Baili, P.; Storm, H.; Siesling, S.; Torrella-Ramos, A.; Voogd, A.C.; Aareleid, T. Breast cancer survival in the US and Europe: A CONCORD high-resolution study. Int. J. Cancer 2013, 132, 1170–1181. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-K.; Jeon, H.-Y.; Kim, H. The molecular mechanisms underlying the therapeutic resistance of cancer stem cells. Arch. Pharmacal Res. 2015, 38, 389–401. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Graham, P.H.; Ni, J.; Hao, J.; Bucci, J.; Cozzi, P.J.; Li, Y. Targeting PI3K/Akt/mTOR signaling pathway in the treatment of prostate cancer radioresistance. Crit. Rev. Oncol. Hematol. 2015, 96, 507–517. [Google Scholar] [CrossRef]
- Shapiro, C.L.; Recht, A. Side effects of adjuvant treatment of breast cancer. N. Engl. J. Med. 2001, 344, 1997–2008. [Google Scholar] [CrossRef] [PubMed]
- Nix, P.; Cawkwell, L.; Patmore, H.; Greenman, J.; Stafford, N. Bcl-2 expression predicts radiotherapy failure in laryngeal cancer. Br. J. Cancer 2005, 92, 2185–2189. [Google Scholar] [CrossRef]
- Meehan, J.; Gray, M.; Martínez-Pérez, C.; Kay, C.; Pang, L.Y.; Fraser, J.A.; Poole, A.V.; Kunkler, I.H.; Langdon, S.P.; Argyle, D.; et al. Precision Medicine and the Role of Biomarkers of Radiotherapy Response in Breast Cancer. Front. Oncol. 2020, 10, 628. [Google Scholar] [CrossRef] [PubMed]
- Paik, S.; Shak, S.; Tang, G.; Kim, C.; Baker, J.; Cronin, M.; Baehner, F.L.; Walker, M.G.; Watson, D.; Park, T. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 2004, 351, 2817–2826. [Google Scholar] [CrossRef] [PubMed]
- Tutt, A.; Wang, A.; Rowland, C.; Gillett, C.; Lau, K.; Chew, K.; Dai, H.; Kwok, S.; Ryder, K.; Shu, H. Risk estimation of distant metastasis in node-negative, estrogen receptor-positive breast cancer patients using an RT-PCR based prognostic expression signature. BMC Cancer 2008, 8, 339. [Google Scholar] [CrossRef] [PubMed]
- Van De Vijver, M.J.; He, Y.D.; Van’t Veer, L.J.; Dai, H.; Hart, A.A.; Voskuil, D.W.; Schreiber, G.J.; Peterse, J.L.; Roberts, C.; Marton, M.J. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 2002, 347, 1999–2009. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.S.; Mullins, M.; Cheang, M.C.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009, 27, 1160. [Google Scholar] [CrossRef]
- Ellis, M.J.; Suman, V.J.; Hoog, J.; Lin, L.; Snider, J.; Prat, A.; Parker, J.S.; Luo, J.; DeSchryver, K.; Allred, D.C. Randomized phase II neoadjuvant comparison between letrozole, anastrozole, and exemestane for postmenopausal women with estrogen receptor–rich stage 2 to 3 breast cancer: Clinical and biomarker outcomes and predictive value of the baseline PAM50-based intrinsic subtype—ACOSOG Z1031. J. Clin. Oncol. 2011, 29, 2342. [Google Scholar]
- Hall, W.A.; Bergom, C.; Thompson, R.F.; Baschnagel, A.M.; Vijayakumar, S.; Willers, H.; Li, X.A.; Schultz, C.J.; Wilson, G.D.; West, C.M. Precision oncology and genomically guided radiation therapy: A report from the American Society for radiation oncology/American association of physicists in medicine/national cancer institute precision medicine conference. Int. J. Radiat. Oncol. Biol. Phys. 2018, 101, 274–284. [Google Scholar] [CrossRef] [PubMed]
- Bernier, J. Precision medicine for early breast cancer radiotherapy: Opening up new horizons? Crit. Rev. Oncol. Hematol. 2017, 113, 79–82. [Google Scholar] [CrossRef]
- Speers, C.; Zhao, S.; Liu, M.; Bartelink, H.; Pierce, L.J.; Feng, F.Y. Development and Validation of a Novel Radiosensitivity Signature in Human Breast Cancer. Clin. Cancer Res. 2015, 21, 3667–3677. [Google Scholar] [CrossRef]
- Forrest, A.P.; Stewart, H.J.; Everington, D.; Prescott, R.J.; McArdle, C.S.; Harnett, A.N.; Smith, D.C.; George, W.D. Scottish Cancer Trials Breast Group. Randomised controlled trial of conservation therapy for breast cancer: 6-year analysis of the Scottish trial. Lancet 1996, 348, 708–713. [Google Scholar] [CrossRef]
