HPV Status and Individual Characteristics of Human Papillomavirus Infection as Predictors for Clinical Outcome of Locally Advanced Cervical Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Polymerase Chain Reaction (PCR)
- -
- The number of E7 gene copies is less than 103 per 100 thousand cells (lgE7 < 3)—low viral load;
- -
- The number of E7 gene copies is equal to or more than 103, but less than 105 per 100 thousand cells (3 ≤ lgE7 < 5)—moderate viral load;
- -
- The number of E7 gene copies is equal to or more than 105 per 100 thousand cells (lgE7 ≥ 5)—high viral load.
2.3. Radiation and Chemoradiation Therapy
2.4. Statistical Analysis
3. Results
3.1. Prevalence and Molecular Genetic Parameters of HPV
3.2. Univariate Analysis of Clinical Outcome According to Candidate Predictor Variables
3.3. Kaplan–Meier Analysis of Disease Free Survival
3.4. Multivariate Analysis of Clinical Outcome
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 1–41. [Google Scholar] [CrossRef]
- Kosary, C.L. FIGO stage, histology, histologic grade, age and race as prognostic factors in determining survival for cancers of the female gynecological system: An analysis of 1973–87 SEER cases of cancers of the endometrium, cervix, ovary, vulva and vagina. Semin. Surg. Oncol. 1994, 10, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Da Mata, S.; Ferreira, J.; Nicolás, I.; Esteves, S.; Esteves, G.; Lérias, S.; Silva, F.; Saco, A.; Cochicho, D.; Cunha, M.; et al. P16 and HPV Genotype Significance in HPV-Associated Cervical Cancer-A Large Cohort of Two Tertiary Referral Centers. Int. J. Mol. Sci. 2021, 22, 2294. [Google Scholar] [CrossRef]
- Mendaza, S.; Fernández-Irigoyen, J.; Santamaría, E.; Zudaire, T.; Guarch, R.; Guerrero-Setas, D.; Vidal, A.; Santos-Salas, J.; Matias-Guiu, X.; Ausín, K.; et al. Absence of Nuclear p16 Is a Diagnostic and Independent Prognostic Biomarker in Squamous Cell Carcinoma of the Cervix. Int. J. Mol. Sci. 2021, 21, 2125. [Google Scholar] [CrossRef]
- Eldakhakhny, S.; Zhou, Q.; Crosbie, E.J.; Sayan, B.S. Human papillomavirus E7 induces p63 expression to modulate DNA damage response. Cell Death Dis. 2018, 9, 127. [Google Scholar] [CrossRef] [PubMed]
- Sitz, J.; Blanchet, S.A.; Gameiro, S.F.; Biquand, E.; Morgan, T.M.; Galloy, M.; Dessapt, J.; Lavoie, E.G.; Blondeau, A.; Smith, B.C.; et al. Human papillomavirus E7 oncoprotein targets RNF168 to hijack the host DNA damage response. Proc. Natl. Acad. Sci. USA 2019, 116, 19552–19562. [Google Scholar] [CrossRef]
- Hamid, N.A.; Brown, C.; Gaston, K. The regulation of cell proliferation by the papillomavirus early proteins. Cell Mol. Life Sci. 2009, 66, 1700–1717. [Google Scholar] [CrossRef]
- Caldeira, S.; Dong, W.; Tommasino, M. Analysis of E7/Rb associations. Methods Mol. Med. 2005, 199, 363–379. [Google Scholar] [CrossRef]
- Tungteakkhum, S.S.; Duerksen-Hughes, P.J. Cellular binding partners of human papillomavirus protein. Arch. Virol. 2008, 153, 397–408. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhang, J.; Rao, Z. Ribozyme targeting HPV16 E6E7 transcripts in cervical cancer cells suppresses cell growth and sensitizes cells to chemotherapy and radiotherapy. Cancer Biol. Ther. 2004, 3, 1129–1134. [Google Scholar] [CrossRef]
