Multi-Institutional Implementation of Clinical Decision Support for APOL1, NAT2, and YEATS4 Genotyping in Antihypertensive Management
Abstract
1. Introduction
2. Materials and Methods
3. Results
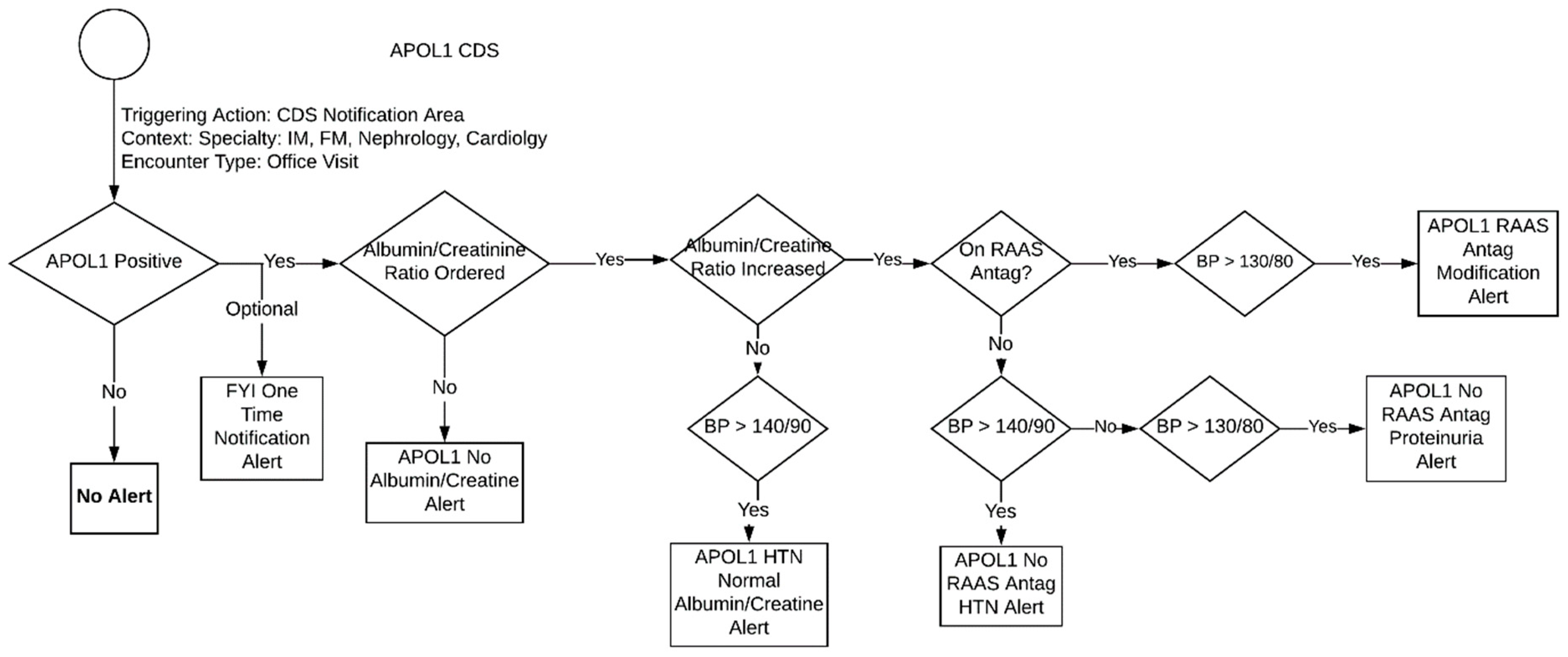
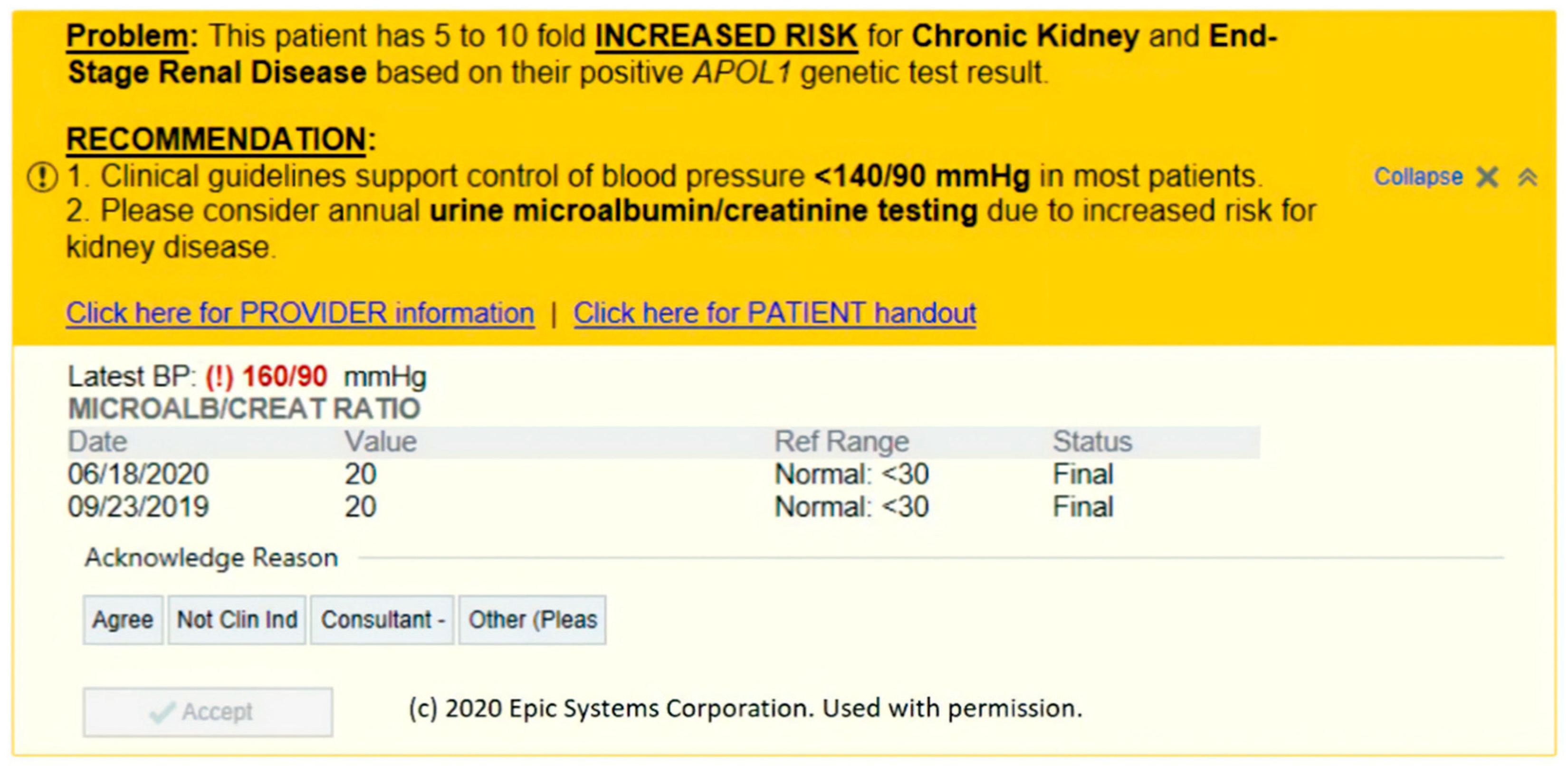
3.1. APOL1 Genotype-Guided Interventions
- One Time Notification Alert
- ∘
- General for your information (FYI) alert (no specific criteria) that triggers once, informing the clinic of the patient’s high-risk phenotype for CKD and the need for annual screening.
- APOL1 No Albumin/Creatinine Ratio Alert
- ∘
- This alert triggers at every encounter if there is no albumin/creatinine ratio result. It tells the provider that ordering an albumin/creatinine ratio is recommended due the patient’s high-risk allele.