- Sjöström, M.; Chang, S.L.; Fishbane, N.; Davicioni, E.; Zhao, S.G.; Hartman, L.; Holmberg, E.; Feng, F.Y.; Speers, C.W.; Pierce, L.J.; et al. Clinicogenomic Radiotherapy Classifier Predicting the Need for Intensified Locoregional Treatment After Breast-Conserving Surgery for Early-Stage Breast Cancer. J. Clin. Oncol. 2019, 37, 3340–3349. [Google Scholar] [CrossRef] [PubMed]
- Eschrich, S.A.; Fulp, W.J.; Pawitan, Y.; Foekens, J.A.; Smid, M.; Martens, J.W.M.; Echevarria, M.; Kamath, V.; Lee, J.-H.; Harris, E.E.; et al. Validation of a Radiosensitivity Molecular Signature in Breast Cancer. Clin. Cancer Res. 2012, 18, 5134–5143. [Google Scholar] [CrossRef]
- Mbeunkui, F.; Metge, B.J.; Shevde, L.A.; Pannell, L.K. Identification of Differentially Secreted Biomarkers Using LC-MS/MS in Isogenic Cell Lines Representing a Progression of Breast Cancer. J. Proteome Res. 2007, 6, 2993–3002. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Huuskonen, J.; Hajivandi, M.; Manzanedo, R.; Predki, P.; Amshey, J.R.; Pope, R.M. Identification and quantification of proteins differentially secreted by a pair of normal and malignant breast-cancer cell lines. Proteomics 2009, 9, 182–193. [Google Scholar] [CrossRef]
- Lai, T.-C.; Chou, H.-C.; Chen, Y.-W.; Lee, T.-R.; Chan, H.-T.; Shen, H.-H.; Lee, W.-T.; Lin, S.-T.; Lu, Y.-C.; Wu, C.-L.; et al. Secretomic and Proteomic Analysis of Potential Breast Cancer Markers by Two-Dimensional Differential Gel Electrophoresis. J. Proteome Res. 2010, 9, 1302–1322. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Zhang, Y.; Chen, K.; Hu, X.; Xu, L.X. Discovery of IL-18 As a Novel Secreted Protein Contributing to Doxorubicin Resistance by Comparative Secretome Analysis of MCF-7 and MCF-7/Dox (Secretome Comparison of MCF-7 and MCF-7/Dox). PLoS ONE 2011, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, F.; Depagne, J.; Hem, S.; Chevillard, S.; Bensimon, J.; Bertrand, P.; Lebeau, J. Accumulation of cyclophilin A isoforms in conditioned medium of irradiated breast cancer cells. Proteomics 2012, 12, 1756–1766. [Google Scholar] [CrossRef]
- Forker, L.-J.; Choudhury, A.; Kiltie, A. Biomarkers of tumour radiosensitivity and predicting benefit from radiotherapy. Clin. Oncol. 2015, 27, 561–569. [Google Scholar] [CrossRef] [PubMed]
- Gray, M.; Turnbull, A.K.; Ward, C.; Meehan, J.; Martinez-Perez, C.; Bonello, M.; Pang, L.Y.; Langdon, S.P.; Kunkler, I.H.; Murray, A.; et al. Development and characterisation of acquired radioresistant breast cancer cell lines. Radiat. Oncol. 2019, 14, 64–83. [Google Scholar] [CrossRef]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, 80–96. [Google Scholar] [CrossRef] [PubMed]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014, 15, 286–293. [Google Scholar] [CrossRef] [PubMed]
- Howe, E.; Holton, K.; Nair, S.; Schlauch, D.; Sinha, R.; Quackenbush, J. MeV: MultiExperiment Viewer. In Biomedical Informatics for Cancer Research; Ochs, M.F., Casagrande, J.T., Davuluri, R.V., Eds.; Springer: Boston, MA, USA, 2010; pp. 267–277. [Google Scholar]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, 362–368. [Google Scholar] [CrossRef] [PubMed]
- Vlasblom, J.; Wodak, S.J. Markov clustering versus affinity propagation for the partitioning of protein interaction graphs. BMC Bioinform. 2009, 10, 99–113. [Google Scholar] [CrossRef]
- Martens, L.; Hermjakob, H.; Jones, P.; Adamski, M.; Taylor, C.; States, D.; Gevaert, K.; Vandekerckhove, J.; Apweiler, R. PRIDE: The proteomics identifications database. Proteomics 2005, 5, 3537–3545. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Leek, J.T.; Johnson, W.E.; Parker, H.S.; Jaffe, A.E.; Storey, J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012, 28, 882–883. [Google Scholar] [CrossRef] [PubMed]
- Turnbull, A.K.; Kitchen, R.R.; Larionov, A.A.; Renshaw, L.; Dixon, J.M.; Sims, A.H. Direct integration of intensity-level data from Affymetrix and Illumina microarrays improves statistical power for robust reanalysis. BMC Med. Genom. 2012, 5, 5–35. [Google Scholar] [CrossRef]