- Hampson, L.; El Hady, E.S.; Moore, J.V.; Kitchener, H.; Hampson, I.N. The HPV16 E6 and E7 proteins and the radiation resistance of cervical carcinoma. FASEB J. 2001, 15, 1445–1463. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.G.; Zhao, L.N.; Liu, Y.W.; Li, T.T.; Fan, D.M.; Chen, J.J. Activation of Cdc2 contributes to apoptosis in HPV E6 expressing human keratinocytes in response to therapeutic agents. J. Mol. Biol. 2007, 374, 334–345. [Google Scholar] [CrossRef] [PubMed]
- Pang, E.; Delic, N.C.; Hong, A.; Zhang, M.; Rose, B.R.; Lyons, J.G. Radiosensitization of oropharyngeal squamous cell carcinoma cells by human papillomavirus 16 oncoprotein E6∗I. Int. J. Radiat. Oncol. Biol. Phys. 2011, 79, 860–865. [Google Scholar] [CrossRef]
- Balasubramaniam, S.D.; Balakrishnan, V.; Oon, C.E.; Kaur, G. Key Molecular Events in Cervical Cancer Development. Medicina 2019, 55, 384. [Google Scholar] [CrossRef]
- Ganguly, N.; Parihar, S.P. Human papillomavirus E6 and E7 oncoproteins as risk factors for tumorigenesis. J. Biosci. 2009, 34, 113–123. [Google Scholar] [CrossRef]
- Jeon, S.; Lambert, P.F. Integration of human papillomavirus type 16 DNA into the human genome leads to increased stability of E6 and E7 mRNAs: Implications for cervical carcinogenesis. Proc. Natl. Acad. Sci. USA 1995, 92, 1654–1658. [Google Scholar] [CrossRef]
- Lopez, J.; Poitevin, A.; Mendoza-Martinez, V.; Perez-Plasencia, C.; Garcia-Carranca, A. Cancer-initiating cells derived from established cervical cell lines exhibit stem-cell markers and increased radioresistance. BMC Cancer 2012, 12, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zeng, J.; Luo, L.; Yang, J.; Chen, J.; Li, B.; Shen, K. Identification of a cancer stem cell-like side population in the HeLa human cervical carcinoma cell line. Oncol. Lett. 2013, 6, 1673–1680. [Google Scholar] [CrossRef]
- Zamulaeva, I.; Selivanova, E.; Matchuk, O.; Kiseleva, V.; Mkrtchyan, L.; Krikunova, L. Radiation Response of Cervical Cancer Stem Cells Is Associated with Pretreatment Proportion of These Cells and Physical Status of HPV DNA. Int. J. Mol. Sci. 2021, 22, 1445. [Google Scholar] [CrossRef]
- Reid, P.; Staudacher, A.H.; Marcu, L.G.; Olver, I.; Moghaddasi, L.; Brown, M.P.; Bezak, E. Influence of the human papillomavirus on the radio-responsiveness of cancer stem cells in head and neck cancers. Sci. Rep. 2020, 10, 2716. [Google Scholar] [CrossRef]
- Zhang, M.; Kumar, B.; Piao, L.; Xie, X.; Schmitt, A.; Arradaza, N.; Cippola, M.; Old, M.; Agrawal, A.; Ozer, E.; et al. Elevated intrinsic cancer stem cell population in human papillomavirus-associated head and neck squamous cell carcinoma. Cancer 2014, 120, 992–1001. [Google Scholar] [CrossRef]
- Vishnoi, K.; Mahata, S.; Tyagi, A.; Pandey, A.; Verma, G.; Jadli, M.; Singh, T.; Singh, S.M.; Bharti, A.C. Cross-talk between Human Papillomavirus Oncoproteins and Hedgehog Signaling Synergistically Promotes Stemness in Cervical Cancer Cells. Sci. Rep. 2016, 6, 1–15. [Google Scholar] [CrossRef]