- APOL1 HTN Normal Ratio Alert
- ∘
- Triggers at every encounter when the albumin/creatinine ratio is normal but the patient’s blood pressure is high. It then provides general blood pressure control recommendations.
- APOL1 NOT On RAAS Antagonist HTN Alert
- ∘
- This alert triggers at every encounter when the patient is (1) not on a RAAS antagonist (2) has a blood pressure that is >140/90 and (3) has an increased albumin/creatinine ratio. It then recommends that the provider add RAAS antagonist therapy to control HTN.
- APOL1 NOT On RAAS Antagonist Proteinuria Alert
- ∘
- This alert triggers at every encounter when the patient is (1) not on a RAAS antagonist and (2) has a blood pressure that is <140/90 but greater than >130/80. It then recommends that provider to add RAAS antagonist therapy to prevent proteinuria.
- APOL1 On RAAS Antagonist Modification Alert
- ∘
- This alert triggers at every encounter if the patient is (1) on a RAAS antagonist, (2) has a blood pressure that is >130/80, and (3) has an increased albumin/creatinine ratio. It then recommends the provider to modify the current RAAS antagonist therapy.
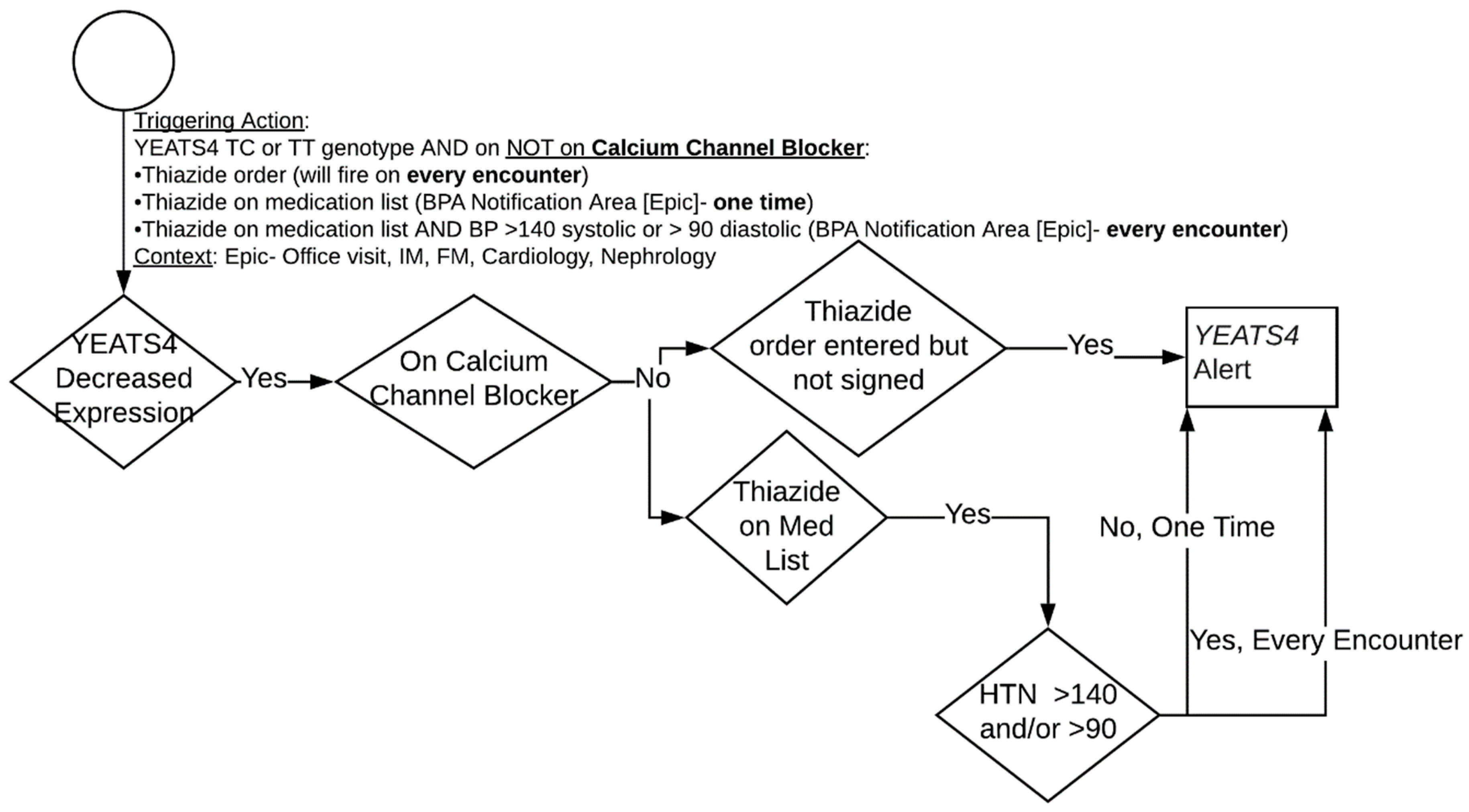
3.2. Hydrochlorothiazide and YEATS4
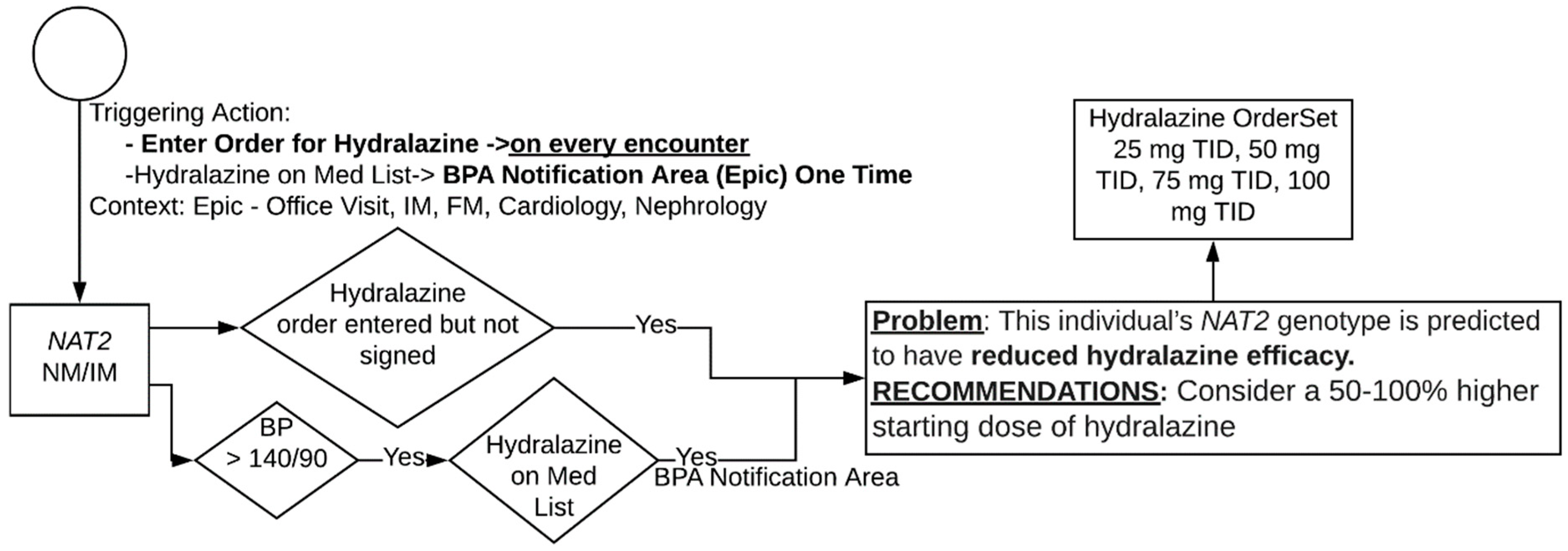
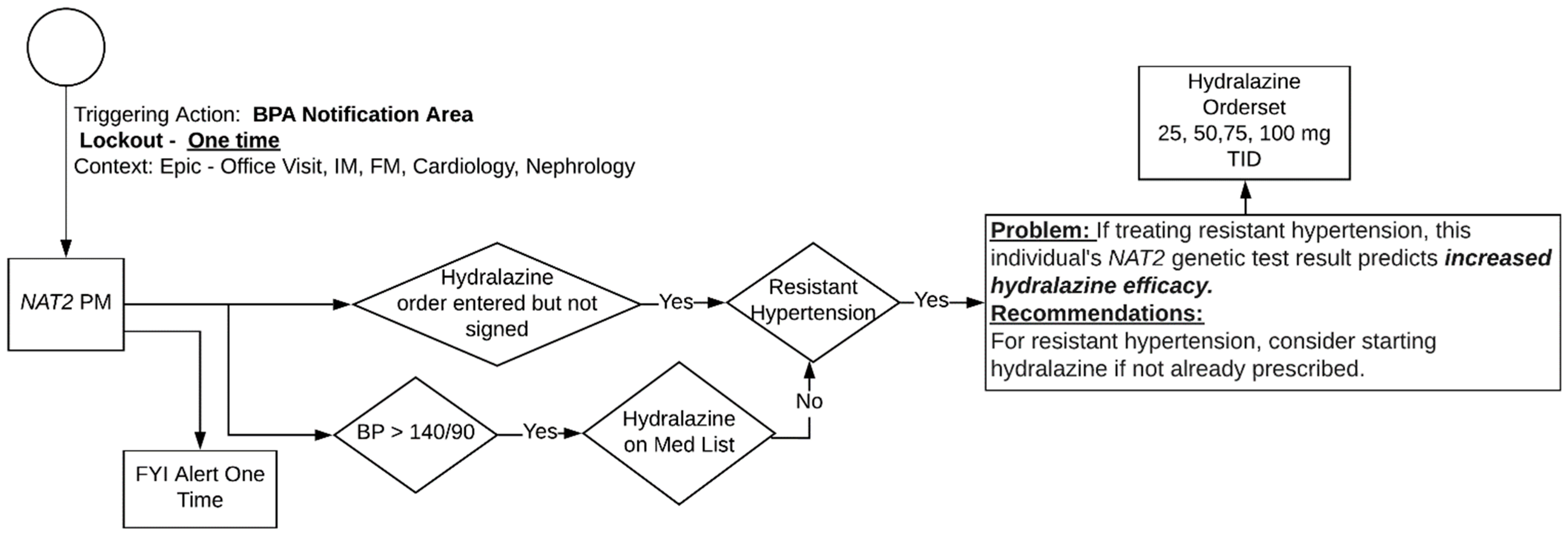
3.3. Hydralazine and NAT2
3.4. Portability to Other Sites
3.5. Other Sites Experience
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
NAT2 Algorithm
| Institution | Location | Clinical Group | EHR Vendor/Version |
|---|---|---|---|
| Icahn School of Medicine at Mount Sinai | New York City, NY | Mount Sinai | EpicCare Ambulatory Aug. 2019 |
| The Institute for Family Health | New York City, NY | Mount Sinai | EpicCare Ambulatory Feb. 2020 |
| University of Florida | Gainesville and Jacksonville, FL | University of Florida | EpicCare Ambulatory Nov. 2019 |
| Indiana University Health | Indianapolis, IN | Indiana University | Cerner Solutions 2018.01 |