- Sherman, B.T.; Tan, Q.; Collins, J.R.; Alvord, W.G.; Roayaei, J.; Stephens, R.; Baseler, M.W.; Lane, H.C.; Lempicki, R.A. The DAVID Gene Functional Classification Tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007, 8, 183. [Google Scholar]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Croft, D.; O’kelly, G.; Wu, G.; Haw, R.; Gillespie, M.; Matthews, L.; Caudy, M.; Garapati, P.; Gopinath, G.; Jassal, B. Reactome: A database of reactions, pathways and biological processes. Nucleic Acids Res. 2010, 39 (Suppl. 1), D691–D697. [Google Scholar] [CrossRef] [PubMed]
- Fabregat, A.; Jupe, S.; Matthews, L.; Sidiropoulos, K.; Gillespie, M.; Garapati, P.; Haw, R.; Jassal, B.; Korninger, F.; May, B. The reactome pathway knowledgebase. Nucleic Acids Res. 2018, 46, D649–D655. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef]
- Meehan, J.; Ward, C.; Turnbull, A.; Bukowski-Wills, J.; Finch, A.J.; Jarman, E.J.; Xintaropoulou, C.; Martinez-Perez, C.; Gray, M.; Pearson, M. Inhibition of pH regulation as a therapeutic strategy in hypoxic human breast cancer cells. Oncotarget 2017, 8, 42857–42875. [Google Scholar] [CrossRef]
- Kunkler, I.H.; Kerr, G.R.; Thomas, J.S.; Jack, W.J.L.; Bartlett, J.M.S.; Pedersen, H.C.; Cameron, D.A.; Dixon, J.M.; Chetty, U. Impact of Screening and Risk Factors for Local Recurrence and Survival After Conservative Surgery and Radiotherapy for Early Breast Cancer: Results From a Large Series With Long-Term Follow-Up. Int. J. Radiat. Oncol. Biol. Phys. 2012, 83, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Mathivanan, S.; Fahner, C.J.; Reid, G.E.; Simpson, R.J. ExoCarta 2012: Database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012, 40, D1241–D1244. [Google Scholar] [CrossRef] [PubMed]
- Pathan, M.; Fonseka, P.; Chitti, S.V.; Kang, T.; Sanwlani, R.; Van Deun, J.; Hendrix, A.; Mathivanan, S. Vesiclepedia 2019: A compendium of RNA, proteins, lipids and metabolites in extracellular vesicles. Nucleic Acids Res. 2019, 47, D516–D519. [Google Scholar] [CrossRef] [PubMed]
- Pontén, F.; Schwenk, J.M.; Asplund, A.; Edqvist, P.H.D. The Human Protein Atlas as a proteomic resource for biomarker discovery. J. Intern. Med. 2011, 270, 428–446. [Google Scholar] [CrossRef]
- Armenteros, J.J.A.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Käll, L.; Krogh, A.; Sonnhammer, E.L. Advantages of combined transmembrane topology and signal peptide prediction—The Phobius web server. Nucleic Acids Res. 2007, 35 (Suppl. 2), W429–W432. [Google Scholar] [CrossRef] [PubMed]
- Viklund, H.; Bernsel, A.; Skwark, M.; Elofsson, A. SPOCTOPUS: A combined predictor of signal peptides and membrane protein topology. Bioinformatics 2008, 24, 2928–2929. [Google Scholar] [CrossRef]
- Kim, K.H.; Yoo, H.Y.; Joo, K.M.; Jung, Y.; Jin, J.; Kim, Y.; Yoon, S.J.; Choi, S.H.; Seol, H.J.; Park, W.-Y.; et al. Time-course analysis of DNA damage response-related genes after in vitro radiation in H460 and H1229 lung cancer cell lines. Exp. Mol. Med. 2011, 43, 419–426. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Torres-Roca, J.F.; Eschrich, S.; Zhao, H.; Bloom, G.; Sung, J.; McCarthy, S.; Cantor, A.B.; Scuto, A.; Li, C.; Zhang, S.; et al. Prediction of Radiation Sensitivity Using a Gene Expression Classifier. Cancer Res. 2005, 65, 7169–7176. [Google Scholar] [CrossRef]
- Choi, C.; Park, S.; Cho, W.K.; Choi, D.H. Cyclin D1 is Associated with Radiosensitivity of Triple-Negative Breast Cancer Cells to Proton Beam Irradiation. Int. J. Mol. Sci. 2019, 20, 4943. [Google Scholar] [CrossRef]
- Zhang, C.; Girard, L.; Das, A.; Chen, S.; Zheng, G.; Song, K. Nonlinear Quantitative Radiation Sensitivity Prediction Model Based on NCI-60 Cancer Cell Lines. Sci. World J. 2014, 2014, 903602. [Google Scholar] [CrossRef]