- Vlashi, E.; Chen, A.M.; Boyrie, S.; Yu, G.; Nguyen, A.; Brower, P.A.; Hess, C.B.; Pajonk, F. Radiation-Induced Dedifferentiation of Head and Neck Cancer Cells into Cancer Stem Cells Depends on HPV status. Int. J. Radiat. Oncol. Biol. Phys. 2016, 94, 1198–1206. [Google Scholar] [CrossRef]
- Hall, J.S.; Iype, R.; Armenoult, L.S.; Taylor, J.; Miller, C.J.; Davidson, S.; de Sanjose, S.; Bosch, X.; Stern, P.L.; West, C.M. Poor prognosis associated with human papillomavirus α7 genotypes in cervical carcinoma cannot be explained by intrinsic radiosensitivity. Int. J. Radiat. Oncol. Biol. Phys. 2013, 85, 223–229. [Google Scholar] [CrossRef]
- Hang, D.; Jia, M.; Ma, H.; Zhou, J.; Feng, X.; Lyu, Z.; Yin, J.; Cui, H.; Yin, Y.; Jin, G.; et al. Independent prognostic role of human papillomavirus genotype in cervical cancer. BMC Infect. Dis. 2017, 17, 391. [Google Scholar] [CrossRef]
- Okonogi, N.; Kobayashi, D.; Suga, T.; Imai, T.; Wakatsuki, M.; Ohno, T.; Kato, S.; Nakano, T.; Kamada, T. Human papillomavirus genotype affects metastatic rate following radiotherapy in patients with uterine cervical cancer. Oncol. Lett. 2018, 15, 459–466. [Google Scholar] [CrossRef]
- Cuschieri, K.; Brewster, D.H.; Graham, C.; Nicoll, S.; Williams, A.R.W.; Murray, G.I.; Millan, D.; Johannessen, I.; Hardie, A.; Cubie, H.A. Influence of HPV type on prognosis in patients diagnosed with invasive cervical cancer. Int. J. Cancer 2014, 135, 2721–2726. [Google Scholar] [CrossRef]
- Tong, S.Y.; Lee, Y.S.; Park, J.S.; Namkoong, S.E. Human papillomavirus genotype as a prognostic factor in carcinoma of the uterine cervix. Int. J. Gynecol. Cancer 2007, 17, 1307–1313. [Google Scholar] [CrossRef]
- Kim, J.Y.; Park, S.; Nam, B.H.; Roh, J.W.; Lee, C.H.; Kim, Y.H.; Shin, H.J.; Lee, S.K.; Kong, S.Y.; Seong, M.W.; et al. Low initial human papilloma viral load implicates worse prognosis in patients with uterine cervical cancer treated with radiotherapy. J. Clin. Oncol. 2009, 27, 5088–5093. [Google Scholar] [CrossRef]
- Deng, T.; Feng, Y.; Zheng, J.; Huang, Q.; Liu, J. Low initial human papillomavirus viral load may indicate worse prognosis in patients with cervical carcinoma treated with surgery. J. Gynecol. Oncol. 2015, 26, 111–117. [Google Scholar] [CrossRef]
- Song, Y.J.; Kim, J.Y.; Lee, S.K.; Lim, H.S.; Lim, M.C.; Seo, S.S.; Kang, S.; Lee, D.O.; Park, S.Y. Persistent human papillomavirus DNA is associated with local recurrence after radiotherapy of uterine cervical cancer. Int. J. Cancer 2011, 15, 896–902. [Google Scholar] [CrossRef]
- Cao, M.; Shah, W.; Qi, J.; Zhou, Y.; Wang, Y.; Chen, H. Prognostic significance of human papillomavirus viral load in correlation with different therapeutic modalities in cervical cancer patients. Pathol. Res. Pract. 2016, 212, 804–810. [Google Scholar] [CrossRef]
- Singh, R.K.; Maulik, S.; Mitra, S.; Mondal, R.; Basu, P.; Roychowdhury, S.; Panda, C. Human papillomavirus prevalence in postradiotherapy uterine cervical carcinoma patients: Correlation with recurrence of the disease. Int. J. Gynecol. Cancer 2006, 16, 1048–1054. [Google Scholar] [CrossRef] [PubMed]