| Eskenazi Health | Indianapolis, IN | Indiana University | EpicCare Ambulatory May 2019 |
| Vanderbilt University Medical Center | Nashville, TN | Vanderbilt University | EpicCare Ambulatory Nov. 2019 |
| Meharry Medical College and Nashville General Hospital | Nashville, TN | Vanderbilt University | EpicCare Ambulatory * |
| Baylor Research Institute (Baylor, Scott and White Health) | Dallas, TX | Duke University | EpicCare Ambulatory * |
| University of North Carolina at Pembroke | Pembroke, NC | Duke University | EpicCare Ambulatory * |
| University Medical Center—New Orleans | New Orleans, LA | Duke University | EpicCare Ambulatory * |
| Southeastern Healthcare | Lumberton, NC | Duke University | EpicCare Ambulatory * |
References
- Freimuth, R.R.; Formea, C.M.; Hoffman, J.M.; Matey, E.; Peterson, J.F.; Boyce, R.D. Implementing Genomic Clinical Decision Support for Drug-Based Precision Medicine. CPT Pharmacomet. Syst. Pharmacol. 2017, 6, 153–155. [Google Scholar] [CrossRef] [PubMed]
- Levy, K.D.; Blake, K.; Fletcher-Hoppe, C.; Franciosi, J.; Goto, D.; Hicks, J.K.; Holmes, A.M.; Kanuri, S.H.; Madden, E.B.; Musty, M.D.; et al. Opportunities to implement a sustainable genomic medicine program: Lessons learned from the IGNITE Network. Genet. Med. 2019, 21, 743–747. [Google Scholar] [CrossRef]
- Horowitz, C.; Abul-Husn, N.; Ellis, S.; Ramos, M.; Negron, R.; Suprun, M.; Zinberg, R.; Sabin, T.; Hauser, D.; Calman, N. Determining the effects and challenges of incorporating genetic testing into primary care management of hypertensive patients with African ancestry. Contemp. Clin. Trials 2016, 47, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Foster, M.C.; Coresh, J.; Fornage, M.; Astor, B.C.; Grams, M.; Franceschini, N.; Boerwinkle, E.; Parekh, R.S.; Kao, W.L. APOL1 variants associate with increased risk of CKD among African Americans. J. Am. Soc. Nephrol. 2013, 24, 1484–1491. [Google Scholar] [CrossRef]
