Abstract
Dihydropyrimidine dehydrogenase deficiency is a major cause of severe fluoropyrimidine-induced toxicity and could lead to interruption of chemotherapy or life-threatening adverse reactions. This study aimed to characterize the DPYD exon sequence, mRNA expression and in vivo DPD activity by plasma uracil concentration. It was carried out in two groups of patients with extreme phenotypes (toxicity versus control) newly treated with a fluoropyrimidine, during the first three cycles of treatment. A novel nonsense gene variant (c.2197insA) was most likely responsible for fluoropyrimidine-induced toxicity in one patient, while neither DPYD mRNA expression nor plasma uracil concentration was globally associated with early toxicity. Our present work may help improve pharmacogenetic testing to avoid severe and undesirable adverse reactions to fluoropyrimidine treatment and it also supports the idea of looking beyond DPYD.
1. Introduction
Severe adverse reactions to fluoropyrimidines are some of the main problems related to chemotherapy treatment in solid cancers [1]. These reactions occur in about one in five patients [2]. Dihydropyrimidine dehydrogenase (DPD) is the key metabolizer of 5-fluorouracil. Partial DPD deficiency is estimated in 3–15% of the Caucasian population, and 0.1–0.5% have a complete deficiency [3,4]. The relationship between DPD deficiency and the increase in fluoropyrimidine-induced toxicity in patients who take 5-fluorouracil or its prodrug capecitabine is well established [5]. These undesirable adverse reactions may compromise treatment outcomes, due to delays in administration, dose reductions and even necessary withdrawals of the drug.
Multiple factors, such as age and sex, are associated with severe adverse reactions to fluoropyrimidines and have been used for therapy individualization [6,7]. However, genetic factors, mainly in the DPYD gene, have been revealed as the most common cause of severe and life-threatening events induced by fluoropyrimidines [8]. Several drug regulatory agencies recommend genotyping of some variants in DPYD or phenotyping of DPD activity before starting treatment with fluoropyrimidines [9].
At least four variants are widely recommended for genotyping prior to the first administration of fluoropyrimidines, due to their well-established relationship with toxicity [10]. These variants include c.1905 + 1G > A (rs3918290), which causes the exon 14 deletion, two missense variants, c.1679T>G (rs55886062) and c.2846A>T (rs67376798), and the intron variant c.1129–5923 (rs75017182), which causes a splice defect. Although the incorporation of this test into clinical practice can avoid 25–50% of severe adverse reactions, there is still room for improvement [11,12]. Other rare genetic variants of DPYD are also clearly associated with fluoropyrimidine-induced toxicity and many others are probably still unknown [13]. In addition, new variants have been identified after widespread DPYD genetic testing in clinical practice [14,15]. Furthermore, some patients without any genetic variant of DPYD can suffer severe adverse reactions to fluoropyrimidines [14], which suggest that other factors must play a role.
Variation in copy number of intragenic regions has also been described as affecting DPD functionality [16]. Recently, a novel intragenic deletion in DPYD which includes exon 4 was found with a high prevalence in Finnish cancer patients receiving capecitabine. This deletion causes a defect in the splicing and thus the loss of exon 4, leading to the generation of a truncated DPD (p.Cys79Thrfs*8).
DPD phenotyping is an alternative to DPYD genotyping. DPD participates in the conversion of uracil (U) to dihydrouracil (UH2). The European Medicine Agency (EMA) recommends DPD testing prior to treatment with fluoropyrimidines [17]. Meanwhile, the Spanish Medicine Agency (AEMPS) established that plasma U levels of 16–150 ng/mL correspond to a partial deficiency and levels above 150 ng/mL indicate a complete deficiency [18].
Another mechanism of DPD activity variation and changes in risk induced by fluoropyrimidines is thorough transcriptional regulation of DPYD. Epigenetic alterations may affect DPYD expression, as shown in the RKO colorectal cancer cell line [19]. These changes in DPYD mRNA expression could affect the quantity of DPD generated in some tissues and potentially correlate with fluoropyrimidine-induced toxicity.
The approach of genotyping patients with an extreme phenotype has been shown to be useful in pharmacogenetics to identify new biomarkers [20,21]. In this work, we compare the whole exon sequence of the DPYD gene, DPD activity and DPYD mRNA expression in patients with toxicity grade ≥3 during the first three cycles versus a control cohort with toxicity <3 during at least eight cycles of fluoropyrimidine treatment.
2. Materials and Methods
2.1. Patients
This was a retrospective, observational, longitudinal case–control study. It included patients aged ≥ 18 years, diagnosed with a solid tumor and newly treated with a chemotherapy regime based on fluoropyrimidines. An exhaustive review of 464 medical records from patients treated with fluoropyrimidines since January of 2018 was carried out to select two extreme phenotypes. First, 28 patients presenting severe adverse reactions (grade ≥ 3 following the CTCAE v5 classification) during the three first cycles of fluoropyrimidine-based treatment and, second, 14 patients who did not present any toxicity grade ≤ 1 during at least eight cycles of treatment. The previous periods were the minimum to classify the patients in one of the two groups. Patients were recruited from January 2020 to March 2021. In our hospital, all patients are usually genotyped for c.1905 + 1G > A (rs3918290), c.1679T>G (rs55886062), c.2846A>T (rs67376798) and c.1129–5923 (rs75017182) prior to the first administration of the fluoropyrimidine. Those participants who were not previously genotyped for these variants were screened and, if positive, excluded from the study.
The following clinical and demographic variables were recorded: age, sex, fluoropyrimidine used and concomitant medication and type of cancer.
Adverse events during every cycle were compiled from the hospital’s medical records. These included GI disorders such as diarrhea, nausea and/or vomiting; hand–foot syndrome, mucositis, hematologic toxicity and other biochemical parameters. Classification and severity rates were based on National Cancer Institute common terminology criteria for adverse events v5.
2.2. Ethics
This study was approved by the Institutional Ethics Committee of Hospital Gregorio Marañón (protocol code FG-2019-02, date of approval 23 September 2019). All patients signed a written informed consent.
2.3. DPYD Exon Sequencing
Genomic DNA was isolated from whole blood using a QIAamp DNA Blood Mini Kit following the manufacturer’s instructions (Qiagen, Hiden, Germay). All of the 23 exons from the DPYD gene and flanking intron regions were amplified as described in García-González et al. [15]. PCR products (5 µL) were purified using PureIT ExoZAP (2 µL) (Ampliqon, Odense, Denmark). Sequences were obtained at the Genomics Unit of Hospital Gregorio Marañón by Sanger technology and analyzed using SnapGene v5.2. DPD activity was predicted from genotype using SIFT, Polyphen2 and ClinVar [22,23,24]. Linkage disequilibrium was measured using the LDmatrix tool (National Cancer Institute) [25].
