3.1. Revealing Differences between Cohorts in Terms of Phenotype Information
We applied Cohort Analyzer to the three different patient cohorts to show how it can be used to investigate phenotype information in datasets of very different designs.
As shown in
Table 3, the DECIPHER dataset shows a much larger number of unique HPO terms and patients than the others, as expected given the resource was designed to collect data for a wide range of phenotypically heterogeneous patients as part of an international initiative [
14]. The ID/MCA and PMM2-CDG datasets show much closer numbers of unique HPO terms; however, they differ greatly in terms of the number of patients. The average numbers of HPO terms per patient differ greatly between groups, being several times higher for the PMM2-CDG dataset. Even more striking is the difference in terms of the number of phenotypes at the 90th percentile, indicating that 10% of DECIPHER and ID/MCA patients have only one HPO term to define their clinical profile in contrast to PMM2-CDG cohort, for which 90th percentile patients have 15 HPO terms.
In terms of phenotype depth, for most of the HPO terms used to describe the patients in the ID/MCA dataset, more specific child terms were available. This also occurred with the DECIPHER and PMM2-CDG datasets but to a much lesser extent, in fact almost half of the HPO terms used in the PMM2-CDG dataset were the most specific terms available. Furthermore, the patient profile length in PMM2-CDG dataset is very large, ~5 times and ~10 times the size of DECIPHER and ID/MCA datasets, respectively. Of particular note was that the ID/MCA cohort patients were assigned less than three phenotypes on average, and that almost all of the phenotypes had more specific ancestors.
These summary statistics provide a clear overview of the properties of the different datasets in terms of how thoroughly the patients have been phenotyped in terms of both breadth and depth; moreover, they give an idea of how consistent the phenotyping is across patients.
Cohort Analyzer can be used to assess the most frequent HPO terms among patients within a dataset. It was applied to the three datasets used in this study (
Table 4).
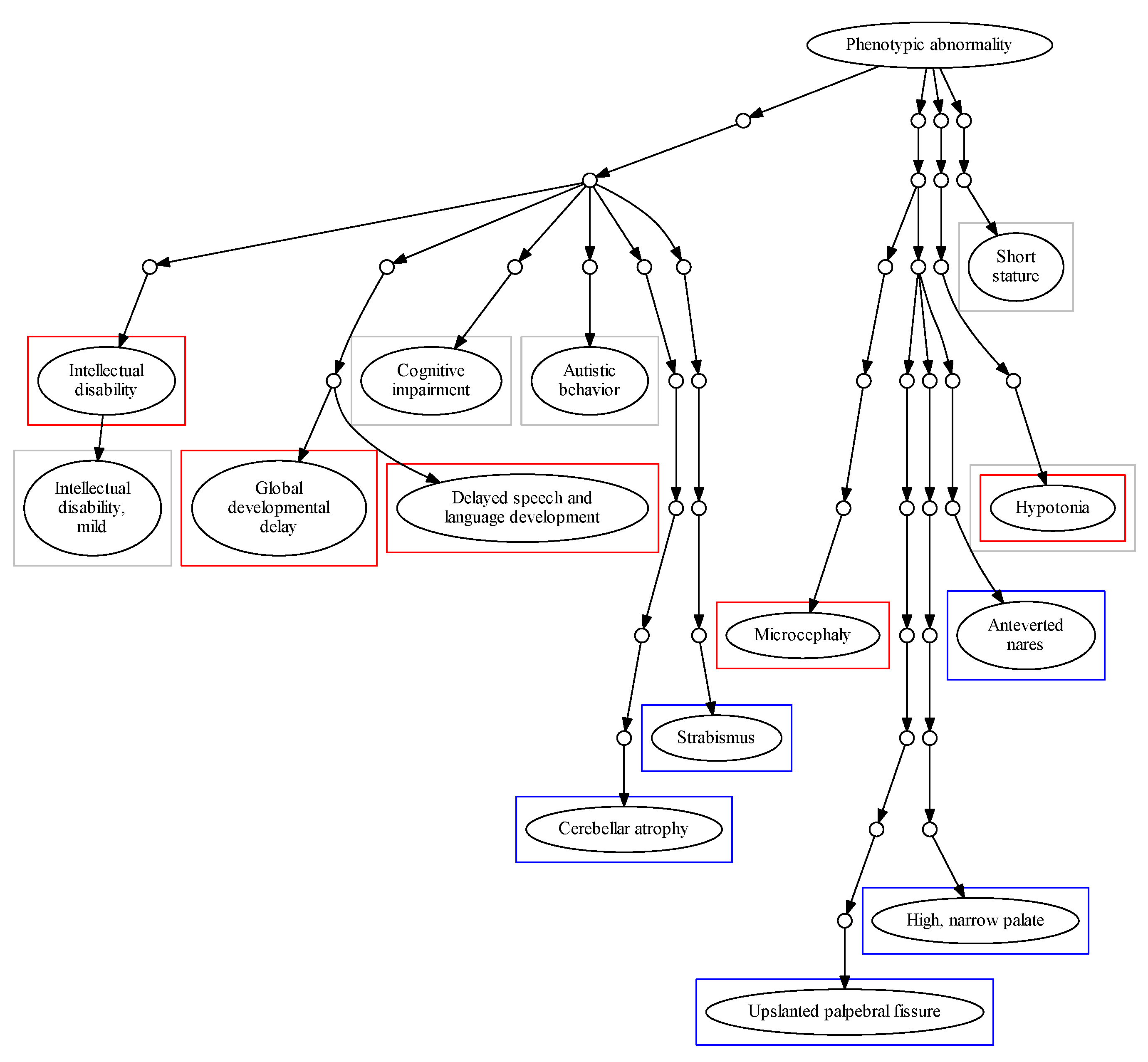
Figure 2 shows the position of these phenotypes within the HPO hierarchical structure. The term HP: “Intellectual disability” and its degrees were highly frequent among patients in the DECIPHER and ID/MCA datasets, with 34.98% of patients in DECIPHER dataset being ascribed this term, and 17.67% of patients in the ID/MCA cohort ascribed its child term, HP: “Intellectual disability, mild”. Whilst the high prevalence of such a term in these datasets might be expected, and it may be a useful phenotype in conjunction with a highly detailed phenotypic profile, by itself it is less useful; moreover, it is found at level 5 of the HPO and its child terms only describe different severity grades; as such, it represents somewhat of a phenotyping dead end within the HPO. This pathological trait is complex and encompasses multiple cognitive deficits expressed to several degrees, with multiple potential causes. Therefore, its precise description can be overly diffuse [
34]. Similar problems occur with other frequent, general terms, such as HP: “Global developmental delay”. This limits the ability of the practitioner to provide a more specific diagnosis in this branch of the HPO. For other phenotype, such as HP: “Cognitive impairment”, ascribed to 80.96% of the ID/MCA cohort members, there are myriad child terms available, including HP: “Mental deterioration” and HP: “Memory impairment”, suggesting unexplored phenotypic space within the cohort. This is also the case for HP: “Delayed speech and language development”, which is at the sixth HPO level but has several child levels.