- Amundson, S.A.; Do, K.T.; Vinikoor, L.C.; Lee, R.A.; Koch-Paiz, C.A.; Ahn, J.; Reimers, M.; Chen, Y.; Scudiero, D.A.; Weinstein, J.N. Integrating global gene expression and radiation survival parameters across the 60 cell lines of the National Cancer Institute Anticancer Drug Screen. Cancer Res. 2008, 68, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Björk-Eriksson, T.; West, C.; Karlsson, E.; Mercke, C. Tumor radiosensitivity (SF2) is a prognostic factor for local control in head and neck cancers. Int. J. Radiat. Oncol. Biol. Phys. 2000, 46, 13–19. [Google Scholar] [CrossRef]
- Lamerz, R.; Stieber, P.; Fateh-Moghadam, A. Serum marker combinations in human breast cancer. In Vivo 1993, 7, 607–613. [Google Scholar] [PubMed]
- Dnistrian, A.M.; Schwartz, M.K.; Greenberg, E.J.; Smith, C.A.; Schwartz, D.C. Evaluation of CA M26, CA M29, CA 15-3 and CEA as circulating tumor markers in breast cancer patients. Tumor Biol. 1991, 12, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Ebeling, F.G.; Stieber, P.; Untch, M.; Nagel, D.; Konecny, G.E.; Schmitt, U.M.; Fateh-Moghadam, A.; Seidel, D. Serum CEA and CA 15-3 as prognostic factors in primary breast cancer. Br. J. Cancer 2002, 86, 1217–1222. [Google Scholar] [CrossRef] [PubMed]
- Stieber, P.; Nagel, D.; Ritzke, C.; Rössler, N.; Kirsch, C.; Eiermann, W.; Fateh-Moghadam, A. Significance of bone alkaline phosphatase, CA 15-3 and CEA in the detection of bone metastases during the follow-up of patients suffering from breast carcinoma. Clin. Chem. Lab. Med. 1992, 30, 809–814. [Google Scholar] [CrossRef]
- Vizcarra, E.; Lluch, A.; Cibrian, R.; Jarque, F.; Garcia-Conde, J. CA15. 3, CEA and TPA tumor markers in the early diagnosis of breast cancer relapse. Oncology 1994, 51, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Duffy, M.J.; Evoy, D.; McDermott, E.W. CA 15-3: Uses and limitation as a biomarker for breast cancer. Clin. Chim. Acta 2010, 411, 1869–1874. [Google Scholar] [CrossRef]
- Ludovini, V.; Gori, S.; Colozza, M.; Pistola, L.; Rulli, E.; Floriani, I.; Pacifico, E.; Tofanetti, F.R.; Sidoni, A.; Basurto, C.; et al. Evaluation of serum HER2 extracellular domain in early breast cancer patients: Correlation with clinicopathological parameters and survival. Ann. Oncol. 2008, 19, 883–890. [Google Scholar] [CrossRef] [PubMed]
- Molina, R.; Augé, J.M.; Escudero, J.M.; Filella, X.; Zanon, G.; Pahisa, J.; Farrus, B.; Muñoz, M.; Velasco, M. Evaluation of tumor markers (HER-2/neu oncoprotein, CEA, and CA 15.3) in patients with locoregional breast cancer: Prognostic value. Tumor Biol. 2010, 31, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Asgeirsson, K.S.; Agrawal, A.; Allen, C.; Hitch, A.; Ellis, I.O.; Chapman, C.; Cheung, K.L.; Robertson, J.F. Serum epidermal growth factor receptor and HER2 expression in primary and metastatic breast cancer patients. Breast Cancer Res. 2007, 9, R75. [Google Scholar] [CrossRef] [PubMed]
- Leyland-Jones, B.; Smith, B.R. Serum HER2 testing in patients with HER2-positive breast cancer: The death knell tolls. Lancet Oncol. 2011, 12, 286–295. [Google Scholar] [CrossRef]
- Villarreal, L.; Méndez, O.; Salvans, C.; Gregori, J.; Baselga, J.; Villanueva, J. Unconventional secretion is a major contributor of cancer cell line secretomes. Mol. Cell. Proteom. MCP 2013, 12, 1046–1060. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, Y.S.; Moresco, J.J.; Yates, J.R., III; Nardulli, A.M. Integration of Breast Cancer Secretomes with Clinical Data Elucidates Potential Serum Markers for Disease Detection, Diagnosis, and Prognosis. PLoS ONE 2016, 11, e0158296. [Google Scholar] [CrossRef]
- Shin, J.; Kim, G.; Lee, J.W.; Lee, J.E.; Kim, Y.S.; Yu, J.H.; Lee, S.T.; Ahn, S.H.; Kim, H.; Lee, C. Identification of ganglioside GM2 activator playing a role in cancer cell migration through proteomic analysis of breast cancer secretomes. Cancer Sci. 2016, 107, 828–835. [Google Scholar] [CrossRef]