2.4. DPD Activity Measure
Blood samples were collected from patients in EDTA tubes between 8:00 a.m. and 11:00 a.m. Plasma was isolated by centrifugation at 3000 r.p.m. for 10 minutes. U concentration was measured in all patients as described previously [4]. A patient was catalogued as partially deficient if U > 16.
2.5. DPYD mRNA Expression
2.5.1. RNA Isolation cDNA Synthesis
Total RNA was isolated from whole blood and preserved in PAXgene® tubes using a PAXgene™ Blood RNA kit (PreAnalytics, Hombrechtikon, Switzerland). Complementary DNA (cDNA) was synthetized from 300 ng of total RNA using a High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA) in a final volume of 20 µL using random hexamers. A 1/10 dilution was used in real-time PCR. All of these procedures were performed as recommended by the manufacturers.
2.5.2. Real-Time PCR and Quantification Method
DPYD was relatively quantified by RT-PCR in a StepOne (Applied Biosystems, Foster City, CA, USA) using HPRT1 for normalization. RT-PCR was performed in triplicate using the following TaqMan probes: DPYD (Hs115750_m1) and HPRT1 (Hs02800695_m1) (Applied Biosystems, Waltham, MA, USA) in 20µL of final volume (10 µL Universal Master Mix, Applied Biosystems; 2µL cDNA 1/10 dilution; 1µlL TaqMan probe, 7µL free-nuclease water). Relative DPYD expression in toxicity versus control group was quantified using the 2−∆∆Ct method, taking as relative expression the sample D031, and in StepOnePlus v-2.3.2 (Applied Biosystems). Relative values of mRNA DPYD expression were represented in Prism version 8 (San Diego, CA, USA).
2.6. Characterization of 2197insA
2.6.1. mRNA Sequencing
Blood was collected in Paxgene Blood RNA tubes (Becton Dickinson, Franklin Lakes, NJ, USA) and RNA isolated using a PaxGene RNA blood RNA kit (Qiagen, Valencia, CA, USA). cDNA was synthesized and diluted as described in Section 2.5.1. The cDNA region containing the 2197insA was amplified in 10 µL (NZY Master Mix 5 µL, 1 µM of primer 17F2 (5’-TGA GCA TCG CAA GAG CTG CA), 1 µM of primer Sp20R (5’ TGG ACT CTG TCC ATC CCA GTC T) and 10 ng of genomic DNA). Sequences were obtained in the Genomics Unit of our hospital using primer Sp20R as a template.
2.6.2. 3D Modeling
The DPD sequence generated by 2197insA and wild type DPD were exported from SnapGene v5.2 in Fasta format. The 3D modeling of wild type DPD and DPD 2197insA was carried out using the RaptorX structure prediction server [26]. Protein database files were downloaded and visualized using Chimera [27].
2.7. Analysis of Exon 4 Skipping
The cDNA region from exon E1-2 to E5 was amplified in all participants to identify the loss of exon 4. PCR amplifications were performed in 10 µL (NZY Master Mix 5 µL, 1 µM of primer E1-2-F (5’-TCG GCG GAC ATC GAG AGT AT), 1 µM of primer E5-R (5’ AGA GGT TGG ACA TAC CAT TCC A) and 2µL of 1/20 cDNA dilution). PCR was performed with the following conditions: 94 °C 8 min, 40 cycles of 94 °C 30 s, 60 °C 30 s, 72 °C 1 min and a final step of 72 °C 5 min. Fragment length was analyzed using a DNA1000 kit in a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). A fragment of 375 nucleotides in length was expected for wild type individuals and a fragment of 287 nucleotides for individuals who skip exon 4.
3. Results
3.1. Clinical and Demographic Characteristics of Patients
After review of 464 clinical records, 41 patients met the inclusion criteria. They were separated into two groups: 14 patients were included in the control group (fluoropyrimidine-related adverse reactions ≤ 1) and 28 patients in the toxicity group (adverse reactions grade ≥3 during the first three cycles). Characteristics of participants are summarized in Table 1. No statistical differences were observed in gender, age, type of cancer, clinical stage, surgery or concomitant chemotherapy. However, the control group included more capecitabine-treated patients (85% vs. 48.1%) and, as expected, due to classification of the groups, a higher fluoropyrimidine cumulative dose, and a lower need for dose reductions due to toxicity after the first cycle (p value < 0.05).

Table 1.
Characteristics of patients.
Table 2 shows the incidence of adverse events <2 and ≥3. As expected, all adverse reactions of any degree were more common in the toxicity group, but the difference between groups was statistically significant only for leukopenia, neutropenia, lymphopenia, diarrhea and nausea/vomiting.

Table 2.
Adverse event comparison between control and toxicity groups.
3.2. DPYD Genetic Variants in Coding Regions and Splice Sites
The whole DPYD sequencing of the coding exon and flanking intron regions (20 bp on both sides) in the 41 patients revealed the presence of 14 SNPs: three synonymous, eight non-synonymous (seven missense and one nonsense), one in the 3′ untranslated region (UTR) and two in the intron region close to an exon. The variants found in every patient can be found in Figure 1. No great differences were observed, in the number of SNPs (12 vs. 8), in SNPs per patient (1.62 vs. 1.43) or in the number of patients without any SNP in the DPYD gene between controls and patients with early severe toxicity (six vs. two, respectively).
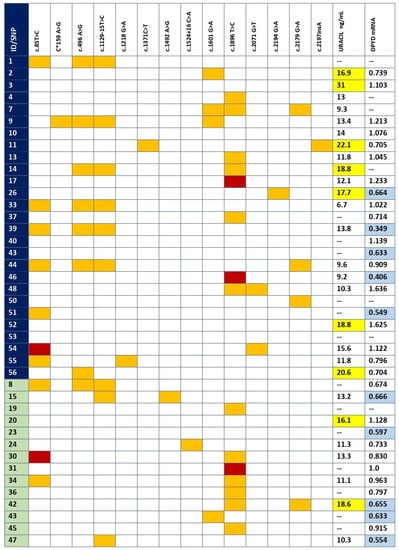
Figure 1.
SNPs, mRNA expression and DPD activity per patient. SNPs found in coding and flanking intronic regions for each patient. Dark blue, toxicity group; green, control group; orange, heterozygous; red, homozygous mutant; yellow, U > 16 ng/mL; light blue, below 25th percentile in mRNA expression. The variant c*159A>G is a new variant of unknown significance placed in the 3′UTR.
The SNPs were categorized into three groups: specific to the toxicity group, shared by toxicity and control groups and specific to the control group (Table 3). The most relevant variants found were two new SNPs that were specific to the patients with toxicity. The variant c.2197insA contains an insertion that provokes amino acid changes in 14 positions until the arrival of a stop codon, consequently generating a shortened DPD (p.Thr733AsnfsTer14). The variant c*159A>G is a new variant of unknown significance placed in the 3′UTR. Another two very infrequent SNPs were found in the toxicity group (p.Met406Ile and p.Val691Leu), both with controversial results from several information sources, such as the Clinical Variation Consortium, Sort Intolerant from Tolerant (SIFT) and Polymorphism Phenotyping (PolyPhen) databases.