- Kopp, J.B.; Nelson, G.W.; Sampath, K.; Johnson, R.C.; Genovese, G.; An, P.; Friedman, D.; Briggs, W.; Dart, R.; Korbet, S. APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J. Am. Soc. Nephrol. 2011, 22, 2129–2137. [Google Scholar] [CrossRef]
- Parsa, A.; Kao, W.L.; Xie, D.; Astor, B.C.; Li, M.; Hsu, C.Y.; Feldman, H.I.; Parekh, R.S.; Kusek, J.W.; Greene, T.H. APOL1 risk variants, race, and progression of chronic kidney disease. N. Engl. J. Med. 2013, 369, 2183–2196. [Google Scholar] [CrossRef] [PubMed]
- Ku, E.; Lipkowitz, M.S.; Appel, L.J.; Parsa, A.; Gassman, J.; Glidden, D.V.; Smogorzewski, M.; Hsu, C.Y. Strict blood pressure control associates with decreased mortality risk by APOL1 genotype. Kidney Int. 2017, 91, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Group, S.R. A randomized trial of intensive versus standard blood-pressure control. N. Engl. J. Med. 2015, 373, 2103–2116. [Google Scholar] [CrossRef]
- Robinson, T.W.; Freedman, B.I. The Impact of APOL1 on Chronic Kidney Disease and Hypertension. Adv. Chronic Kidney Dis. 2019, 26, 131–136. [Google Scholar] [CrossRef]
- Cunningham, P.N.; Wang, Z.; Grove, M.L.; Cooper-DeHoff, R.M.; Beitelshees, A.L.; Gong, Y.; Gums, J.G.; Johnson, J.A.; Turner, S.T.; Boerwinkle, E. Hypertensive APOL1 risk allele carriers demonstrate greater blood pressure reduction with angiotensin receptor blockade compared to low risk carriers. PLoS ONE 2019, 14, e0221957. [Google Scholar] [CrossRef]
- Kopp, J.B.; Winkler, C.A. Genetic Testing for APOL1 Genetic Variants in Clinical Practice: Finally, Starting to Arrive. Clin. J. Am. Soc. Nephrol. 2020, 15, 126–128. [Google Scholar] [CrossRef]
- Duarte, J.D.; Turner, S.T.; Tran, B.; Chapman, A.B.; Bailey, K.R.; Gong, Y.; Gums, J.G.; Langaee, T.Y.; Beitelshees, A.L.; Cooper-Dehoff, R.M. Association of chromosome 12 locus with antihypertensive response to hydrochlorothiazide may involve differential YEATS4 expression. Pharm. J. 2013, 13, 257–263. [Google Scholar] [CrossRef]