For the PMM2-CDG cohort, we found significant differences between the top terms of this dataset in comparison with the DECIPHER and ID/MCA datasets. We found that all patients were described with HP: “Cerebellar atrophy” and most of them with HP: “Upslanted palpebral fissure” (88.88%), two very specific terms (ninth and eleventh HPO levels, respectively). In fact, the frequency of these pathological terms within the cohort is very high, revealing a high level of phenotypic homogeneity in the cohort.
This characteristic could be expected for a monogenic disease dataset. However, it is interesting that the specific HPO terms describing precise attributes of PMM2-CDG dataset are the most prevalent, whereas for the other cohorts the most common terms are far more general. This makes sense given the findings in
Table 3, showing that many of the phenotypes ascribed to these patients are the most specific possible within the HPO, i.e., have no child terms.
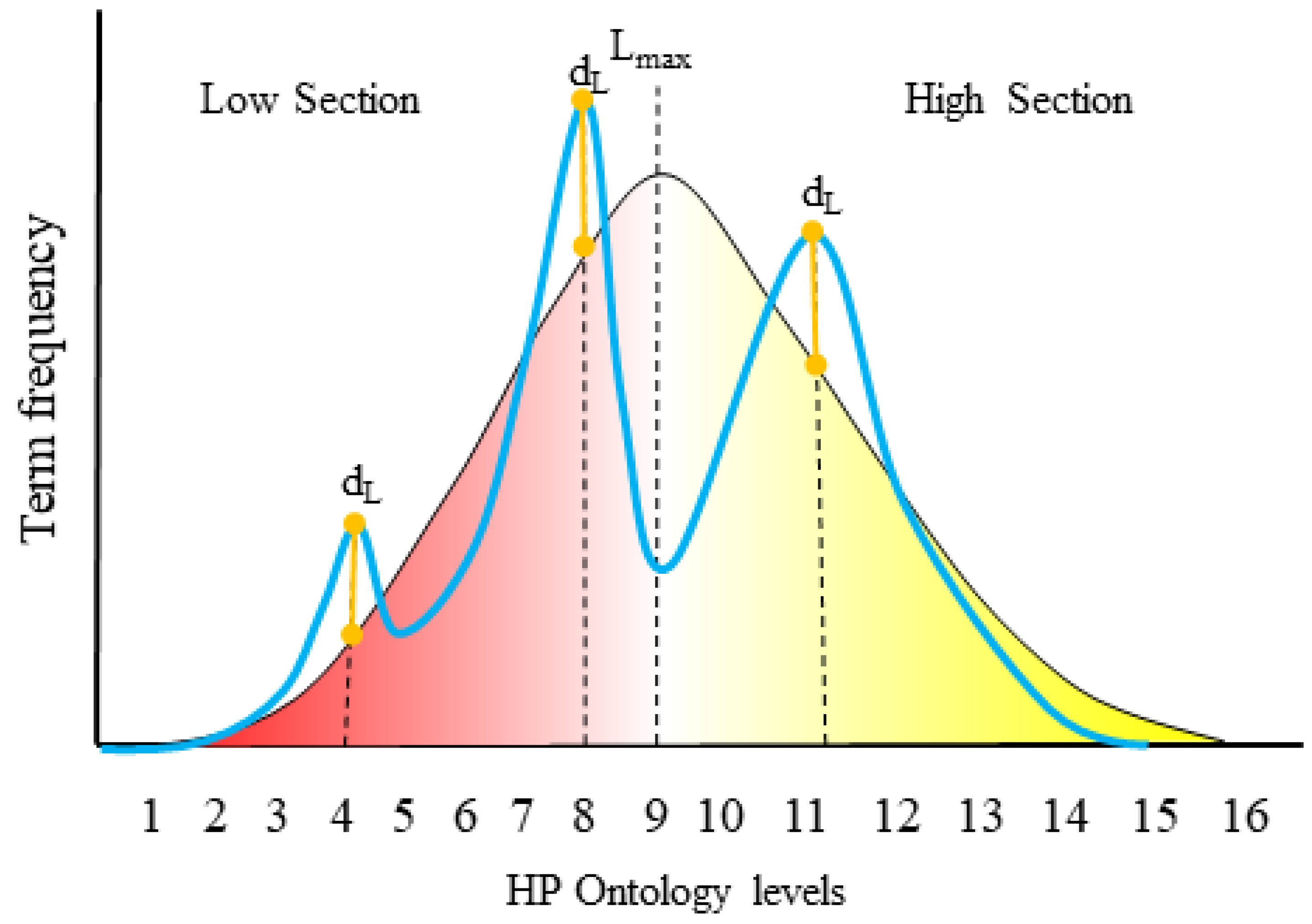
To provide a more detailed overview of phenotype specificity, Cohort Analyzer compares the distribution of HPO term levels used within a given cohort to the distribution of HPO term levels for all terms within the ontology (
Figure 3). This is performed taking into account term frequency (blue curves, “weighted cohort”) or counting each unique term only once (green curves, “unique terms cohort”). This distinction is important as a single, highly phenotyped patient could strongly affect the unique terms cohort, but its effect on the weighted cohort curve would be diluted. The distribution of terms within the HPO is represented as a pink curve.