Table 3.
Single nucleotide variants in DPYD found in the recruited patients.
Regarding the SNPs found both in the control and the toxicity groups, six variants were found but only one was more frequent in the toxicity group compared to the control group, p.Met166Val (25.9% vs. 7.1%, respectively). However, this increase was not statistically significant. Interestingly, the haplotype c.496G/c.1129-15C was more frequently observed in the toxicity group (25.9%) than in the control one (7.1%). Both SNPs are in disequilibrium linkage (D′ = 0.955, R2 = 0.911). This analysis was also performed by tumor type and no statistically significant association was obtained (Supplementary Table S1).
Exon 4 skipping was not detected in any patient after analysis of PCR length fragment from cDNA using oligonucleotides placed in exons 1 and 5. A fragment of 375 nucleotides containing exon 4 was amplified for all patients, while the expected 287 nucleotide fragment generated after exclusion of exon 4 was not found in any sample (Figure 2).

Figure 2.
Analysis of exon 4 skipping. cDNA from exon 1 to 5 was amplified. A wild type fragment of 375 nucleotides was observed in all samples. The expected fragment of 287 nucleotides resulting from exon 4 skipping was not detected in any sample. L, ladder; 10, patient 10.
3.3. DPD Activity by Uracil Concentration
DPD activity was indirectly measured for each patient by quantifying U concentration (see individual values in Figure 1). DPD activity was measured in seven controls and 21 patients of the severe toxicity group. Since DPD activity is dependent on circadian rhythms, seven samples from the control group and six from the toxicity group were discarded because they were collected after 11:00 a.m. Seven patients in the toxicity group (2, 3, 11, 14, 26, 52 and 56) had U > 16 and were classified as DPD deficient, while two patients (20 and 42) from the control group also had U > 16. However, the analysis did not show differences in U between control and severe toxicity groups. Mean plasma U was 14.4 ng/mL (IQR 8.3, 6.7–31.0) in the toxicity group and 13.4 ng/mL (IQR 4.8, 9.8–18.6) in the control group (p value > 0.05) (Figure 3).
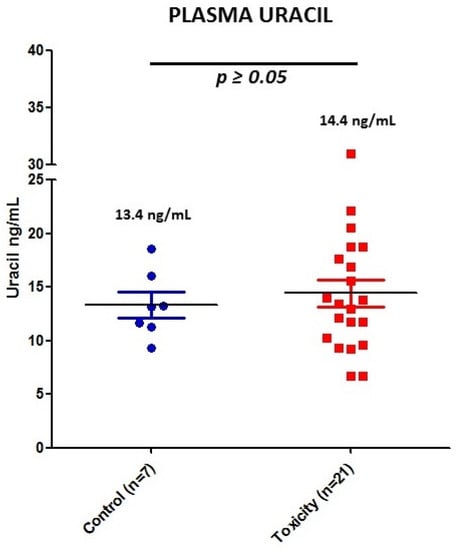
Figure 3.
Plasma U concentrations for the 28 patients with validated measurement of DPD activity. Blue, control patients; red, early toxicity patients.
3.4. mRNA DPYD Expression
PaxGene tubes were collected from all the recruited patients. However, total RNA isolation was not high enough quality in 12 samples. We obtained a new sample for RNA isolation in five patients, but seven declined to provide a new sample or had died. The expression of mRNA of DPYD was measured in 13 controls and 21 patients of the early severe toxicity group (see individual values in Figure 1). The analysis did not show differential expression (p value > 0.05) of DPYD mRNA between controls and patients with early severe toxicity (Figure 4).
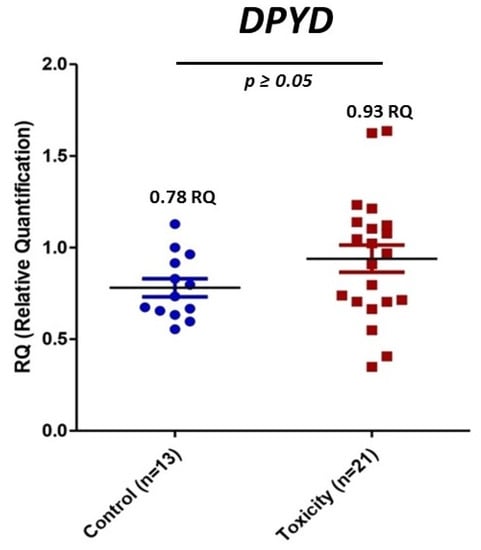
Figure 4.
Relative mRNA expression of DPYD in controls and early severe toxicity patients. Blue, control patients; red, early toxicity patients.
3.5. Characterization of c.2197insA
3.5.1. Sequence Analysis
Patient 11 showed an insertion in position 2197 after the starting codon. Sequencing of exon 18 for this patient showed a clean sequence until this position using a forward primer as well as a reverse primer (Figure 5). The read of duplicated peaks shows an insertion of an adenine at this position.
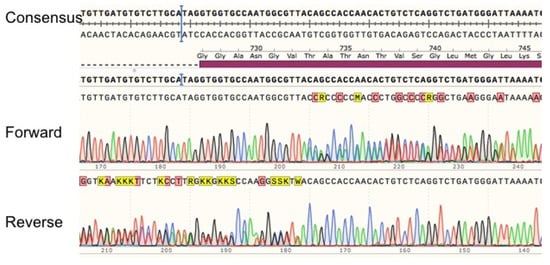
Figure 5.
Sequencing alignment of exon 18 for patient with c.2197insA. Exon 18 PCR amplification was sequenced using forward and reverse primers for this exon. Sequences were aligned with the consensus sequence (NC_000001.11 GRCh38.p13).
The insertion of an A into the DPYD mRNA was verified by sequencing the complementary DNA from whole blood of this patient.
3.5.2. DPD Modeling Generated by c.2197insA Variant
The adenine insertion at position 2197 of DPYD mRNA after the ATG initiation predicts the generation of a truncated protein of 746 amino acids. The truncated DPD protein sequence is the same until amino acid position 732, different from position 733 to 746 and lacks the fragment from 747 to the end (position 1225). This anomaly of DPD breaks the flavin mononucleotide domain, where the pyrimidine binding site is located, and lacks the 4Fe-4S domain. Results of 3D modeling of mutated and wild type DPD were obtained using the RaptorX web server and colored and then visualized using Chimera (Figure 6). Clear differences are shown in the central structure of DPD affecting the missing domains (Supplementary Materials Video S1).