- Rotival, M.; Zeller, T.; Wild, P.S.; Maouche, S.; Szymczak, S.; Schillert, A.; Castagné, R.; Deiseroth, A.; Proust, C.; Brocheton, J. Integrating genome-wide genetic variations and monocyte expression data reveals trans-regulated gene modules in humans. PLoS Genet. 2011, 7, e1002367. [Google Scholar] [CrossRef] [PubMed]
- Turner, S.T.; Bailey, K.R.; Fridley, B.L.; Chapman, A.B.; Schwartz, G.L.; Chai, H.S.; Sicotte, H.; Kocher, J.-P.; Rodin, A.S.; Boerwinkle, E. Genomic association analysis suggests chromosome 12 locus influencing antihypertensive response to thiazide diuretic. Hypertension 2008, 52, 359–365. [Google Scholar] [CrossRef]
- Spinasse, L.B.; Santos, A.R.; Suffys, P.N.; Muxfeldt, E.S.; Salles, G.F. Different phenotypes of the NAT2 gene influences hydralazine antihypertensive response in patients with resistant hypertension. Pharmacogenomics 2014, 15, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Kannry, J.; McCullagh, L.; Kushniruk, A.; Mann, D.; Edonyabo, D.; McGinn, T. A Framework for Usable and Effective Clinical Decision Support: Experience from the iCPR Randomized Clinical Trial. EGEMS 2015, 3, 1150. [Google Scholar] [CrossRef] [PubMed]
- Friedlin, J.; Dexter, P.R.; Overhage, J.M. Details of a successful clinical decision support system. AMIA Annu. Symp. Proc. 2007, 2007, 254–258. [Google Scholar]
- Johnson, J.A.; Burkley, B.M.; Langaee, T.Y.; Clare-Salzler, M.J.; Klein, T.E.; Altman, R.B. Implementing personalized medicine: Development of a cost-effective customized pharmacogenetics genotyping array. Clin. Pharmacol. Ther. 2012, 92, 437–439. [Google Scholar] [CrossRef]
- Rosenman, M.B.; Decker, B.; Levy, K.D.; Holmes, A.M.; Pratt, V.M.; Eadon, M.T. Lessons learned when introducing pharmacogenomic panel testing into clinical practice. Value Health 2017, 20, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Ginsburg, G.S.; Cavallari, L.H.; Chakraborty, H.; Cooper-DeHoff, R.M.; Dexter, P.R.; Eadon, M.T.; Ferket, B.S.; Horowitz, C.R.; Johnson, J.A.; Kannry, J.; et al. Establishing the value of genomics in medicine: The IGNITE Pragmatic Trials Network. Genet. Med. 2021, 1–7. [Google Scholar] [CrossRef]