- Blache, U.; Horton, E.R.; Xia, T.; Schoof, E.M.; Blicher, L.H.; Schönenberger, A.; Snedeker, J.G.; Martin, I.; Erler, J.T.; Ehrbar, M. Mesenchymal stromal cell activation by breast cancer secretomes in bioengineered 3D microenvironments. Life Sci. Alliance 2019, 2, e201900304. [Google Scholar] [CrossRef]
- Guo, W.; Li, H.; Zhu, Y.; Lan, L.; Yang, S.; Drukker, K.; Morris, E.A.; Burnside, E.S.; Whitman, G.J.; Giger, M.L. Prediction of clinical phenotypes in invasive breast carcinomas from the integration of radiomics and genomics data. J. Med. Imaging 2015, 2, 041007. [Google Scholar] [CrossRef]
- Lin, Q.; Tan, H.T.; Lim, H.S.R.; Chung, M.C. Sieving through the cancer secretome. Biochim. Biophys. Acta BBA Proteins Proteom. 2013, 1834, 2360–2371. [Google Scholar] [CrossRef]
- Cooper, S. Reappraisal of serum starvation, the restriction point, G0, and G1 phase arrest points. FASEB J. 2003, 17, 333–340. [Google Scholar] [CrossRef]
- Shin, J.-S.; Hong, S.-W.; Lee, S.-L.O.; Kim, T.-H.; Park, I.-C.; An, S.-K.; Lee, W.-K.; Lim, J.-S.; Kim, K.-I.; Yang, Y. Serum starvation induces G1 arrest through suppression of Skp2-CDK2 and CDK4 in SK-OV-3 cells. Int. J. Oncol. 2008, 32, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Hasan, N.M.; Adams, G.E.; Joiner, M.C. Effect of serum starvation on expression and phosphorylation of PKCα and p53 in V79 cells: Implications for cell death. Int. J. Cancer 1999, 80, 400–405. [Google Scholar] [CrossRef]
- Zander, L.; Bemark, M. Identification of genes deregulated during serum-free medium adaptation of a Burkitt’s lymphoma cell line. Cell Prolif. 2008, 41, 136–155. [Google Scholar] [CrossRef]
- Yamaguchi, N.; Yamamura, Y.; Koyama, K.; Ohtsuji, E.; Imanishi, J.; Ashihara, T. Characterization of new human pancreatic cancer cell lines which propagate in a protein-free chemically defined medium. Cancer Res. 1990, 50, 7008–7014. [Google Scholar] [PubMed]
- Inoue, Y.; Kawamoto, S.; Shoji, M.; Hashizume, S.; Teruya, K.; Katakura, Y.; Shirahata, S. Properties of ras-amplified recombinant BHK-21 cells in protein-free culture. Cytotechnology 2000, 33, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Mbeunkui, F.; Fodstad, O.; Pannell, L.K. Secretory protein enrichment and analysis: An optimized approach applied on cancer cell lines using 2D LC− MS/MS. J. Proteome Res. 2006, 5, 899–906. [Google Scholar] [CrossRef]
- Pellitteri-Hahn, M.; Warren, M.; Didier, D.; Winkler, E.; Mirza, S.; Greene, A.; Olivier, M. Improved mass spectrometric proteomic profiling of the secretome of rat vascular endothelial cells. J. Proteome Res. 2006, 5, 2861–2864. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, R.; Fujiwara, Y.; Ikari, K.; Hamada, K.; Otomo, A.; Yasuda, K.; Noda, M.; Kaburagi, Y. Extracellular proteome of human hepatoma cell, HepG2 analyzed using two-dimensional liquid chromatography coupled with tandem mass spectrometry. Mol. Cell. Biochem. 2007, 298, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Pardo, M.; García, Á.; Antrobus, R.; Blanco, M.J.; Dwek, R.A.; Zitzmann, N. Biomarker Discovery from Uveal Melanoma Secretomes: Identification of gp100 and Cathepsin D in Patient Serum. J. Proteome Res. 2007, 6, 2802–2811. [Google Scholar] [CrossRef] [PubMed]
- Brandi, J.; Manfredi, M.; Speziali, G.; Gosetti, F.; Marengo, E.; Cecconi, D. Proteomic approaches to decipher cancer cell secretome. Semin. Cell Dev. Biol. 2018, 78, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Nagarajan, A.; Uchil, P.D. Analysis of Cell Viability by the Lactate Dehydrogenase Assay. Cold Spring Harb. Protoc. 2018, 2018. [Google Scholar] [CrossRef] [PubMed]
- Willms, E.; Johansson, H.J.; Mäger, I.; Lee, Y.; Blomberg, K.E.M.; Sadik, M.; Alaarg, A.; Smith, C.I.E.; Lehtiö, J.; El Andaloussi, S.; et al. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci. Rep. 2016, 6, 22519. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.C.; Hsu, C.W.; Chen, C.D.; Yu, C.J.; Chang, K.P.; Tai, D.I.; Liu, H.P.; Su, W.H.; Chang, Y.S.; Yu, J.S. Candidate serological biomarkers for cancer identified from the secretomes of 23 cancer cell lines and the human protein atlas. Mol. Cell. Proteom. MCP 2010, 9, 1100–1117. [Google Scholar] [CrossRef]