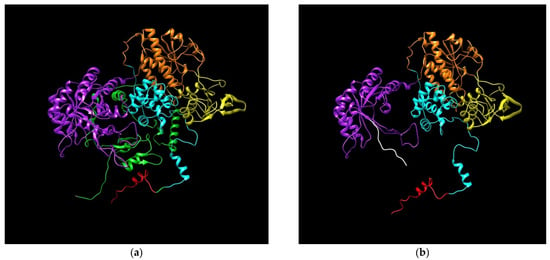
Figure 6.
Predicted models for DPD. (a) Wild type DPD; (b) DPD generated by insA in c.2197. Protein Database (PDB) files were generated using RaptorX web software (raptorX.uchicago.edu) and images were obtained using Chimera v1.13.1. DPD domains: I, N-terminal Fe-S clusters (red); II, flavin mononucleotide (FAD) binding (orange); III, NADPH binding (yellow); IV, flavin adenine dinucleotide (FMN)/pyrimidine binding (purple); and V, C-terminal Fe-S clusters (green). Amino acids added from insertion to stop codon (white).
According to plasma U concentration, the patient carrying this mutation showed a partial DPD deficit (U = 22.1 ng/mL).
4. Discussion
The use of the whole coding DPYD sequence followed by measurement of DPD activity is an efficient approach to identify new pathogenic genetic variants [14,15,16,28,29]. However, this approach is not able to explain all severe fluoropyrimidine-induced toxicities. In this work, we compared genetic variants, protein activity and blood DPYD mRNA expression in 41 cancer patients receiving fluoropyrimidine-based treatment. We have separated them in two groups: those that suffered early severe toxicity (cycles 1–3) and those that did not.
Our study has disclosed and characterized c.2197insA, a new and infrequent genetic variant most likely associated with DPD deficiency and early severe toxicity to fluoropyrimidine-based treatments. The predicted human DPD model showed that the sequence carrying variant c.2197insA generates a truncated protein (p.Thr733AsnfsTer14) affecting the flavin mononucleotide/pyrimidine binding domain and the C-terminal Fe-S clusters [30]. DPD works as a homodimer and these alterations clearly suggest that the truncated protein is not able to bind 5-FU, impeding drug metabolism. Measurement of plasma U levels showed that the patient carrying this mutation was DPD deficient. Accumulation of 5-FU due to inefficient elimination is probably the cause of severe toxicity in the patient carrying this mutation. Another SNP (c.2242+1G>T) leads to skipping of exon 19 and generates a very similar truncated DPD protein which lacks the same domains [15]. This variant was associated with capecitabine-induced severe toxicity in a breast cancer patient. According to this evidence, the presence of c.2197insA is the most probable cause of the toxicity observed in the carrier patient.
A SNP in the position c*159A>G in the 3′UTR was identified in patient 9. Variants in the 3′UTR may affect mRNA stability, translation efficiency, nuclear export and cellular location [31]. This variant does not seem to affect the polyadenylation signal region, one of the most sensitive places in the 3′UTR [32]. However, it may affect microRNA binding sites. Expression of DPYD mRNA in this patient was low in comparison with other samples. No 3′UTR DPYD variants have been associated with toxicity to fluoropyrimidines to date. Nevertheless, a study analyzing 33 germline polymorphisms in the 3′UTR of genes involved in drug absorption, distribution, metabolism and elimination (ADME) showed that DPYD rs291593 was associated with recurrence-free survival in breast cancer patients [33]. No data on the effect of this SNP on DPYD expression have been reported, but the variant position suggests a putative posttranscriptional regulation, a role that would explain its relationship with treatment response. In a similar way, c*159A>G could alter DPYD mRNA stability and decrease DPD translation, leading to the early toxicity observed in this patient. The low frequency of this variant hampers the chance to prove this relationship in bigger cohort studies.
Another two non-synonymous genetic variants were only found in the group of patients with early severe toxicity to fluoropyrimidines, p.Met406Ile (patient 55) and p.Val691Leu (patients 48 and 54). p.Met406Ile was categorized as benign by SIFT and tolerated by PolyPhen, but ClinVar recognized a conflicting interpretation. No damaging effect of this SNP on DPD activity has been reported [34]. Accordingly, patient 55 had U and DPYD mRNA expression levels that were within range. pVal691Leu was considered probably damaging by Polyphen-2 and tolerated by SIFT. In vitro DPD activity was measured for this SNP and considered normal [35]. Both patients carrying this variant in our study had U concentrations within the established limits of normal DPD activity. Consequently, these SNPs are not good candidates to explain the toxicity observed in the carrier patients.
Recently, a Dutch pharmacogenetics group has identified SNPs pMet166Val and c.1129-15 as fully functional with a weak level of evidence [36]. The SNP p.Met166Val was more frequent in the toxicity group than in the control group. This SNP is in linkage disequilibrium with c.1129-15C. These variants have been widely considered as not related to toxicity to fluoropyrimidines [37,38,39]. In addition, DPD in vitro activity has been reported as normal or even higher than normal for p.Met166Val [34]. However, this result is conflicting and other authors have found a relationship of this SNP with fluoropyrimidine-induced toxicity [8,40,41,42,43,44]. Furthermore, recent work showed this SNP to be associated with a lower UH2/U ratio, another indirect way to measure DPD activity [45]. The higher frequency of this SNP in the group of patients with early toxicity to fluoropyrimidines suggests it plays a role in the risk of toxicity. The limited sample size does not allow us to obtain conclusive results for this SNP. Nonetheless, our results suggest that the effect of haplotype c.496C/c.1129-15C on toxicity risk to fluoropyrimidines may be more relevant than previously thought and, hence, should be explored in larger studies.
Unfortunately, the approach followed in this work was not able to explain why most of the recruited patients suffered early severe toxicity as a consequence of their fluoropyrimidine-based treatment. Recently, in a similar study, the skipping of exon 4 was found to be common in patients from Finland with severe toxicity to fluoropyrimidines [16]. This established a promising biomarker to help increase the power of detection of risky patients of severe toxicity induced by fluoropyrimidines. However, none of the patients included in our study had mRNA skipping of exon 4. Thus, the clinical usefulness of this biomarker outside of the Finnish population remains to be elucidated.
High DPYD mRNA expression correlates with poor disease-free survival [46]. Our group observed lower DPYD mRNA expression (data not shown) in descendants of a breast cancer patient suffering from severe toxicity to capecitabine and carrying a genetic variant in DPYD skipping of exon 19 [15]. In this study, we analyzed DPYD mRNA expression in control and toxicity groups of patients treated with fluoropyrimidines. DPYD mRNA expression was similar in control and toxicity groups and failed to identify patients at risk of severe fluoropyrimidine toxicity. On the one hand, previous studies found a strong correlation between DPYD mRNA expression with DPD activity in liver sections [47] and in tumor tissue [48]. On the other hand, it has been suggested that DPYD mRNA might not be reflective of global DPD activity, because it does not distinguish mRNA coding for non-functional DPD [49]. DPYD expression is regulated by STAT3 [50], interferon alpha [51], TWIST1 [52] and several microRNAs or long non-coding RNAs [53,54]. None of these genes have been associated with toxicity to fluoropyrimidines. A limitation of this part of our study is that samples for mRNA expression were collected after patients were classified in the control or toxicity group and, therefore, had already received several cycles of chemotherapy. Perhaps collecting samples prior to the start of therapy would have rendered more significant results. More and larger studies are needed to explore the effect of other variables, such as age and sex.