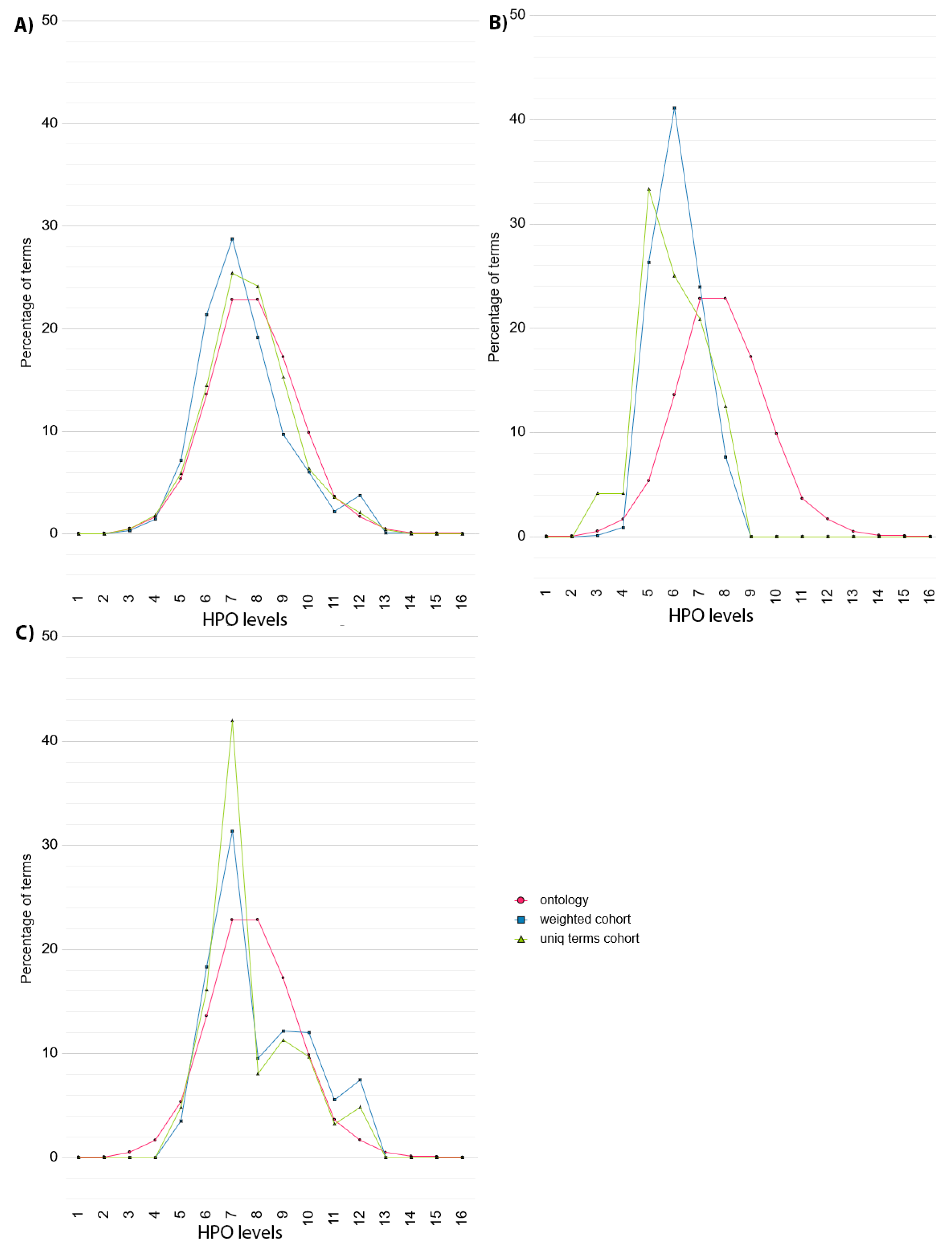
In the case of DECIPHER dataset (
Figure 3A), the HPO terms used in the dataset (green curve) show a similar distribution to the HPO (pink curve), with two peaks at level 7 and 8. When considering the frequency of each term (blue curve), the distribution is shifted slightly towards the initial levels of the HPO, although there is a small increase compared to the HPO at level 12.
In contrast, the distribution shown by the ID/MCA cohort data (
Figure 3B) are skewed far more to the left, towards the initial levels of the HPO (green curve), with peaks at level 3 and 5. There are no terms described from level 8 onwards, showing that the deepest half the HPO has not been used to describe the patients, suggesting unexplored phenotypic space for this dataset.
For the PMM2-CDG dataset, the distribution of unique HPO terms (green curve) has a small increase at level 6 and a high peak at level 7, followed by a smaller peak at level 12. When the HPO term frequency is considered (blue curve), this shifts in favour of deeper levels of the HPO, reducing the high peak at level 7 and increasing the peaks at 10 to 12. This pattern is suggestive of a common phenotype at level 7, but additional, more specific phenotypes at deeper levels. The shift to the right when taking term frequency into account suggests that many of the patients have been phenotyped deeply.
To quantify the extent to which a cohort has been phenotyped in terms of HPO depth, we used the Dataset specificity Index (DsI), applying it to all cohort datasets, both for unique terms and considering term frequency (
Table 5).