- Lindel, K.; de Villiers, E.M.; Burri, P.; Studer, U.; Altermatt, H.J.; Greiner, R.H.; Gruber, G. Impact of viral E2-gene status on outcome after radiotheraphy for patients with human papillomavirus 16-positive cancer of the uterine cervix. Int. J. Radat. Oncol. Biol. Phys. 2006, 65, 760–765. [Google Scholar] [CrossRef]
- Nambaru, L.; Meenakumari, B.; Swaminathan, R.; Rajkumar, T. Prognostic significance of HPV physical satus and integration sites in cervical cancer. Asian Pac. J. Cancer Prev. 2009, 10, 355–360. [Google Scholar] [PubMed]
- Hernádi, Z.; Szarka, K.; Sápy, T.; Krasznai, Z.; Veress, G.; Póka, R. The prognostic significance of HPV-16 genome status of the lymph nodes, the integration status and p53 genotype in HPV-16 positive cervical cancer: A long term follow up. BJOG 2003, 110, 205–209. [Google Scholar] [CrossRef]
- Shin, H.-J.; Joo, J.; Yoon, J.H.; Yoo, C.W.; Kim, J.-Y. Physical Status of Human Papillomavirus Integration in Cervical Cancer Is Associated with Treatment Outcome of the Patients Treated with Radiotherapy. PLoS ONE 2014, 9, e78995. [Google Scholar] [CrossRef]
- Li, P.; Tan, Y.; Zhu, L.X.; Zhou, L.N.; Zeng, P.; Liu, Q.; Chen, M.B.; Tian, Y. Prognostic value of HPV DNA status in cervical cancer before treatment: A systematic review and meta-analysis. Oncotarget 2017, 8, 66352–66359. [Google Scholar] [CrossRef]
- Rodríguez-Carunchio, L.; Soveral, I.; Steenbergen, R.; Torné, A.; Martinez, S.; Fusté, P.; Pahisa, J.; Marimon, L.; Ordi, J.; del Pino, M. HPV-negative carcinoma of the uterine cervix: A distinct type of cervical cancer with poor prognosis. BJOG 2015, 122, 119–127. [Google Scholar] [CrossRef]
- Kurman, R.J.; Carcangiu, M.L.; Harrington, C.S.; Young, R.H. WHO Classification of Tumors of the Female Reproductive Organs, 4th ed.; International Agency for Research on Cancer: Geneva, Switzerland; Lyon, France, 2014; pp. 264–298. ISBN 978-92-8322-435-8. [Google Scholar]
- Sobin, L.H.; Gospodarowicz, M.K.; Wittekind, C. TNM Classification of Malignant Tumours, 7th ed.; Wiley-Blackwell: Chichester, UK; Hoboken, NJ, USA, 2010; pp. 206–212. ISBN 978-14-4433-241-4. [Google Scholar]
- FIGO Committee on Gynecologic Oncology. FIGO staging for carcinoma of the vulva, cervix, and corpus uteri. Int. J. Gynaecol. Obstet. 2014, 125, 97–98. [Google Scholar] [CrossRef]
- Eisenhauera, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef]
- Bohmer, G.; van den Brule, A.J.; Brummer, O.; Meijer, C.L.; Petry, K.U. No confirmed case of human papillomavirus DNA-negative cervical intraepithelial neoplasia grade 3 or invasive primary of cancer of the uterine cervix among 511 patients. Am. J. Obstet. Gynecol. 2003, 189, 118–120. [Google Scholar] [CrossRef] [PubMed]
- Igidbashian, S.; Schettino, M.T.; Boveri, S.; Barberis, M.; Sandri, M.T.; Carinelli, S.; Cannone, M.; Sideri, M. Tissue genotyping of 37 in situ and invasive cervical cancer with a concomitant negative HC2 HPV DNA test. J. Low. Genit. Tract Dis. 2014, 18, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.; Zheng, B.; Yin, F.; Zeng, Z.; Li, Z.; Griffith, C.; Luo, B.; Ding, X.; Zhou, X.; Zhao, C. Polymerase Chain Reaction Human Papillomavirus (HPV) Detection and HPV Genotyping in Invasive Cervical Cancers With Prior Negative HC2 Test Results. Am. J. Clin. Pathol. 2017, 147, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Herrington, C.S. Do HPV-negative cervical carcinomas exist?—Revisited. J. Pathol. 1999, 189, 1–3. [Google Scholar] [CrossRef]