Only one of the newly discovered variants in the toxicity group, c.2197insA, seemed to be clearly associated with DPD deficiency and early toxicity, based on DPD modeling and the patient’s level of plasma U. Furthermore, the absence of inactivating mutations is not the only mechanism involved in DPD activity, for example, methylation has been recognized as a gene silencing mechanism [55]. Ezzeldin et al. found that 100% of patients with DPD deficiency without inactivating mutations in the DPYD gene had aberrant methylation of the DPYD promoter [56]. Moreover, no correlation was observed between U or UH2/U and DPYD mRNA expression, suggesting that DPD activity is not dependent on gene expression in whole blood.
Interestingly, two patients with a DPD-deficient phenotype, as established by plasmatic U in the toxicity group (patients 3 and 52) and one in the control group (patient 20), did not carry any DPYD variant. Intriguingly, no DPYD variants were observed in the patient with the greatest activity deficit (patient 3, U = 31 ng/mL) and their DPYD mRNA expression was above the 25th percentile. Intriguingly, patient 42 in the control group showed partial DPD deficiency by plasma U and presented low DPYD mRNA expression.
The measurement of plasma U is widely accepted for detecting DPD deficiency [4,57]. However, our results do not show a clear association of U with the occurrence of early severe fluoropyrimidine toxicity. It has been recently reported that an artificial increase in U concentration during fluoropyrimidine treatment can lead to DPD deficiency misinterpretation [58]. Competition of U and 5-fluorouracil for DPD may increase U plasma concentration. However, 5-FU was detected only in one patient (data not shown).
Altogether, the present findings strongly suggest that several other variables may influence the tolerability of fluoropyrimidine-based treatments. Nevertheless, a larger comparative case–control study involving thousands of individuals is needed to draw statistically significant conclusions. In addition, these results support the use of approaches including multiple genes and other parameters in future studies in order to try to better predict fluoropyrimidine-induced toxicity.
Future Research Directions
We used a multiple approach, studying DPYD genetic variants, mRNA expression and indirect measuring of DPD activity to try to identify the cause of severe fluoropyrimidine-induced toxicity in a group of cancer patients. For most of the patients, our approach was unable to provide the cause of toxicity. Other authors have used multiparametric approaches for predicting patients at risk of toxicity due to fluoropyrimidine-based treatment [59]. This seems to be the best option to explore in the future. The role of conflicting variants in the literature must be clarified to improve the prediction models. The trend observed with the haplotype c.496C/c.1129-15T and fluoropyrimidine toxicity in our study, along with recent results obtained by other groups, suggests this haplotype should be investigated in larger cohorts. Since no relationship between DPYD mRNA expression in whole blood and early severe toxicity has been observed in this work, in our opinion, the effort to find new biomarkers for toxicity should be focused in genotyping not only DPYD but also other genes, such as TYMS, CDA, ENOSF1 and others related to fluoropyrimidine’s pharmacodynamics and pharmacokinetics, such as CES1, CES2 or UMPS. Implementation of functional studies to prevent toxicity in those patients in whom DPD deficits were detected would be necessary as well.
5. Conclusions
The proposed multiple approach including DPYD gene sequencing, expression and measurement of DPD activity does not explain all the toxicity observed in fluoropyrimidine-treated patients. Thus, it is necessary to investigate other genes and factors. However, the whole DPYD sequencing helped us to identify a new deleterious variant, c.2197insA, that potentially codes for a non-functional DPD protein (p.Thr733AsnfsTer14) which could cause early severe toxicity.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/jpm11080792/s1, Video S1: 3D modeling of p.Thr733AsnfsTer14 truncated and wild type DPD. Table S1: Single nucleotide variants in DPYD found in the recruited patients stratified by tumor type.
Author Contributions
Conceptualization, M.M. and L.A.L.-F.; methodology, P.V., B.M.-A., J.L.R.-H., X.G.-G., S.S.-M., M.Y. and F.T.; formal analysis, P.V., M.Y., F.T. and S.S.-M.; investigation, P.G.-A., S.L.-T., A.C., F.L.-L., I.A., M.S.-S. and M.M.; data curation, P.V., B.M.-A. and J.L.R.-H.; writing—original draft preparation, P.V., B.M.-A., J.L.R.-H., X.G.-G., S.S.-M., F.T. and L.A.L.-F.; writing—review and editing, all; supervision, L.A.L.-F.; funding acquisition, L.A.L.-F. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Instituto de Investigación Sanitaria Gregorio Marañón, grant number II-PI-2-2019.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Ethics Committee of Hospital Gregorio Marañón (protocol code FG-2019-02, date of approval 23 September 2019).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study. Written informed consent has been obtained from the patients to publish this paper.
Data Availability Statement
The data are accessible upon request to the corresponding author.