- Tsai, M.H.; Cook, J.A.; Chandramouli, G.V.; DeGraff, W.; Yan, H.; Zhao, S.; Coleman, C.N.; Mitchell, J.B.; Chuang, E.Y. Gene expression profiling of breast, prostate, and glioma cells following single versus fractionated doses of radiation. Cancer Res. 2007, 67, 3845–3852. [Google Scholar] [CrossRef]
- Bosma, S.C.J.; Hoogstraat, M.; van der Leij, F.; de Maaker, M.; Wesseling, J.; Lips, E.; Loo, C.E.; Rutgers, E.J.; Elkhuizen, P.H.M.; Bartelink, H.; et al. Response to Preoperative Radiation Therapy in Relation to Gene Expression Patterns in Breast Cancer Patients. Int. J. Radiat. Oncol. Biol. Phys. 2020, 106, 174–181. [Google Scholar] [CrossRef]
- Mao, B.; Wu, W.; Davidson, G.; Marhold, J.; Li, M.; Mechler, B.M.; Delius, H.; Hoppe, D.; Stannek, P.; Walter, C.; et al. Kremen proteins are Dickkopf receptors that regulate Wnt/beta-catenin signalling. Nature 2002, 417, 664–667. [Google Scholar] [CrossRef]
- Mariz, K.; Ingolf, J.B.; Daniel, H.; Teresa, N.J.; Erich-Franz, S. The Wnt inhibitor dickkopf-1: A link between breast cancer and bone metastases. Clin. Exp. Metastasis 2015, 32, 857–866. [Google Scholar] [CrossRef]
- Pinzone, J.J.; Hall, B.M.; Thudi, N.K.; Vonau, M.; Qiang, Y.-W.; Rosol, T.J.; Shaughnessy, J.D., Jr. The role of Dickkopf-1 in bone development, homeostasis, and disease. Blood 2009, 113, 517–525. [Google Scholar] [CrossRef] [PubMed]
- Gunn, W.G.; Conley, A.; Deininger, L.; Olson, S.D.; Prockop, D.J.; Gregory, C.A. A crosstalk between myeloma cells and marrow stromal cells stimulates production of DKK1 and interleukin-6: A potential role in the development of lytic bone disease and tumor progression in multiple myeloma. Stem Cells 2006, 24, 986–991. [Google Scholar] [CrossRef]
- Heath, D.J.; Chantry, A.D.; Buckle, C.H.; Coulton, L.; Shaughnessy, J.D., Jr.; Evans, H.R.; Snowden, J.A.; Stover, D.R.; Vanderkerken, K.; Croucher, P.I. Inhibiting Dickkopf-1 (Dkk1) removes suppression of bone formation and prevents the development of osteolytic bone disease in multiple myeloma. J. Bone Miner. Res. 2009, 24, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Kasoha, M.; Bohle, R.M.; Seibold, A.; Gerlinger, C.; Juhasz-Böss, I.; Solomayer, E.F. Dickkopf-1 (Dkk1) protein expression in breast cancer with special reference to bone metastases. Clin. Exp. Metastasis 2018, 35, 763–775. [Google Scholar] [CrossRef]
- Zhou, S.-J.; Zhuo, S.-R.; Yang, X.-Q.; Qin, C.-X.; Wang, Z.-L. Serum Dickkopf-1 expression level positively correlates with a poor prognosis in breast cancer. Diagn. Pathol. 2014, 9, 161. [Google Scholar] [CrossRef]
- Akella, N.M.; Ciraku, L.; Reginato, M.J. Fueling the fire: Emerging role of the hexosamine biosynthetic pathway in cancer. BMC Biol. 2019, 17, 52. [Google Scholar] [CrossRef] [PubMed]
- Chu, J.; Li, N.; Gai, W. Identification of genes that predict the biochemical recurrence of prostate cancer. Oncol. Lett. 2018, 16, 3447–3452. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, A.K.; Shojaie, A.; Panzitt, K.; Sonavane, R.; Venghatakrishnan, H.; Manikkam, M.; Zaslavsky, A.; Putluri, V.; Vasu, V.T.; Zhang, Y.; et al. Inhibition of the hexosamine biosynthetic pathway promotes castration-resistant prostate cancer. Nat. Commun. 2016, 7, 11612. [Google Scholar] [CrossRef]
- Liu, W.; Jiang, K.; Wang, J.; Mei, T.; Zhao, M.; Huang, D. Upregulation of GNPNAT1 Predicts Poor Prognosis and Correlates With Immune Infiltration in Lung Adenocarcinoma. Front. Mol. Biosci. 2021, 8, 605754. [Google Scholar] [CrossRef]
- Zheng, X.; Li, Y.; Ma, C.; Zhang, J.; Zhang, Y.; Fu, Z.; Luo, H. Independent Prognostic Potential of GNPNAT1 in Lung Adenocarcinoma. Biomed. Res. Int. 2020, 2020, 8851437. [Google Scholar] [CrossRef]