- Chong, G.O.; Lee, Y.H.; Han, H.S.; Lee, H.J.; Park, J.Y.; Hong, D.G.; Lee, Y.S.; Cho, Y.L. Prognostic value of pre-treatment human papilloma virus DNA status in cervical cancer. Gynecol. Oncol. 2018, 148, 97–102. [Google Scholar] [CrossRef]
- Okuma, K.; Yamashita, H.; Yokoyama, T.; Nakagawa, K.; Kawana, K. Undetected human papillomavirus DNA and uterine cervical carcinoma: Association with cancer recurrence. Strahlenther Onkol. 2016, 192, 55–62. [Google Scholar] [CrossRef]
- Wang, C.C.; Lai, C.H.; Huang, H.J.; Chao, A.; Chang, C.J.; Chang, T.C.; Chou, H.H.; Hong, J.H. Clinical effect of human papillomavirus genotypes in patients with cervical cancer undergoing primary radiotherapy. Int. J. Radiat. Oncol. Biol. Phys. 2010, 78, 1111–1120. [Google Scholar] [CrossRef]
- Wang, C.C.; Lai, C.H.; Huang, Y.T.; Chao, A.; Chou, H.H.; Hong, J.H. HPV genotypes predict survival benefits from concurrent chemotherapy and radiation therapy in advanced squamous cell carcinoma of the cervix. Int. J. Radiat. Oncol. Biol. Phys. 2012, 84, 499–506. [Google Scholar] [CrossRef]
- De Boer, M.A.; Jordanova, E.S.; Kenter, G.G.; Peters, A.A.; Corver, W.E.; Trimbos, J.B.; Fleuren, G.J. High human papillomavirus oncogene mRNA expression and not viral DNA load is associated with poor prognosis in cervical cancer patients. Clin. Cancer Res. 2007, 13, 132–138. [Google Scholar] [CrossRef]
- Vernon, S.D.; Unger, E.R.; Miller, D.L.; Lee, D.R.; Reeves, W.C. Association of human papillomavirus type 16 integration in the E2 gene with poor disease-free survival from cervical cancer. Int. J. Cancer 1997, 74, 50–56. [Google Scholar] [CrossRef]
- Waggoner, S.E. Cervical cancer. Lancet 2003, 361, 2217–2225. [Google Scholar] [CrossRef]
- Datta, N.R.; Kumar, P.; Singh, S.; Gupta, D.; Srivastava, A.; Dhole, T.N. Does pretreatment human papillomavirus (HPV) titers predict radiation response and survival outcomes in cancer cervix?—A pilot study. Gynecol Oncol. 2006, 103, 100–105. [Google Scholar] [CrossRef]
- Kahla, S.; Kochbati, L.; Sarraj, S.; Daya, I.B.; Maalej, M.; Oueslati, R. Molecular detection of human papillomavirus and viral DNA load after radiotherapy for cervical carcinomas. Tumori 2016, 102, 521–526. [Google Scholar] [CrossRef] [PubMed]
- Bourgioti, C.; Chatoupis, K.; Moulopoulos, L.A. Current imaging strategies for the evaluation of uterine cervical cancer. World J. Radiol. 2016, 8, 342–354. [Google Scholar] [CrossRef]
- Kapp, D.S.; Fischer, D.; Gutierrez, E.; Kohorn, E.I.; Schwartz, P.E. Pretreatment prognostic factors in carcinoma of the uterine cervix: A multivariable analysis of the effect of age, stage, histology and blood counts on survival. Int. J. Radiat. Oncol. Biol. Phys. 1983, 9, 445–455. [Google Scholar] [CrossRef]
- Kim, T.E.; Park, B.J.; Kwack, H.S.; Kwon, J.Y.; Kim, J.H.; Yoon, S.C. Outcomes and prognostic factors of cervical cancer after concurrent chemoradiation. J. Obstet. Gynaecol. Res. 2012, 38, 1315–1320. [Google Scholar] [CrossRef]