Acknowledgments
We would like to thank Paula Ruiz and Alejandra Melgarejo for their help during their time in the laboratory, and to Alicia López and Sandra Fabiola Velasco for English revision. Molecular graphics were made and analyses were performed with UCSF Chimera, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from NIH P41-GM103311.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Brito, R.A.; Medgyesy, D.; Zukowski, T.H.; Royce, M.E.; Ravandi-Kashani, F.; Hoff, P.M.; Pazdur, R. Fluoropyrimidines: A critical evaluation. Oncology 1999, 57, 2–8. [Google Scholar] [CrossRef]
- Barin-Le Guellec, C.; Lafay-Chebassier, C.; Ingrand, I.; Tournamille, J.-F.; Boudet, A.; Lanoue, M.-C.; Defossez, G.; Ingrand, P.; Perault-Pochat, M.-C.; Etienne-Grimaldi, M.-C. Toxicities associated with chemotherapy regimens containing a fluoropyrimidine: A real-life evaluation in France. Eur. J. Cancer 2020, 124, 37–46. [Google Scholar] [CrossRef] [Green Version]
- Pallet, N.; Hamdane, S.; Garinet, S.; Blons, H.; Zaanan, A.; Paillaud, E.; Taieb, J.; Laprevote, O.; Loriot, M.-A.; Narjoz, C. A comprehensive population-based study comparing the phenotype and genotype in a pretherapeutic screen of dihydropyrimidine dehydrogenase deficiency. Br. J. Cancer 2020, 123, 811–818. [Google Scholar] [CrossRef]
- Loriot, M.-A.; Ciccolini, J.; Thomas, F.; Barin-Le-Guellec, C.; Royer, B.; Milano, G.; Picard, N.; Becquemont, L.; Verstuyft, C.; Narjoz, C.; et al. Dihydropyrimidine déhydrogenase (DPD) deficiency screening and securing of fluoropyrimidine-based chemotherapies: Update and recommendations of the French GPCO-Unicancer and RNPGx networks. Bull. Cancer 2018, 105, 397–407. [Google Scholar] [CrossRef]
- Katona, C.; Kralovánszky, J.; Rosta, A.; Pandi, E.; Fónyad, G.; Tóth, K.; Jeney, A. Putative role of dihydropyrimidine dehydrogenase in the toxic side effect of 5-fluorouracil in colorectal cancer patients. Oncology 1998, 55, 468–474. [Google Scholar] [CrossRef]
- Stein, B.N.; Petrelli, N.J.; Douglass, H.O.; Driscoll, D.L.; Arcangeli, G.; Meropol, N.J. Age and sex are independent predictors of 5-fluorouracil toxicity. Analysis of a large scale phase III trial. Cancer 1995, 75, 11–17. [Google Scholar] [CrossRef]
- Wagner, A.D.; Grothey, A.; Andre, T.; Dixon, J.G.; Wolmark, N.; Haller, D.G.; Allegra, C.J.; de Gramont, A.; VanCutsem, E.; Alberts, S.R.; et al. Sex and Adverse Events of Adjuvant Chemotherapy in Colon Cancer: An Analysis of 34 640 Patients in the ACCENT Database. J. Natl. Cancer Inst. 2021, 113, 400–407. [Google Scholar] [CrossRef] [PubMed]
- Ruzzo, A.; Graziano, F.; Galli, F.; Galli, F.; Rulli, E.; Lonardi, S.; Ronzoni, M.; Massidda, B.; Zagonel, V.; Pella, N.; et al. Dihydropyrimidine dehydrogenase pharmacogenetics for predicting fluoropyrimidine-related toxicity in the randomised, phase III adjuvant TOSCA trial in high-risk colon cancer patients. Br. J. Cancer 2017, 117, 1269–1277. [Google Scholar] [CrossRef] [PubMed]
- Agency, E.M. European Medicines Agency—Multidisciplinary—Multidisciplinary: Pharmacogenomics. Available online: http://www.ema.europa.eu/ema/index.jsp?curl=pages/regulation/general/general_content_000411.jsp&mid=WC0b01ac058002958e (accessed on 5 June 2015).
- Shakeel, F.; Fang, F.; Kwon, J.W.; Koo, K.; Pasternak, A.L.; Henry, N.L.; Sahai, V.; Kidwell, K.M.; Hertz, D.L. Patients carrying DPYD variant alleles have increased risk of severe toxicity and related treatment modifications during fluoropyrimidine chemotherapy. Pharmacogenomics 2021, 22, 145–155. [Google Scholar] [CrossRef]
- Amstutz, U.; Henricks, L.M.; Offer, S.M.; Barbarino, J.; Schellens, J.H.M.; Swen, J.J.; Klein, T.E.; McLeod, H.L.; Caudle, K.E.; Diasio, R.B.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Dihydropyrimidine Dehydrogenase Genotype and Fluoropyrimidine Dosing: 2017 Update. Clin. Pharmacol. Ther. 2018, 103, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Guío, A.; Bernabéu-Martínez, Á.; Corno-Caparrós, A.; Aznar-Saliente, T.; Bonete-Sánchez, M.; Calleja-Hernández, M.Á. DPYD variant testing in candidates for fluoropyrimidine treatment: A study protocol. Farm. Hosp. 2021, 45, 155–159. [Google Scholar]
- Collie-Duguid, E.S.; Etienne, M.C.; Milano, G.; McLeod, H.L. Known variant DPYD alleles do not explain DPD deficiency in cancer patients. Pharmacogenetics 2000, 10, 217–223. [Google Scholar] [CrossRef]
- García-González, X.; Kaczmarczyk, B.; Abarca-Zabalía, J.; Thomas, F.; García-Alfonso, P.; Robles, L.; Pachón, V.; Vaz, Á.; Salvador-Martín, S.; Sanjurjo-Sáez, M.; et al. New DPYD variants causing DPD deficiency in patients treated with fluoropyrimidine. Cancer Chemother. Pharmacol. 2020, 86, 45–54. [Google Scholar] [CrossRef] [PubMed]
- García-González, X.; López-Tarruella, S.; García, M.I.; González-Haba, E.; Blanco, C.; Salvador-Martin, S.; Jerez, Y.; Thomas, F.; Jarama, M.; Sanjurjo Sáez, M.; et al. Severe toxicity to capecitabine due to a new variant at a donor splicing site in the dihydropyrimidine dehydrogenase (DPYD) gene. Cancer Manag. Res. 2018, 10, 4517–4522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saarenheimo, J.; Wahid, N.; Eigeliene, N.; Ravi, R.; Salomons, G.S.; Ojeda, M.F.; Vijzelaar, R.; Jekunen, A.; van Kuilenburg, A.B.P. Preemptive screening of DPYD as part of clinical practice: High prevalence of a novel exon 4 deletion in the Finnish population. Cancer Chemother. Pharmacol. 2021, 87, 657–663. [Google Scholar] [CrossRef] [PubMed]
- Agency, E.M. EMA Recommendations on DPD Testing Prior to Treatment with Fluorouracil, Capecitabine, Tegafur and Flucytosine. Available online: https://www.ema.europa.eu/en/documents/press-release/ema-recommendations-dpd-testing-prior-treatment-fluorouracil-capecitabine-tegafur-flucytosine_en.pdf (accessed on 12 May 2021).
- Spanish Medicine and Health Products Agency (AEMPS) Recommendation on DPD Testing. Available online: https://www.aemps.gob.es/informa/notasinformativas/medicamentosusohumano-3/seguridad-1/2020-seguridad-1/fluorouracilo-capecitabina-tegafur-y-flucitosina-en-pacientes-con-deficit-de-dihidropirimidina-deshidrogenasa/?lang=en (accessed on 12 May 2021).