- Chokchaitaweesuk, C.; Kobayashi, T.; Izumikawa, T.; Itano, N. Enhanced hexosamine metabolism drives metabolic and signaling networks involving hyaluronan production and O-GlcNAcylation to exacerbate breast cancer. Cell Death Dis. 2019, 10, 803. [Google Scholar] [CrossRef] [PubMed]
- Aufderklamm, S.; Todenhöfer, T.; Gakis, G.; Kruck, S.; Hennenlotter, J.; Stenzl, A.; Schwentner, C. Thymidine kinase and cancer monitoring. Cancer Lett. 2012, 316, 6–10. [Google Scholar] [CrossRef]
- Eriksson, S.; Munch-Petersen, B.; Johansson, K.; Ecklund, H. Structure and function of cellular deoxyribonucleoside kinases. Cell. Mol. Life Sci. CMLS 2002, 59, 1327–1346. [Google Scholar] [CrossRef] [PubMed]
- Hurwitz, S.N.; Rider, M.A.; Bundy, J.L.; Liu, X.; Singh, R.K.; Meckes, D.G. Proteomic profiling of NCI-60 extracellular vesicles uncovers common protein cargo and cancer type-specific biomarkers. Oncotarget 2016, 7, 86999–87015. [Google Scholar] [CrossRef] [PubMed]
- Hong, B.S.; Cho, J.H.; Kim, H.; Choi, E.J.; Rho, S.; Kim, J.; Kim, J.H.; Choi, D.S.; Kim, Y.K.; Hwang, D.; et al. Colorectal cancer cell-derived microvesicles are enriched in cell cycle-related mRNAs that promote proliferation of endothelial cells. BMC Genom. 2009, 10, 556–578. [Google Scholar] [CrossRef] [PubMed]
- Skog, J.; Wurdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Sena-Esteves, M.; Curry, W.T., Jr.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef] [PubMed]
- Sinha, A.; Ignatchenko, V.; Ignatchenko, A.; Mejia-Guerrero, S.; Kislinger, T. In-depth proteomic analyses of ovarian cancer cell line exosomes reveals differential enrichment of functional categories compared to the NCI 60 proteome. Biochem. Biophys. Res. Commun. 2014, 445, 694–701. [Google Scholar] [CrossRef] [PubMed]
- Topolcan, O.; Holubec, L., Jr. The role of thymidine kinase in cancer diseases. Expert Opin. Med. Diagn. 2008, 2, 129–141. [Google Scholar] [CrossRef]
- Li, H.; Lei, D.; Wang, X.; Skog, S.; He, Q. Serum thymidine kinase 1 is a prognostic and monitoring factor in patients with non-small cell lung cancer. Oncol. Rep. 2005, 13, 145–149. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Wu, J.; Wang, N.; He, L.; Wu, C.; He, Q.; Skog, S. A Comparative Study: Immunohistochemical Detection of Cytosolic Thymidine Kinase and Proliferating Cell Nuclear Antigen in Breast Cancer. Cancer Investig. 2002, 20, 922–931. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Zou, L.; Zhang, P.; Lui, J.; Skog, S.; Fornander, T. The clinical significance of thymidine kinase 1 measurement in serum of breast cancer patients using anti-TK1 antibody. Int. J. Biol. Markers 2000, 15, 139–146. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Fornander, T.; Johansson, H.; Johansson, U.; Hu, G.Z.; Rutqvist, L.-E.; Skog, S. Thymidine kinase 1 in serum predicts increased risk of distant or loco-regional recurrence following surgery in patients with early breast cancer. Anticancer Res. 2006, 26, 4753–4759. [Google Scholar]
- Lerouge, D.; Touboul, E.; Lefranc, J.P.; Genestie, C.; Moureau-Zabotto, L.; Blondon, J. Combined chemotherapy and preoperative irradiation for locally advanced noninflammatory breast cancer: Updated results in a series of 120 patients. Int. J. Radiat. Oncol. Biol. Phys. 2004, 59, 1062–1073. [Google Scholar] [CrossRef]
- Semiglazov, V.F.; Topuzov, E.E.; Bavli, J.L.; Moiseyenko, V.M.; Ivanova, O.A.; Seleznev, I.K.; Orlov, A.A.; Barash, N.Y.; Golubeva, O.M.; Chepic, O.F. Primary (neoadjuvant) chemotherapy and radiotherapy compared with primary radiotherapy alone in stage IIb-IIIa breast cancer. Ann. Oncol. 1994, 5, 591–595. [Google Scholar] [CrossRef]
- Bondiau, P.Y.; Courdi, A.; Bahadoran, P.; Chamorey, E.; Queille-Roussel, C.; Lallement, M.; Birtwisle-Peyrottes, I.; Chapellier, C.; Pacquelet-Cheli, S.; Ferrero, J.M. Phase 1 clinical trial of stereotactic body radiation therapy concomitant with neoadjuvant chemotherapy for breast cancer. Int. J. Radiat. Oncol. Biol. Phys. 2013, 85, 1193–1199. [Google Scholar] [CrossRef] [PubMed]