In the case of DECIPHER dataset, the DsI value is 0.13 for the unique HPO terms used to describe the cohort, in accordance to the distribution shift to the shallower levels of the HPO observed in
Figure 3A. When DsI is computed taking the frequency of each term within the cohort into account, the value slightly increases to 0.195, due to the peak at level 12. This suggests that DECIPHER patients are described using a wide range of HPO terms, representative of the HPO itself, in line with the nature of the resource. However, when we consider term frequency, the reduction in DsI suggests that many patients are actually phenotyped using much less specific terms.
In the case of ID/MCA cohort, DsI values for both unique HPO terms and the frequency of each term within the cohort is zero because this dataset has zero phenotypes in the
High section of the HPO, in line with
Figure 3B.
Higher DsI values where found for the PMM2-CDG dataset. Considering the unique HPO terms used to describe the cohort, the DsI value was 0.27; however, when calculating the frequency of each term within the cohort, this increased to 1.06. This increase in score suggests that many of the patients have been deeply phenotyped, in line with the change in distributions seen in
Figure 3B—peaks at levels 10, 11 and 12 explain this increment. Again, this suggests that not only are highly informative phenotypes used for this dataset, they have been used to described a relatively large number of patients.
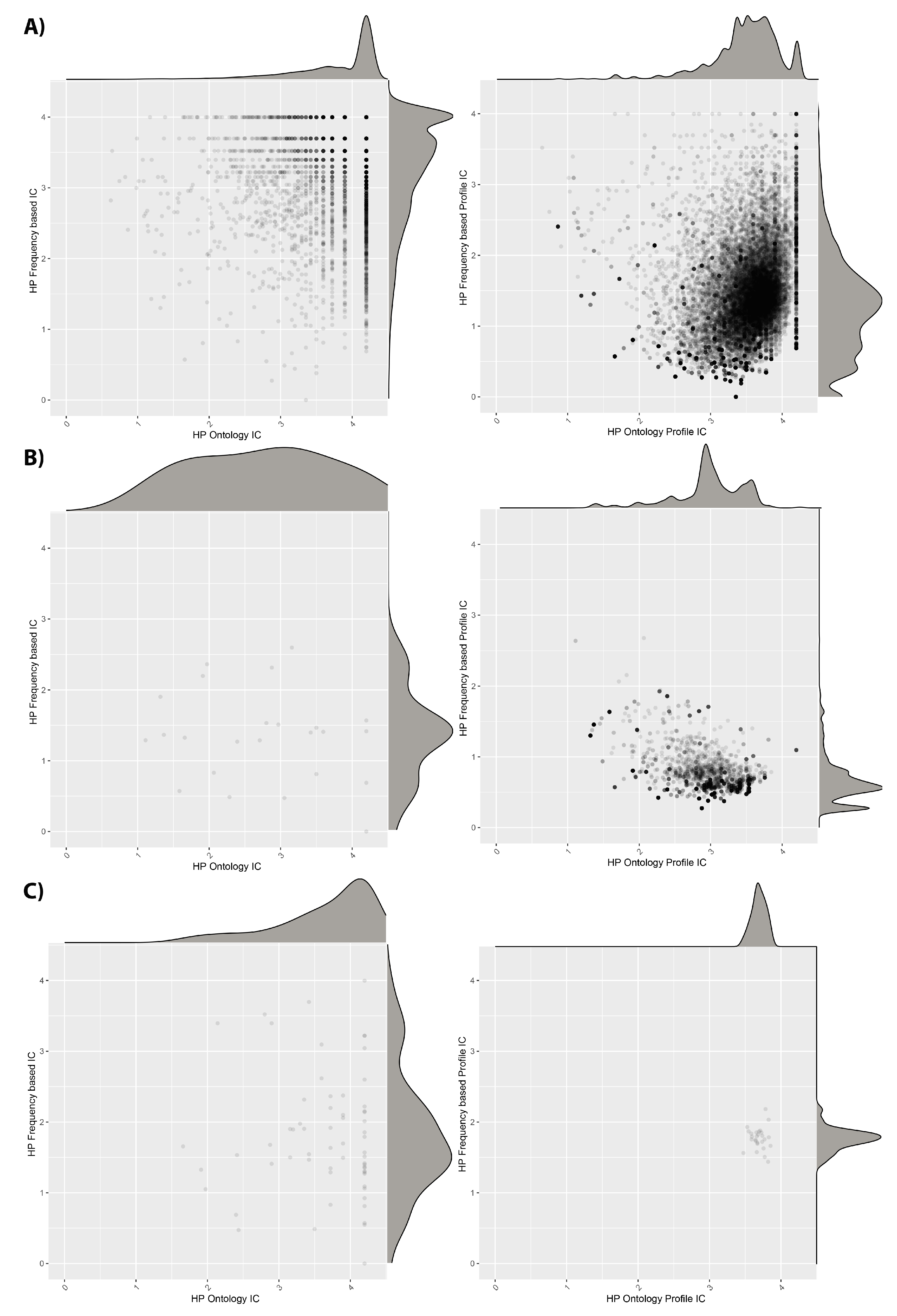
The information content (IC) values for individual HPO terms and phenotypic profiles, in terms of their frequency within the HPO and the cohorts, are shown in
Figure 4. We see that the DECIPHER dataset uses HPO terms with relatively high IC according to both the ontology and the dataset calculations. However, when we look at the IC averaged across patient profiles, the dataset-frequency IC drops dramatically. This suggests that, whilst there are many informative HPO terms used in DECIPHER, the majority of the patients have combinations of less specific ones, in line with the reduction in DsI shown in
Table 5 between unique and weighted values. For the ID/MCA dataset, the individual ICs are less informative, as is also the case for the patient profile ICs, also in line with
Table 5. However, in the case of the PMM2-CDG dataset, although the individual ICs are quite low, when IC is calculated for the patient profiles, it improves, leading to higher values than for the other datasets. This also fits with the DsI values, and fits with the idea that the patients within this dataset have been consistently phenotyped to a deep level.