- Bates, D.W.; Kuperman, G.J.; Wang, S.; Gandhi, T.; Kittler, A.; Volk, L.; Spurr, C.; Khorasani, R.; Tanasijevic, M.; Middleton, B. Ten commandments for effective clinical decision support: Making the practice of evidence-based medicine a reality. J. Am. Med. Inform. Assoc. 2003, 10, 523–530. [Google Scholar] [CrossRef]
- Collins, K.S.; Raviele, A.L.; Elchynski, A.L.; Woodcock, A.M.; Zhao, Y.; Cooper-DeHoff, R.M.; Eadon, M.T. Genotype-Guided Hydralazine Therapy. Am. J. Nephrol. 2020, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Unertl, K.M.; Jaffa, H.; Field, J.R.; Price, L.; Peterson, J.F. Clinician perspectives on using pharmacogenomics in clinical practice. Pers. Med. 2015, 12, 339–347. [Google Scholar] [CrossRef] [PubMed]
- St Sauver, J.L.; Bielinski, S.J.; Olson, J.E.; Bell, E.J.; Mc Gree, M.E.; Jacobson, D.J.; McCormick, J.B.; Caraballo, P.J.; Takahashi, P.Y.; Roger, V.L.; et al. Integrating Pharmacogenomics into Clinical Practice: Promise vs. Reality. Am. J. Med. 2016, 129, 1093–1099. [Google Scholar] [CrossRef]
- Olander, M.; Waring, S.; Stenehjem, D.D.; Taran, A.; Raneli, P.L.; Brown, J. Primary Care Clinicians Attitudes and Knowledge of Pharmacogenetics in a Large, Multi-state, Healthcare System. Innov. Pharm. 2018, 9, 6. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Haga, S.B.; Mills, R.; Moaddeb, J.; Allen LaPointe, N.; Cho, A.; Ginsburg, G.S. Primary care providers’ use of pharmacist support for delivery of pharmacogenetic testing. Pharmacogenomics 2017, 18, 359–367. [Google Scholar] [CrossRef] [PubMed]
- McCoy, A.B.; Waitman, L.R.; Lewis, J.B.; Wright, J.A.; Choma, D.P.; Miller, R.A.; Peterson, J.F. A framework for evaluating the appropriateness of clinical decision support alerts and responses. J. Am. Med. Inform. Assoc. 2011, 19, 346–352. [Google Scholar] [CrossRef] [PubMed]