- Sousa, C.; Cruz, M.; Neto, A.; Pereira, K.; Peixoto, M.; Bastos, J.; Henriques, M.; Roda, D.; Marques, R.; Miranda, C.; et al. Neoadjuvant radiotherapy in the approach of locally advanced breast cancer. ESMO Open 2020, 4 (Suppl. 2), e000640. [Google Scholar] [CrossRef]
- Hughes, K.; Neoh, D. Neoadjuvant Radiotherapy: Changing the Treatment Sequence to Allow Immediate Free Autologous Breast Reconstruction. J. Reconstr. Microsurg. 2018, 34, 624–631. [Google Scholar] [PubMed]
- Singh, P.; Hoffman, K.; Schaverien, M.V.; Krause, K.J.; Butler, C.; Smith, B.D.; Kuerer, H.M. Neoadjuvant Radiotherapy to Facilitate Immediate Breast Reconstruction: A Systematic Review and Current Clinical Trials. Ann. Surg. Oncol. 2019, 26, 3312–3320. [Google Scholar] [CrossRef] [PubMed]
- Pazos, M.; Corradini, S.; Dian, D.; von Bodungen, V.; Ditsch, N.; Wuerstlein, R.; Schönecker, S.; Harbeck, N.; Scheithauer, H.; Belka, C. Neoadjuvant radiotherapy followed by mastectomy and immediate breast reconstruction: An alternative treatment option for locally advanced breast cancer. Strahlenther. Onkol. 2017, 193, 324–331. [Google Scholar] [CrossRef] [PubMed]
- Calitchi, E.; Kirova, Y.M.; Otmezguine, Y.; Feuilhade, F.; Piedbois, Y.; Le Bourgeois, J.P. Long-term results of neoadjuvant radiation therapy for breast cancer. Int. J. Cancer 2001, 96, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Blitzblau, R.C.; Arya, R.; Yoo, S.; Baker, J.A.; Chang, Z.; Palta, M.; Duffy, E.; Horton, J.K. A phase 1 trial of preoperative partial breast radiation therapy: Patient selection, target delineation, and dose delivery. Pract. Radiat. Oncol. 2015, 5, e513–e520. [Google Scholar] [CrossRef] [PubMed]
- Horton, J.K.; Blitzblau, R.C.; Yoo, S.; Geradts, J.; Chang, Z.; Baker, J.A.; Georgiade, G.S.; Chen, W.; Siamakpour-Reihani, S.; Wang, C.; et al. Preoperative Single-Fraction Partial Breast Radiation Therapy: A Novel Phase 1, Dose-Escalation Protocol With Radiation Response Biomarkers. Int. J. Radiat. Oncol. Biol. Phys. 2015, 92, 846–855. [Google Scholar] [CrossRef] [PubMed]
- Poleszczuk, J.; Luddy, K.; Chen, L.; Lee, J.K.; Harrison, L.B.; Czerniecki, B.J.; Soliman, H.; Enderling, H. Neoadjuvant radiotherapy of early-stage breast cancer and long-term disease-free survival. Breast Cancer Res. 2017, 19, 75. [Google Scholar] [CrossRef] [PubMed]
- Riet, F.G.; Fayard, F.; Arriagada, R.; Santos, M.A.; Bourgier, C.; Ferchiou, M.; Heymann, S.; Delaloge, S.; Mazouni, C.; Dunant, A.; et al. Preoperative radiotherapy in breast cancer patients: 32 years of follow-up. Eur. J. Cancer 2017, 76, 45–51. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meehan, J.; Gray, M.; Martínez-Pérez, C.; Kay, C.; Wills, J.C.; Kunkler, I.H.; Dixon, J.M.; Turnbull, A.K. A Novel Approach for the Discovery of Biomarkers of Radiotherapy Response in Breast Cancer. J. Pers. Med. 2021, 11, 796. https://doi.org/10.3390/jpm11080796
Meehan J, Gray M, Martínez-Pérez C, Kay C, Wills JC, Kunkler IH, Dixon JM, Turnbull AK. A Novel Approach for the Discovery of Biomarkers of Radiotherapy Response in Breast Cancer. Journal of Personalized Medicine. 2021; 11(8):796. https://doi.org/10.3390/jpm11080796
Chicago/Turabian StyleMeehan, James, Mark Gray, Carlos Martínez-Pérez, Charlene Kay, Jimi C. Wills, Ian H. Kunkler, J. Michael Dixon, and Arran K. Turnbull. 2021. "A Novel Approach for the Discovery of Biomarkers of Radiotherapy Response in Breast Cancer" Journal of Personalized Medicine 11, no. 8: 796. https://doi.org/10.3390/jpm11080796
APA StyleMeehan, J., Gray, M., Martínez-Pérez, C., Kay, C., Wills, J. C., Kunkler, I. H., Dixon, J. M., & Turnbull, A. K. (2021). A Novel Approach for the Discovery of Biomarkers of Radiotherapy Response in Breast Cancer. Journal of Personalized Medicine, 11(8), 796. https://doi.org/10.3390/jpm11080796