- Zhang, X.; Soong, R.; Wang, K.; Li, L.; Davie, J.R.; Guarcello, V.; Diasio, R.B. Suppression of DPYD expression in RKO cells via DNA methylation in the regulatory region of the DPYD promoter: A potentially important epigenetic mechanism regulating DPYD expression. Biochem. Cell Biol. 2007, 85, 337–346. [Google Scholar] [CrossRef]
- Dong, S.-Q.; Wang, T.-M.; Zhang, J.-B.; He, Y.-Q.; Xue, W.-Q.; Wu, Z.-Y.; Yang, D.-W.; Cao, L.-J.; Huang, J.-W.; Li, X.-Z.; et al. Polymorphisms in TYMS for Prediction of Capecitabine-Induced Hand-Foot Syndrome in Chinese Patients with Colorectal Cancer. Cancer Res. Treat. 2021, 53, 724–732. [Google Scholar] [CrossRef]
- Ruiz-Pinto, S.; Pita, G.; Martín, M.; Nuñez-Torres, R.; Cuadrado, A.; Shahbazi, M.N.; Caronia, D.; Kojic, A.; Moreno, L.T.; de la Torre-Montero, J.C.; et al. Regulatory CDH4 Genetic Variants Associate With Risk to Develop Capecitabine-Induced Hand-Foot Syndrome. Clin. Pharmacol. Ther. 2021, 109, 462–470. [Google Scholar] [CrossRef]
- Kumar, P.; Henikoff, S.; Ng, P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009, 4, 1073–1081. [Google Scholar] [CrossRef]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef] [Green Version]
- Landrum, M.J.; Chitipiralla, S.; Brown, G.R.; Chen, C.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; Kaur, K.; Liu, C.; et al. ClinVar: Improvements to accessing data. Nucleic Acids Res. 2020, 48, D835–D844. [Google Scholar] [CrossRef] [PubMed]
- National Cancer Institute. LDmatrix tool. Available online: https://ldlink.nci.nih.gov/?tab=ldmatrix (accessed on 12 August 2021).
- Källberg, M.; Margaryan, G.; Wang, S.; Ma, J.; Xu, J. RaptorX server: A resource for template-based protein structure modeling. Methods Mol. Biol. 2014, 1137, 17–27. [Google Scholar] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Deenen, M.J.; Tol, J.; Burylo, A.M.; Doodeman, V.D.; de Boer, A.; Vincent, A.; Guchelaar, H.-J.; Smits, P.H.M.; Beijnen, J.H.; Punt, C.J.A.; et al. Relationship between single nucleotide polymorphisms and haplotypes in DPYD and toxicity and efficacy of capecitabine in advanced colorectal cancer. Clin. Cancer Res. 2011, 17, 3455–3468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Etienne-Grimaldi, M.C.; Boyer, J.C.; Beroud, C.; Mbatchi, L.; Van Kuilenburg, A.; Bobin-Dubigeon, C.; Thomas, F.; Chatelut, E.; Merlin, J.L.; Pinguet, F.; et al. New advances in DPYD genotype and risk of severe toxicity under capecitabine. PLoS ONE 2017, 12, e0175998. [Google Scholar] [CrossRef] [PubMed]
- Dobritzsch, D.; Schneider, G.; Schnackerz, K.D.; Lindqvist, Y. Crystal structure of dihydropyrimidine dehydrogenase, a major determinant of the pharmacokinetics of the anti-cancer drug 5-fluorouracil. EMBO J. 2001, 20, 650–660. [Google Scholar] [CrossRef] [Green Version]
- Fabian, M.R.; Sonenberg, N.; Filipowicz, W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010, 79, 351–379. [Google Scholar] [CrossRef] [Green Version]
- Shulman, E.D.; Elkon, R. Systematic identification of functional SNPs interrupting 3’UTR polyadenylation signals. PLoS Genet. 2020, 16, e1008977. [Google Scholar] [CrossRef]
- Pamuła-Piłat, J.; Tęcza, K.; Kalinowska-Herok, M.; Grzybowska, E. Genetic 3’UTR variations and clinical factors significantly contribute to survival prediction and clinical response in breast cancer patients. Sci. Rep. 2020, 10, 5736. [Google Scholar] [CrossRef]
- Offer, S.M.; Fossum, C.C.; Wegner, N.J.; Stuflesser, A.J.; Butterfield, G.L.; Diasio, R.B. Comparative functional analysis of DPYD variants of potential clinical relevance to dihydropyrimidine dehydrogenase activity. Cancer Res. 2014, 74, 2545–2554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shrestha, S.; Zhang, C.; Jerde, C.R.; Nie, Q.; Li, H.; Offer, S.M.; Diasio, R.B. Gene-Specific Variant Classifier (DPYD-Varifier) to Identify Deleterious Alleles of Dihydropyrimidine Dehydrogenase. Clin. Pharmacol. Ther. 2018, 104, 709–718. [Google Scholar] [CrossRef]
- Lunenburg, C.A.T.C.; van der Wouden, C.H.; Nijenhuis, M.; Crommentuijn-van Rhenen, M.H.; de Boer-Veger, N.J.; Buunk, A.M.; Houwink, E.J.F.; Mulder, H.; Rongen, G.A.; van Schaik, R.H.N.; et al. Dutch Pharmacogenetics Working Group (DPWG) guideline for the gene-drug interaction of DPYD and fluoropyrimidines. Eur. J. Hum. Genet. 2020, 28, 508–517. [Google Scholar] [CrossRef] [PubMed]
- Toffoli, G.; Giodini, L.; Buonadonna, A.; Berretta, M.; De Paoli, A.; Scalone, S.; Miolo, G.; Mini, E.; Nobili, S.; Lonardi, S.; et al. Clinical validity of a DPYD-based pharmacogenetic test to predict severe toxicity to fluoropyrimidines. Int. J. Cancer 2015, 137, 2971–2980. [Google Scholar] [CrossRef] [PubMed]
- Schwab, M.; Zanger, U.M.; Marx, C.; Schaeffeler, E.; Klein, K.; Dippon, J.; Kerb, R.; Blievernicht, J.; Fischer, J.; Hofmann, U.; et al. Role of genetic and nongenetic factors for fluorouracil treatment-related severe toxicity: A prospective clinical trial by the German 5-FU toxicity study group. J. Clin. Oncol. 2008, 26, 2131–2138. [Google Scholar] [CrossRef] [PubMed]
- Pellicer, M.; García-González, X.; García, M.I.; Blanco, C.; García-Alfonso, P.; Robles, L.; Grávalos, C.; Rueda, D.; Martínez, J.; Pachón, V.; et al. Use of exome sequencing to determine the full profile of genetic variants in the fluoropyrimidine pathway in colorectal cancer patients affected by severe toxicity. Pharmacogenomics 2017, 18, 1215–1223. [Google Scholar] [CrossRef]
- Hariprakash, J.M.; Vellarikkal, S.K.; Keechilat, P.; Verma, A.; Jayarajan, R.; Dixit, V.; Ravi, R.; Senthivel, V.; Kumar, A.; Sehgal, P.; et al. Pharmacogenetic landscape of DPYD variants in south Asian populations by integration of genome-scale data. Pharmacogenomics 2018, 19, 227–241. [Google Scholar] [CrossRef]