- Lanciano, R.M.; Won, M.; Coia, L.R.; Hanks, G.E. Pretreatment and treatment factors associated with improved outcome in squamous cell carcinoma of the uterine cervix: A final report of the 1973 and 1978 Patterns of Care studies. Int. J. Radiat. Oncol. Biol. Phys. 1991, 20, 667–676. [Google Scholar] [CrossRef]
- Bush, R.S. Current status of treatment of localized disease and future aspects. Int. J. Radiat. Oncol. Biol. Phys. 1984, 10, 1165–1174. [Google Scholar] [CrossRef]
- Kudaka, W.; Nagai, Y.; Toita, T.; Inamine, M.; Asato, K.; Nakamoto, T.; Wakayama, A.; Ooyama, T.; Tokura, A.; Murayama, S.; et al. Long-term results and prognostic factors in patients with stage III-IVA squamous cell carcinoma of the cervix treated with concurrent chemoradiotherapy from a single institution study. Int. J. Clin. Oncol. 2013, 18, 916–921. [Google Scholar] [CrossRef]
- Dyer, B.A.; Feng, C.H.; Eskander, R.; Sharabi, A.B.; Mell, L.K.; McHale, M.; Mayadev, J.S. Current Status of Clinical Trials for Cervical and Uterine Cancer Using Immunotherapy Combined with Radiation. Int. J. Radiat. Oncol. Biol. Phys. 2021, 109, 396–412. [Google Scholar] [CrossRef]
- Nalliah, S.; Karikalan, B.; Kademane, K. Multifaceted usage of HPV related tests and products in the management of cervical cancer—A review. Asian Pac. J. Cancer Prev. 2015, 16, 2145–2150. [Google Scholar] [CrossRef][Green Version]
- Nguyen, N.D.; Deshpande, V.; Luebeck, J.; Mischel, P.S.; Bafna, V. ViFi: Accurate detection of viral integration and mRNA fusion reveals indiscriminate and unregulated transcription in proximal genomic regions in cervical cancer. Nucleic Acids Res. 2018, 46, 3309–3325. [Google Scholar] [CrossRef]
- Lippert, J.; Bonlokke, S.; Utke, A.; Knudsen, B.R.; Sorensen, B.S.; Steiniche, T.; Stougaard, M. Targeted next generation sequencing panel for HPV genotyping in cervical cancer. Exp. Mol. Pathol. 2021, 118, 104568. [Google Scholar] [CrossRef] [PubMed]
- Brant, A.C.; Menezes, A.N.; Felix, S.P.; de Almeida, L.M.; Sammeth, M.; Moreira, M.A.M. Characterization of HPV integration, viral gene expression and E6E7 alternative transcripts by RNA-Seq: A descriptive study in invasive cervical cancer. Genomics 2019, 111, 1853–1861. [Google Scholar] [CrossRef] [PubMed]
- Burchardt, E.; Roszak, A. Hyperthermia in cervical cancer—Current status. Rep. Pract. Oncol. Radiother. 2018, 23, 595–603. [Google Scholar] [CrossRef]
- Orbegoso, C.; Murali, K.; Banerjee, S. The current status of immunotherapy for cervical cancer. Rep. Pract. Oncol. Radiother. 2018, 23, 580–588. [Google Scholar] [CrossRef] [PubMed]
- Regalado Porras, G.O.; Chávez Nogueda, J.; Poitevin Chacón, A. Chemotherapy and molecular therapy in cervical cancer. Rep. Pract. Oncol. Radiother. 2018, 23, 533–539. [Google Scholar] [CrossRef]
- Rödel, F.; Martin, D.; Balermpas, P.; Wieland, U.; Winkelmann, R.; Riekmann, T.; Falk, S.; Rödel, C.; Fokas, E. Modulation of radiation sensitivity and antitumor immunity by viral pathogenic factors: Implications for radio-immunotherapy. Biochim. Biophys. Acta Rev. Cancer 2019, 1871, 126–137. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Mu, X.; He, H.; Zhang, X.-D. Cancer Radiosensitizers. Trends Pharmacol. Sci. 2018, 39, 24–48. [Google Scholar] [CrossRef] [PubMed]