3.2. Identifying Patient Subgroups with Low Information Profiles
Cohort Analyzer also performs clustering analysis to assess the phenotypic information in a cohort and identify patients with less informative phenotypic profiles. This initial procedure ignores the ontological attributes of the HPO terms; as such, we have named it Naïve clustering. We assume that if patients within a cohort are well-phenotyped, their profiles will include multiple, specific HPO terms. Conversely, the profiles of uninformative patients will include smaller profiles with more general HPO terms. As such, the profiles of these patients are more likely to be similar across a cohort and, therefore, to cluster together.
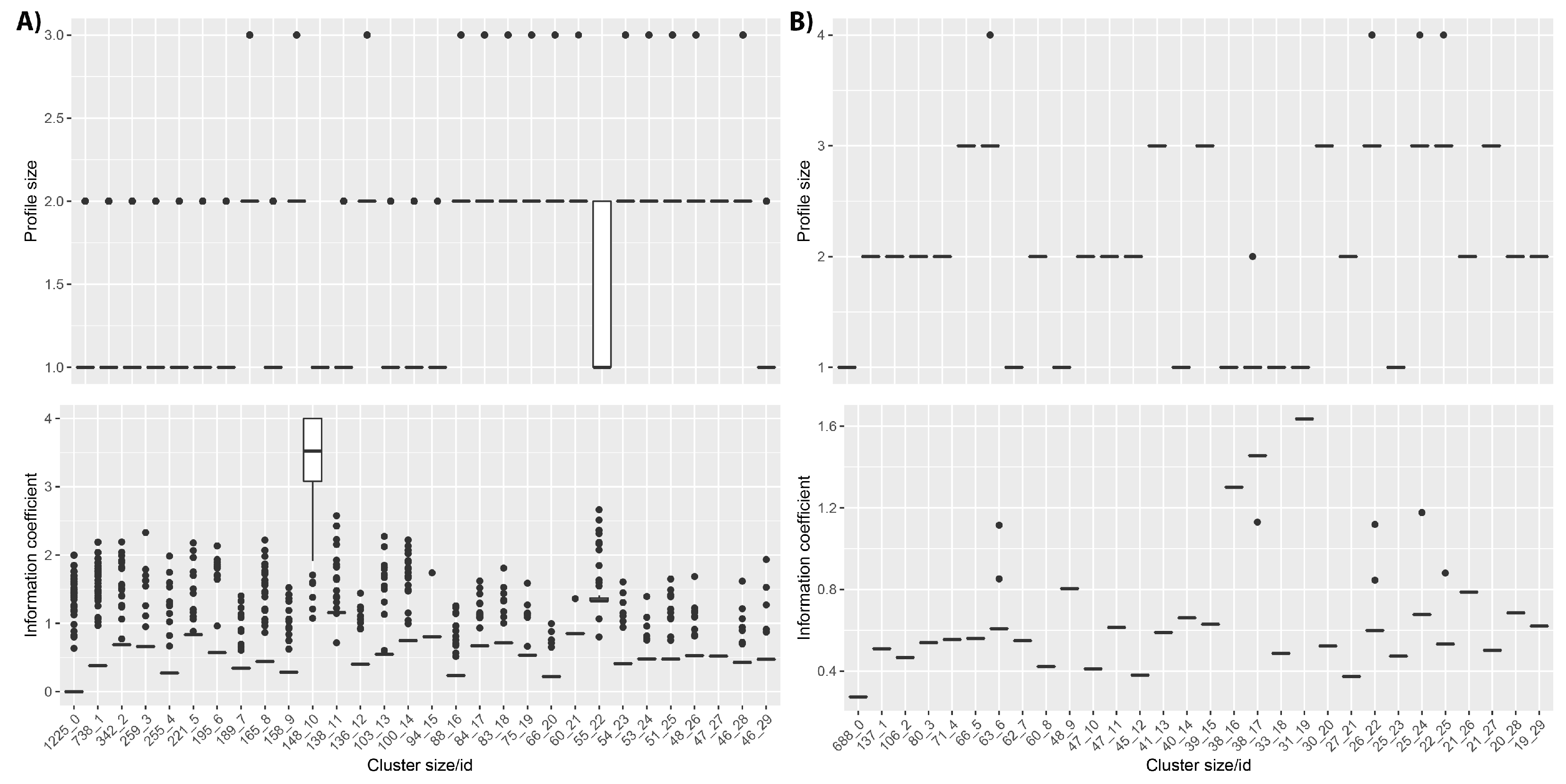
This is shown for the the DECIPHER cohort (
Figure 5), for which the first four clusters include more than 2500 patients with profile IC values between 0 and 1. These clusters contain patients with profiles describing only one or two HPO terms and frequently contain the same combination of HPO terms repeated for all patients, or possibly including only one different HPO term as shown in
Supplementary Table S1. Notably, cluster 10 has an average IC greater than 3.5. The patients within this cluster do often have high IC profiles; however, this is because they have only been phenotyped with a single HPO term, and this HPO term has not been ascribed to any other patients within the database. In fact, this cluster includes 148 different HPO terms described for 148 patients.
In the case of the ID/MCA cohort, clusters include lower number of phenotypes in comparison to the DECIPHER dataset and all members in each cluster have identical phenotypes, except for clusters 6, 17, 22 and 24. These clusters contain patients with profiles containing only one or two HPO terms assigned to all patients within cluster, as shown in
Supplementary Table S2. For the PMM2-CDG dataset, the Naïve clustering produces almost as many clusters as patients (data not shown). This is to be expected, given that the patients have been ascribed a large number of phenotypes, with no two patients having the same phenotypic profile.