- Kleibl, Z.; Fidlerova, J.; Kleiblova, P.; Kormunda, S.; Bilek, M.; Bouskova, K.; Sevcik, J.; Novotny, J. Influence of dihydropyrimidine dehydrogenase gene (DPYD) coding sequence variants on the development of fluoropyrimidine-related toxicity in patients with high-grade toxicity and patients with excellent tolerance of fluoropyrimidine-based chemotherapy. Neoplasma 2009, 56, 303–316. [Google Scholar] [CrossRef] [Green Version]
- Gross, E.; Busse, B.; Riemenschneider, M.; Neubauer, S.; Seck, K.; Klein, H.-G.; Kiechle, M.; Lordick, F.; Meindl, A. Strong association of a common dihydropyrimidine dehydrogenase gene polymorphism with fluoropyrimidine-related toxicity in cancer patients. PLoS ONE 2008, 3, e4003. [Google Scholar] [CrossRef] [Green Version]
- Del Re, M.; Cinieri, S.; Michelucci, A.; Salvadori, S.; Loupakis, F.; Schirripa, M.; Cremolini, C.; Crucitta, S.; Barbara, C.; Di Leo, A.; et al. DPYD*6 plays an important role in fluoropyrimidine toxicity in addition to DPYD*2A and c.2846A>T: A comprehensive analysis in 1254 patients. Pharm. J. 2019, 19, 556–563. [Google Scholar] [CrossRef]
- Falvella, F.S.; Cheli, S.; Martinetti, A.; Mazzali, C.; Iacovelli, R.; Maggi, C.; Gariboldi, M.; Pierotti, M.A.; Di Bartolomeo, M.; Sottotetti, E.; et al. DPD and UGT1A1 deficiency in colorectal cancer patients receiving triplet chemotherapy with fluoropyrimidines, oxaliplatin and irinotecan. Br. J. Clin. Pharmacol. 2015, 80, 581–588. [Google Scholar] [CrossRef] [Green Version]
- Hamzic, S.; Schärer, D.; Offer, S.M.; Meulendijks, D.; Nakas, C.; Diasio, R.B.; Fontana, S.; Wehrli, M.; Schürch, S.; Amstutz, U.; et al. Haplotype structure defines effects of common DPYD variants c.85T > C (rs1801265) and c.496A > G (rs2297595) on dihydropyrimidine dehydrogenase activity: Implication for 5-fluorouracil toxicity. Br. J. Clin. Pharmacol. 2021, 87, 3234–3243. [Google Scholar] [CrossRef]
- Gokare, P.; Finnberg, N.K.; Abbosh, P.H.; Dai, J.; Murphy, M.E.; El-Deiry, W.S. P53 represses pyrimidine catabolic gene dihydropyrimidine dehydrogenase (DPYD) expression in response to thymidylate synthase (TS) targeting. Sci. Rep. 2017, 7, 9711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Z.; Zhang, R.; Diasio, R.B. Population characteristics of hepatic dihydropyrimidine dehydrogenase activity, a key metabolic enzyme in 5-fluorouracil chemotherapy. Clin. Pharmacol. Ther. 1995, 58, 512–522. [Google Scholar] [CrossRef]
- Uetake, H.; Ichikawa, W.; Takechi, T.; Fukushima, M.; Nihei, Z.; Sugihara, K. Relationship between intratumoral dihydropyrimidine dehydrogenase activity and gene expression in human colorectal cancer. Clin. Cancer Res. 1999, 5, 2836–2839. [Google Scholar]
- Wigle, T.J.; Tsvetkova, E.V.; Welch, S.A.; Kim, R.B. DPYD and Fluorouracil-Based Chemotherapy: Mini Review and Case Report. Pharmaceutics 2019, 11, 199. [Google Scholar] [CrossRef] [Green Version]
- Kato, H.; Naiki-Ito, A.; Suzuki, S.; Inaguma, S.; Komura, M.; Nakao, K.; Naiki, T.; Kachi, K.; Kato, A.; Matsuo, Y.; et al. DPYD, down-regulated by the potentially chemopreventive agent luteolin, interacts with STAT3 in pancreatic cancer. Carcinogenesis 2021, 42, 940–950. [Google Scholar] [CrossRef]
- Milano, G.; Fischel, J.L.; Etienne, M.C.; Renée, N.; Formento, P.; Thyss, A.; Gaspard, M.H.; Thill, L.; Cupissol, D. Inhibition of dihydropyrimidine dehydrogenase by alpha-interferon: Experimental data on human tumor cell lines. Cancer Chemother. Pharmacol. 1994, 34, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Sakowicz-Burkiewicz, M.; Przybyla, T.; Wesserling, M.; Bielarczyk, H.; Maciejewska, I.; Pawelczyk, T. Suppression of TWIST1 enhances the sensitivity of colon cancer cells to 5-fluorouracil. Int. J. Biochem. Cell Biol. 2016, 78, 268–278. [Google Scholar] [CrossRef]
- Lin, Y.; Gao, Z.-X.; Shen, X.; Chen, M.-J.; Li, Y.-T.; Li, S.-L.; Lin, H.-L.; Zhao, Q.-F.; Liu, F.; Niu, J.-J. Correlation between polymorphisms in toll-like receptor genes and the activity of hepatitis B virus among treatment-naïve patients: A case-control study in a Han Chinese population. BMC Infect. Dis. 2018, 18, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Offer, S.M.; Butterfield, G.L.; Jerde, C.R.; Fossum, C.C.; Wegner, N.J.; Diasio, R.B. MicroRNAs miR-27a and miR-27b directly regulate liver dihydropyrimidine dehydrogenase expression through two conserved binding sites. Mol. Cancer Ther. 2014, 13, 742–751. [Google Scholar] [CrossRef] [Green Version]
- Esteller, M. Relevance of DNA methylation in the management of cancer. Lancet. Oncol. 2003, 4, 351–358. [Google Scholar] [CrossRef]
- Ezzeldin, H.H.; Lee, A.M.; Mattison, L.K.; Diasio, R.B. Methylation of the DPYD promoter: An alternative mechanism for dihydropyrimidine dehydrogenase deficiency in cancer patients. Clin. Cancer Res. 2005, 11, 8699–8705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chavani, O. 5-Fluorouracil Response Prediction and Blood Level-Guided Therapy in Oncology: Existing Evidence Fundamentally Supports Instigation. Ther. Drug Monit. 2020, 42, 660–664. [Google Scholar] [CrossRef]
- Thomas, F.; Maillard, M.; Launay, M.; Tron, C.; Etienne-Grimaldi, M.-C.; Gautier-Veyret, E.; Haufroid, V.; Pallet, N.; Royer, B.; Narjoz, C.; et al. Artificial increase of uracilemia during fluoropyrimidine treatment can lead to DPD deficiency misinterpretation. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2021, 32, 810–811. [Google Scholar] [CrossRef] [PubMed]
- Knikman, J.E.; Gelderblom, H.; Beijnen, J.H.; Cats, A.; Guchelaar, H.-J.; Henricks, L.M. Individualized Dosing of Fluoropyrimidine-Based Chemotherapy to Prevent Severe Fluoropyrimidine-Related Toxicity: What Are the Options? Clin. Pharmacol. Ther. 2021, 109, 591–604. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).