- Kareliotis, G.; Tremi, I.; Kaitatzi, M.; Drakaki, E.; Serafetinides, A.A.; Makropoulou, M.; Georgakilas, A.G. Combined radiation strategies for novel and enhanced cancer treatment. Int. J. Radiat. Biol. 2020, 96, 1087–1103. [Google Scholar] [CrossRef]
- Huang, R.X.; Zhou, P.K. DNA damage response signaling pathways and targets for radiotherapy sensitization in cancer. Signal Transduct. Target Ther. 2020, 5, 60. [Google Scholar] [CrossRef]




| Clinical and Morphological Characteristics of Patients with CC | Number of Patients (%) |
|---|---|
| Age, years | |
| <30 | 6 (4.4) |
| 30–44 | 44 (32.6) |
| 45–55 | 45 (33.3) |
| 56–65 | 30 (22.2) |
| >65 | 10 (7.5) |
| Stage of the disease (FIGO) | |
| II | 62 (45.9) |
| III | 73 (54.1) |
| Histological type | |
| squamous cell carcinoma | 119 (88.1) |
| adenocarcinoma | 11 (8.2) |
| adenocarcinoma with mixed subtypes | 5 (3.7) |
| Grade | |
| low | 26 (19.3) |
| intermediate | 25 (18.5) |
| high | 83 (62.2) |
| Form of tumor growth | |
| exophytic | 16 (11.8) |
| endophytic | 37 (27.4) |
| mixed | 82 (60.8) |
| Infiltration of parametrium | |
| absence(T1b2, T2a) | 49 (36.3) |
| presence (T2b, T3b) | 86 (63.7) |
| Metastases in regional lymph nodes | |
| absence(T1b2-3N0M0) | 65 (63.0) |
| presence(T1b2-3N1M0) | 50 (37.0) |
| Molecular Genetic Parameters of HPV Infection | Number of Patients (%) |
|---|---|
| Genotypes | |
| HPV16 | 95 (63.3) |
| HPV18 | 20 (36.7) |
| Number of genotypes | |
| mono infection (HPV 16 or 18) | 102 (88.7) |
| multiple infection (HPV16, 18, 31, 45, and other types) | 13 (11.3) |
| Viral load | |
| lgE7 < 3 | 4 (3.5) |
| 3 ≤ lgE7 < 5 | 23 (20.0) |
| lgE7 ≥ 5 | 88 (76.5) |
| Physical state of viral DNA | |
| absence of integration (episomal form) | 49 (42.6) |
| partial or complete integration (integrated form) | 66 (57.4) |
| Integration degree, n = 66 | |
| <50% | 20 (30.3) |
| ≥50% | 46 (69.7) |
| Variables | Outcome of the Disease | OR (95% CI) p-Value | ||
|---|---|---|---|---|
| Favorable, Patient Number (%) | Unfavorable, Patient Number (%) | |||
| HPV status | HPV 16/18-positivity | 84 (73.0) | 31 (27.0) | 3.31 (1.23–8.93) p = 0.018 |
| HPV-negativity | 9 (45.0) | 11 (55.0) | ||
| Physical state of HPV 16/18 DNA | Episomal form | 45 (91.8) | 4 (8.2) | 3.66 (1.96–6.83) p= 4.7 × 10−5 |
| Integrated form | 39 (59.1) | 27 (40.9) | ||
| Genotype | 16 | 72 (75.8) | 23 (24.2) | 2.09 (0.74–5.85) p = 0.170 |
| 18 | 12 (60.0) | 8 (40.0) | ||
| Viral load | lgE7 < 3 | 3 (75.0) | 1 (25.0) | 0.90 (0.41–1.97) p = 0.793 |
| 3 ≤ lgE7 < 5 | 16 (69.6) | 7 (30.4) | ||
| lgE7 ≥ 5 | 65 (73.9) | 23 (26.1) | ||
| Number of genotypes | Mono infection | 75 (73.5) | 27 (26.5) | 1.23 (0.34–4.45) p = 0.745 |
| Multiple infection | 9 (69.2) | 4 (30.8) | ||
| Age category (years) | <30 | 3 (50.0) | 3 (50.0) | 0.75 (0.52–1.10) p = 0.137 |
| 30–44 | 27 (61.4) | 17 (38.6) | ||
| 45–55 | 32 (71.1) | 13 (28.9) | ||
| 56–65 | 25 (83.3) | 5 (16.7) | ||
| >65 | 4 (40.0) | 6 (60.0) | ||