We conclude that Naïve clustering can identify large groups of patients with very small phenotype profiles (one or two terms per patient) that also have low IC values. These patients do not provide enough information to be used in downstream analysis such as clustering-based semantic similarity to find subgroups of phenotypically similar patients. Consequently, we should consider removing these patients. Patient removal must be performed carefully, since the total number of unique phenotypes used to characterize the cohort can also be affected and some specific phenotypes can be removed. As such, the effects of filtering on the dataset should be examined.
3.3. Removing Patients with Limited Phenotypic Information from the Cohort and Its Effect on the Dataset Properties
Given that DECIPHER and ID/MCA datasets contain large numbers of patient with very small phenotype profiles, we investigated the consequences of removing these patients on the summary statistics and other cohort properties.
We see in the rightmost columns of
Table 3 that for both datasets, this filter barely reduces the total number of unique HPO terms; however, it reduces the total number of patients in the dataset by almost half. This shows the phenotypes ascribed to the filtered patients were also found among the remaining patients. As expected, the mean HPO terms per patient and HPO terms for percentile 90 both increased. The percentage of HPO terms with more specific child terms only reduces slightly, in line with the low-information patients representing a subset of the terms held by the high-information patients. This shows that removing these patients has little effect on the phenotypic diversity of the dataset. Interestingly, the most common phenotypes actually became more frequent within the DECIPHER and ID/MCA datasets after filtering, suggesting that these phenotypes were more frequently found within longer phenotypic profiles (
Supplementary Table S3).
As can be seen in
Table 5, for the DECIPHER dataset, the DsI calculated for the unique terms increases very slightly after the filter, showing that the few unique HPO terms that were removed were of lower-information content, this is also reflected by the slight shift to the right in
Supplementary Figure S1 compared to
Figure 3. However, for weighted terms, the increase was slightly more marked, with the DsI increasing by a larger amount and an appreciable shift towards deeper levels in
Supplementary Figure S1. This suggests that the filtered patients not only had few ascribed phenotypes, but that the phenotypes tended to be unspecific. For the ID/MCA dataset, there was no change in DsI—this remained as 0, due to this cohort having HPO terms corresponding to the
High section levels, something that cannot be improved by anything other than more thorough phenotyping of the patients.
In terms of the IC values (
Supplementary Figure S2), we see that for all cohorts, removing the low-information patients has little effect on the distributions of IC values for individual HPO terms, in line with the small reduction in total unique terms across the cohorts (
Table 3). However, when the IC values calculated using the phenotypic profiles of each patient are considered, we see a clear smoothing of the distributions, particular for the cohort-frequency calculated values, for both DECIPHER and ID/MCA cohorts. For these datasets, large peaks corresponding to groups of low IC patients are removed, in line with the idea that many of the patients with few phenotypes have also been assigned unspecific ones. For the DECIPHER dataset, there is also a clear peak of high profile IC values for HPO ontology-based values before filtering; this may be due to the patients with single but unique phenotypes found in cluster 10 in
Figure 5, the patient IC is the same as the phenotype IC for these patients because their profiles only contain single terms.
This suggests that some of the filtered patients had specific phenotypic profiles according to the ontology, but that were less specific within the cohort itself. No appreciable change is apparent for the PMM2-CDG cohort, unsurprising given that no patients were removed, although the cohort-frequency of the HPO terms changed very slightly.
In terms of the Naïve clustering, for the unfiltered DECIPHER dataset (
Figure 5) there were many clusters containing hundreds of patients with identical low IC phenotypes alongside a handful of outlier patients with slightly higher ICs. Removing the very small phenotype patients and repeating the Naïve clustering led to much smaller clusters with a higher range of ICs (
Supplementary Figure S3), as would be expected.
This was less clear for the ID/MCA dataset—although several very large clusters were removed, the remaining ones also showed a small range of ICs. This may be due to the patients having fewer phenotypes, most of which had more specific child terms, even after filtering (
Table 3), in line with the DsI values of 0 and lower patient level IC values (
Supplementary Figure S2), all indicative of these patients having, in general, small phenotypic profiles consisting of unspecific HPO terms.
3.4. Comparing Phenotype Profiles to Cluster Patients into Phenotypically-Related Subgroups
After removing poorly-phenotyped patients, it was possible to analyze the cohorts to identify groups of phenotypically related patients. Cohort Analyzer calculates pairwise semantic similarity values between the phenotypic profiles of patients to generate a similarity matrix. Although three distinct similarity measures can be used (Resnik, Lin and Jiang–Conrath), here, we present results for the Lin similarity measure. It normalizes values between 0 (no similarity) and 1 (maximum similarity), allowing the easy calculation of distance matrices for hierarchical clustering.