| CDS General Principles | CDS Project Specific Principles |
|---|---|
| 1. Medical algorithm consensus | 5. Feasibility |
| 2. Actionability | 6. Interpretability |
| 3. Context-sensitive triggers | 7. Portability |
| 4. Workflow integration | 8. Discrete Results Reporting |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schneider, T.M.; Eadon, M.T.; Cooper-DeHoff, R.M.; Cavanaugh, K.L.; Nguyen, K.A.; Arwood, M.J.; Tillman, E.M.; Pratt, V.M.; Dexter, P.R.; McCoy, A.B.; et al. Multi-Institutional Implementation of Clinical Decision Support for APOL1, NAT2, and YEATS4 Genotyping in Antihypertensive Management. J. Pers. Med. 2021, 11, 480. https://doi.org/10.3390/jpm11060480
Schneider TM, Eadon MT, Cooper-DeHoff RM, Cavanaugh KL, Nguyen KA, Arwood MJ, Tillman EM, Pratt VM, Dexter PR, McCoy AB, et al. Multi-Institutional Implementation of Clinical Decision Support for APOL1, NAT2, and YEATS4 Genotyping in Antihypertensive Management. Journal of Personalized Medicine. 2021; 11(6):480. https://doi.org/10.3390/jpm11060480
Chicago/Turabian StyleSchneider, Thomas M., Michael T. Eadon, Rhonda M. Cooper-DeHoff, Kerri L. Cavanaugh, Khoa A. Nguyen, Meghan J. Arwood, Emma M. Tillman, Victoria M. Pratt, Paul R. Dexter, Allison B. McCoy, and et al. 2021. "Multi-Institutional Implementation of Clinical Decision Support for APOL1, NAT2, and YEATS4 Genotyping in Antihypertensive Management" Journal of Personalized Medicine 11, no. 6: 480. https://doi.org/10.3390/jpm11060480
APA StyleSchneider, T. M., Eadon, M. T., Cooper-DeHoff, R. M., Cavanaugh, K. L., Nguyen, K. A., Arwood, M. J., Tillman, E. M., Pratt, V. M., Dexter, P. R., McCoy, A. B., Orlando, L. A., Scott, S. A., Nadkarni, G. N., Horowitz, C. R., & Kannry, J. L. (2021). Multi-Institutional Implementation of Clinical Decision Support for APOL1, NAT2, and YEATS4 Genotyping in Antihypertensive Management. Journal of Personalized Medicine, 11(6), 480. https://doi.org/10.3390/jpm11060480