| Stage of the disease | II | 51 (83.6) | 10 (16.4) | 3.89 (1.62–9.84) p = 0.001 |
| III | 42 (56.8) | 32 (53.2) | ||
| Histological type | Squamous cell carcinoma | 85 (71.4) | 34 (28.6) | 2.32 (1.06–5.08) p = 0.035 |
| Adenocarcinoma | 7 (63.6) | 4 (36.4) | ||
| Adenocarcinoma with mixed subtypes | 1 (20.0) | 4 (80.0) | ||
| Grade | Low | 21 (80.8) | 5 (19.2) | 1.25 (0.78–2.02) p = 0.356 |
| Intermediate | 15 (60.0) | 10 (40.0) | ||
| High | 57 (67.9) | 27 (32.1) | ||
| Lymph node metastases | N- | 63 (74.1) | 22 (25.9) | 1.91 (0.84–4.29) p = 0.123 |
| N+ | 30 (60.0) | 20 (40.0) | ||
| Parametrial infiltration | Absence | 40 (81.6) | 9 (18.4) | 2.77 (1.17–6.54) p = 0.020 |
| Presence | 53 (61.6) | 33 (38.4) | ||
| Form of tumor growth | Exophytic | 10 (62.5) | 6 (37.5) | 1.03 (0.61–1.75) p = 0.901 |
| Endophytic | 28 (75.7) | 9 (24.3) | ||
| Mixed | 55 (67.1) | 27 (32.9) | ||
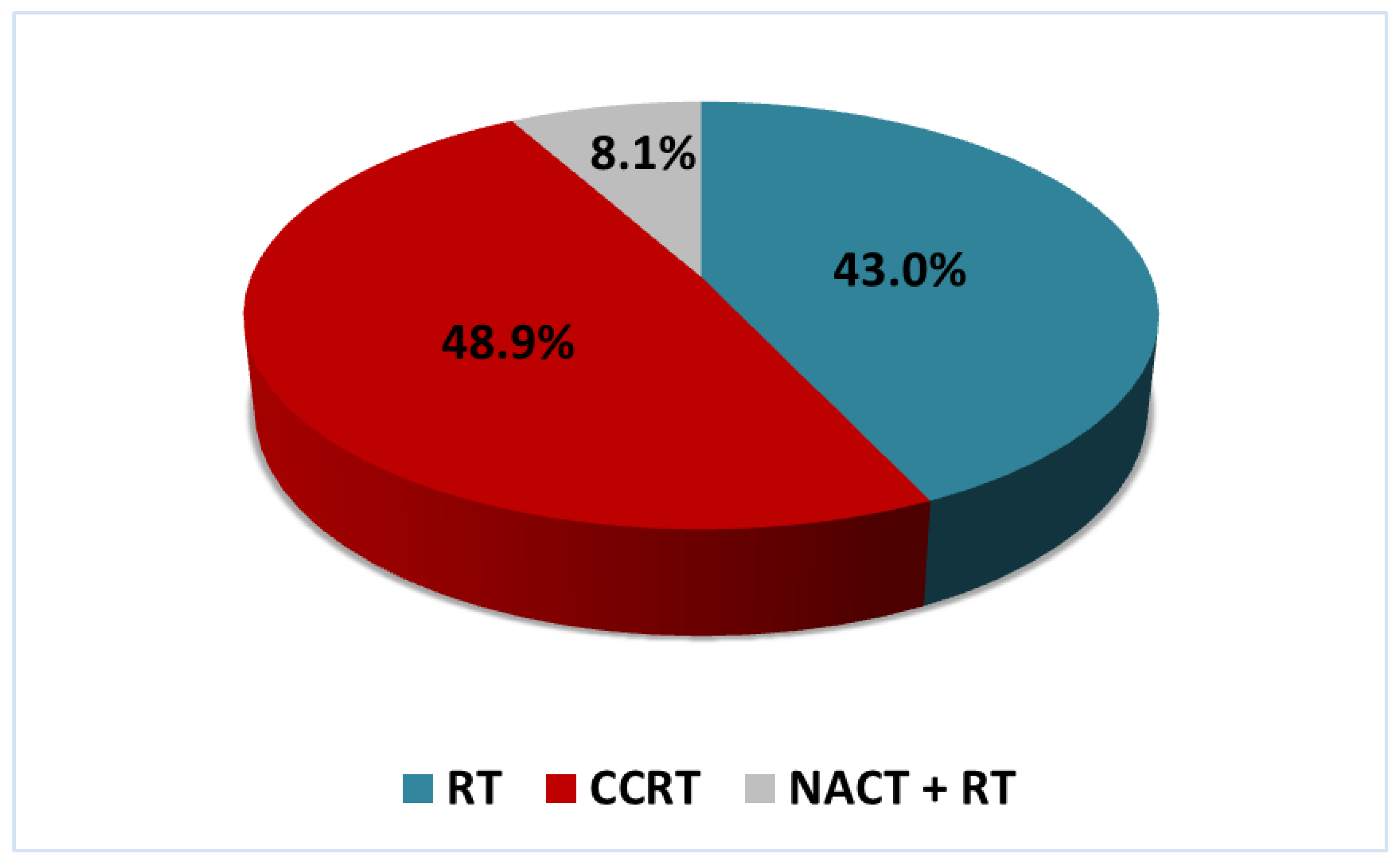
| Method of treatment | RT | 38 (65.5) | 20 (34.5) | 0.88 (0.49–1.59) p = 0.682 |
| CCRT | 48 (72.7) | 18 (27.3) | ||
| NACT + RT | 7 (63.6) | 4 (36.4) | ||
| Predictor | b * | SE | p-Value | OR = exp(b) (95% CI) |
|---|---|---|---|---|
| Presence/absence of the biomarker | 2.269 | 0.590 | 1.2 × 10−4 | 9.67 (3.04−30.75) |
| Stage of disease | 1.546 | 0.471 | 0.001 | 4.69 (1.86−11.81) |
| Patient’s age | −0.497 | 0.222 | 0.025 | 0.61 (0.39−0.94) |
| Constant | −7.350 | 1.701 | 1.6 × 10−4 | 0.001 |
| Observed Cases | Predicted Cases | Percentage of Correct Cases | |
|---|---|---|---|
| Favorable Outcome | Unfavorable Outcome | ||
| Favorable outcome | 70 | 23 | 75.3 |
| Unfavorable outcome | 11 | 31 | 73.8 |
| Overall percentage | 74.8 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mkrtchian, L.; Zamulaeva, I.; Krikunova, L.; Kiseleva, V.; Matchuk, O.; Liubina, L.; Kulieva, G.; Ivanov, S.; Kaprin, A. HPV Status and Individual Characteristics of Human Papillomavirus Infection as Predictors for Clinical Outcome of Locally Advanced Cervical Cancer. J. Pers. Med. 2021, 11, 479. https://doi.org/10.3390/jpm11060479
Mkrtchian L, Zamulaeva I, Krikunova L, Kiseleva V, Matchuk O, Liubina L, Kulieva G, Ivanov S, Kaprin A. HPV Status and Individual Characteristics of Human Papillomavirus Infection as Predictors for Clinical Outcome of Locally Advanced Cervical Cancer. Journal of Personalized Medicine. 2021; 11(6):479. https://doi.org/10.3390/jpm11060479
Chicago/Turabian StyleMkrtchian, Liana, Irina Zamulaeva, Liudmila Krikunova, Valentina Kiseleva, Olga Matchuk, Liubov Liubina, Gunel Kulieva, Sergey Ivanov, and Andrey Kaprin. 2021. "HPV Status and Individual Characteristics of Human Papillomavirus Infection as Predictors for Clinical Outcome of Locally Advanced Cervical Cancer" Journal of Personalized Medicine 11, no. 6: 479. https://doi.org/10.3390/jpm11060479
APA StyleMkrtchian, L., Zamulaeva, I., Krikunova, L., Kiseleva, V., Matchuk, O., Liubina, L., Kulieva, G., Ivanov, S., & Kaprin, A. (2021). HPV Status and Individual Characteristics of Human Papillomavirus Infection as Predictors for Clinical Outcome of Locally Advanced Cervical Cancer. Journal of Personalized Medicine, 11(6), 479. https://doi.org/10.3390/jpm11060479