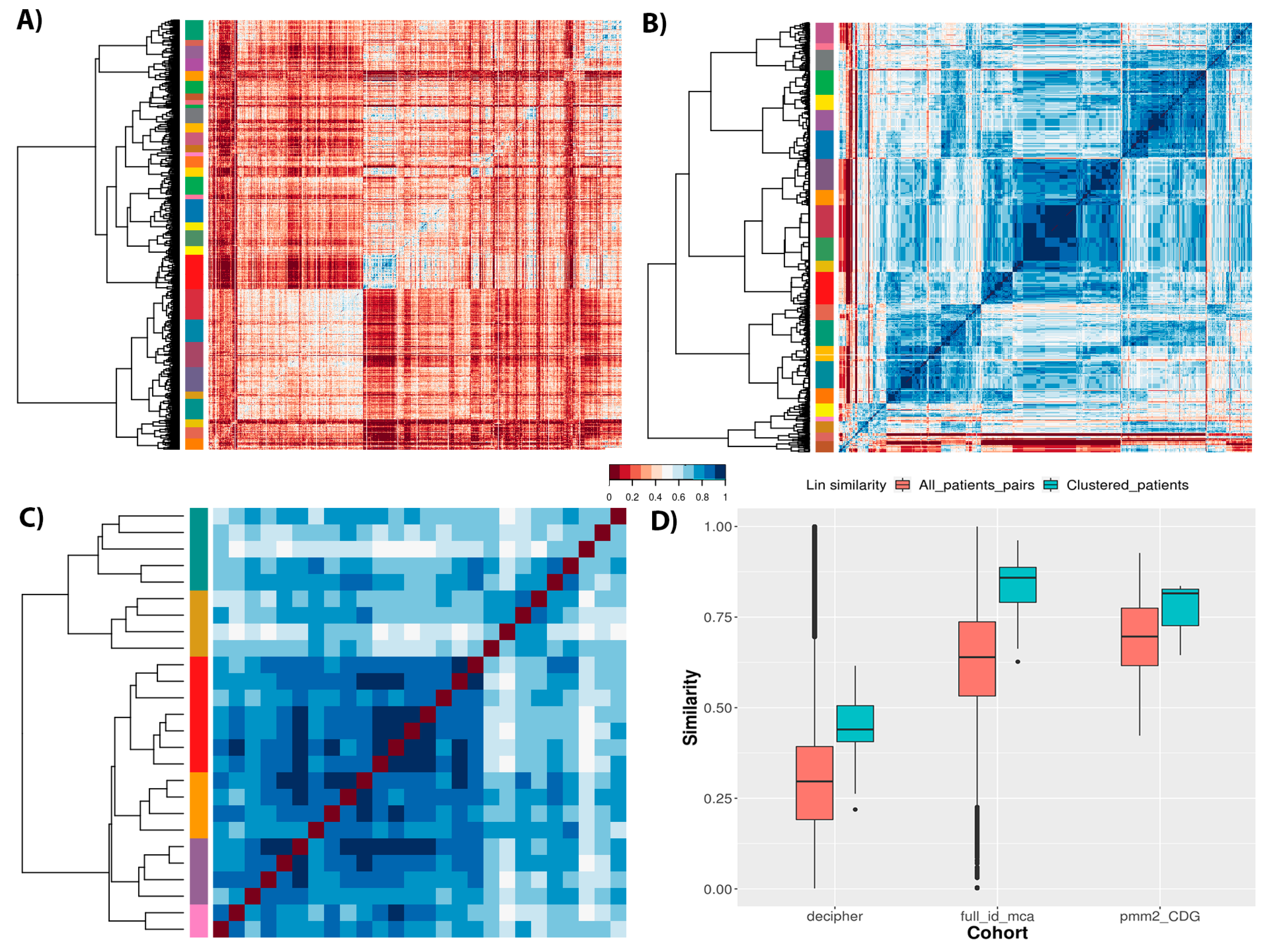
Figure 6 shows the semantic similarity matrices for the different cohorts, revealing the cohort structure and patient clustering for each. There is clearly much less similarity between most patients within the DECIPHER cohort than the others, in line with the distributions of similarity values for each cohort (
Figure 6D, salmon boxes). Notably, both DECIPHER and ID/MCA cohorts show a wide range of similarity values, whilst PMM2-CDG dataset shows a much smaller range, which is unsurprising given the first two are aimed at a wider range of patients, whilst the latter only contains patients diagnosed with the same monogenic disease. It should also be noted that the ID/MCA and PMM2-CDG cohorts have remarkably similar median similarity values (0.63 and 0.69, respectively), despite being very different in most other ways. This highlights the importance of looking at the full distribution of similarity values, and taking into account other cohort-related statistics, rather than simply comparing medians. Returning to the heatmaps, we see clear clusters of similar patients for the different datasets, although it is difficult to compare the datasets directly given the differences in total numbers of patients for each.
Finally, we checked the clustering homogeneity for each cohort calculating the average similarity measure for the members of each patient cluster as shown in
Figure 6D, blue boxes. The DECIPHER dataset showed an increase in average similarity to 0.43, suggesting a large number of phenotypically diverse patients per cluster. However, ID/MCA cohort showed the greatest increase average similarity, increasing from a similarity of 0.63 to 0.85. Conversely, PMM2-CDG cohort showed the smallest increment, from 0.69 to 0.81. These results suggest that ID/MCA cohort forms close clusters easily due to the very narrow phenotype spectrum and the small patient profiles, contrary to PMM2-CDG cohort.
3.5. Genomic Variant Data Analysis
Cohort Analyzer can also perform analysis of genomic variant data. Firstly, it computes various summary statistics, as shown in
Table 6, applied to the three datasets included in this study. We see that variant sizes are much greater for the DECIPHER and ID/MCA datasets; this is because they contain CNV data, whilst the PMM2-CDG dataset contains a range of variants affecting a single gene, as such the variant size refers to the
PMM2 gene coordinates (GRCh37/hg19 human genome assembly). Despite similar variant sizes, the DECIPHER dataset covers a larger proportion of the genome than ID/MCA dataset, in line with it containing a higher number of patients that are more phenotypically distinct.
Cohort Analyzer also includes metrics to analyse the overlap between patient variants. For this, it determines genome windows named Short Overlapping Regions (SOR), which consist of genomic regions shared by at least two patients in a given cohort. In the case of DECIPHER dataset, there are 39,136 genome distinct genomic windows, which are reduced to 39,109 when Cohort Analyzer establishes SORs, i.e., only including regions that overlap between patients. In the case of ID/MCA dataset, there are 1597 genomic windows, of which 1097 can be considered SORs.
With respect to the PMM2-CDG dataset, all metrics present the characteristics of a monogenic disease. Variant size and affected genome nucleotides agree with the PMM2 gene coordinates and there is only one genome window for all patients.
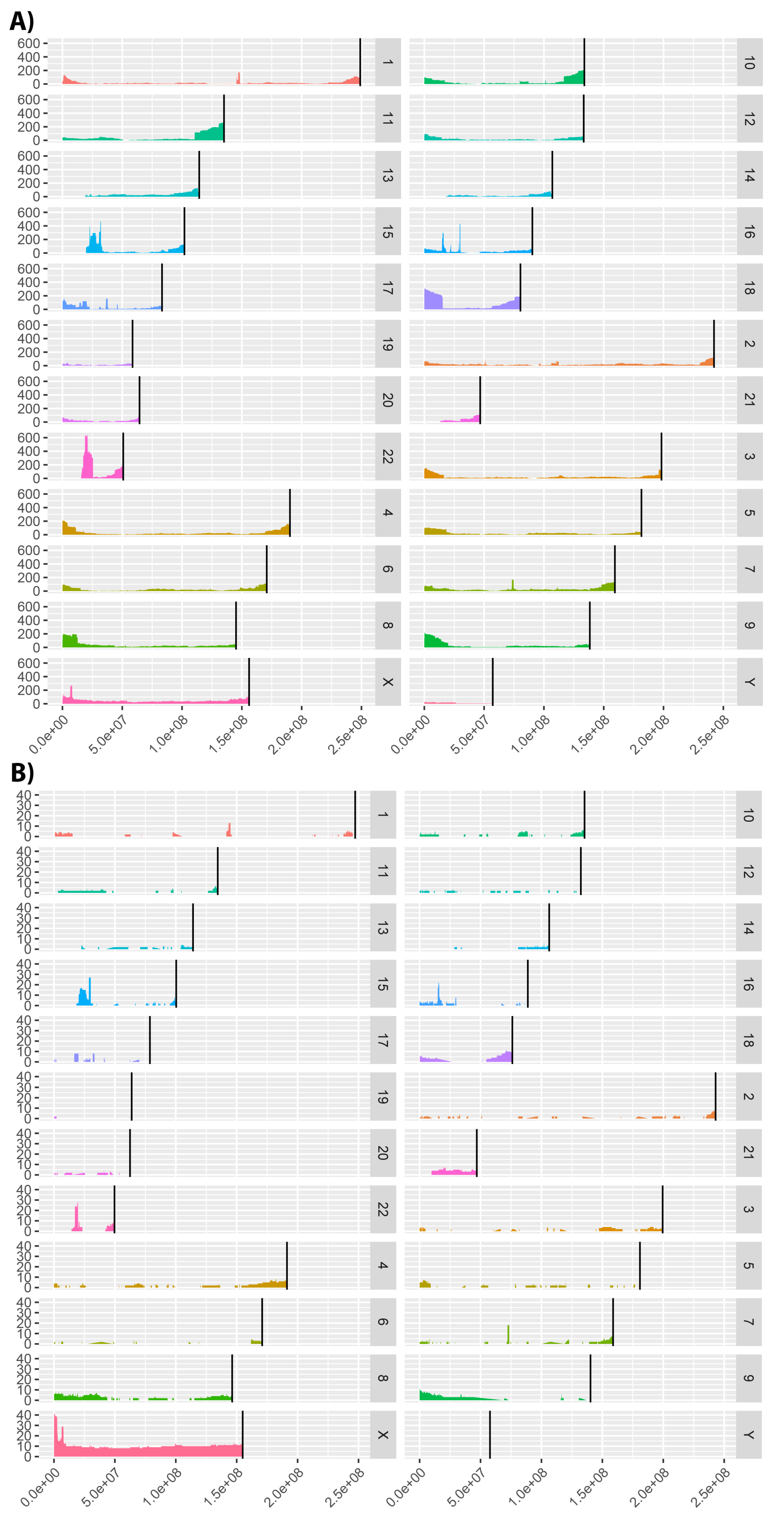
Furthermore, Cohort Analyzer generates a genome coverage graph showing patient variant distribution throughout the genome. We show the coverage for the DECIPHER and ID/MCA cohorts in
Figure 7. The human genome assembly versions were GRCh38/hg38 and NCBI36/hg18, respectively. Analysis was not performed on the PMM2-CDG dataset as only a single gene locus is implicated in these patients.
The DECIPHER dataset contains patients with variants affecting virtually all of the genome, albeit at low coverage is most places, whilst the ID/MCA dataset shows more defined islands of coverage surrounded by uncovered regions.
Interestingly, there are a number of clear peaks common to both datasets. We analyzed a number of these regions to confirm if they were related to known diseases, using the OMIM [
35] and Orphanet [
36] databases. Microdeletions in many of these genomic regions are associated with neurological diseases, such as intellectual disability, autism and schizophrenia [
37]. Specifically, microdeletions in the 15q11.2 and 16p13.11 regions have been associated with idiopathic generalized epilepsy [
37]. Peaks in chromosome 15 are in a genomic region containing variants that have also been associated with Prader–Willi syndrome (15q11-q13 duplication) [
38]. Deletions in the 22q11.21-q11.23 region that corresponds to the peak shown in chromosome 22 have been associated with DiGeorge syndrome [
39]. This is not as marked in the ID/MCA dataset, consistent with DECIPHER cohort containing more phenotypically diverse patients. In relation to peaks observed for ID/MCA dataset on chromosome X, a large number of diseases involving this chromosome have been described with pathological phenotypes including intellectual disability [
40], dystrophinopathies [
41] and cardiopathies [
42] among others [
43].
There are also regions with no coverage in either cohort, for example, the initial base pairs in chromosomes 13, 14, 15, 21 and 22. This may be due to these genomic regions not allowing variation for the viability of the organism, because no patients characterised with mutations in these regions or other limitations. However, more studies are required.