Implementing Personalized Medicine in COVID-19 in Andalusia: An Opportunity to Transform the Healthcare System
Abstract
1. Introduction
2. Impact of Human Genome on COVID-19
3. Role of Viral Genetic Variants in Covid-19
4. Genetic Epidemiology of COVID-19
5. Data Science in Health Data Sheet from Large Populations: An Opportunity for COVID-19
6. Ethics, Data Science and Data Sharing in the Times of COVID-19
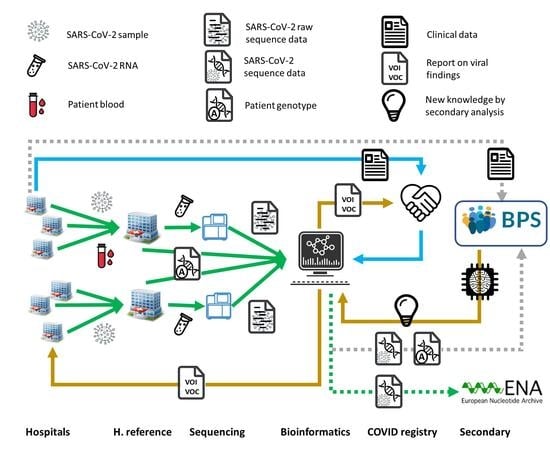
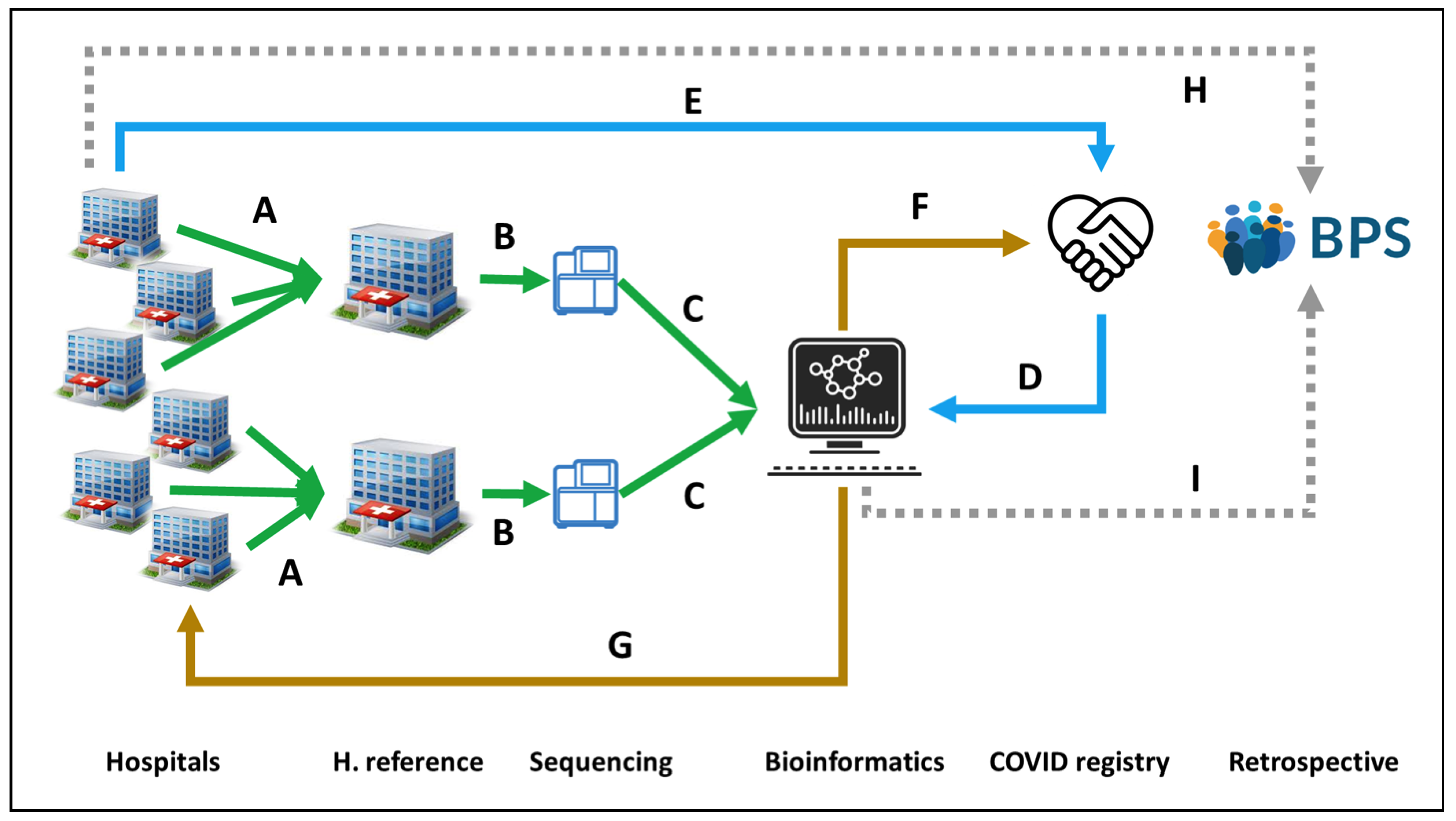
7. Translating Personalized Medicine into Clinical Practice: The Andalusian Experience
8. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Loucera, C.; Esteban-Medina, M.; Rian, K.; Falco, M.M.; Dopazo, J.; Peña-Chilet, M. Drug repurposing for COVID-19 using machine learning and mechanistic models of signal transduction circuits related to SARS-CoV-2 infection. Signal Transduct. Target. Ther. 2020, 5, 290. [Google Scholar] [CrossRef]
- Friedman, J.M.; Jones, K.L.; Carey, J.C. Exome Sequencing as Part of a Multidisciplinary Approach to Diagnosis-Reply. JAMA 2020, 324, 2445–2446. [Google Scholar] [CrossRef]
- Wolff, D.; Nee, S.; Hickey, N.S.; Marschollek, M. Risk factors for Covid-19 severity and fatality: A structured literature review. Infection 2021, 49, 15–28. [Google Scholar] [CrossRef]
- Severe Covid-19 GWAS Group; Ellinghaus, D.; Degenhardt, F.; Bujanda, L.; Buti, M.; Albillos, A.; Invernizzi, P.; Fernández, J.; Prati, D.; Baselli, G.; et al. Genomewide Association Study of Severe Covid-19 with Respiratory Failure. N. Engl. J. Med. 2020, 383, 1522–1534. [Google Scholar] [CrossRef]
- Shelton, J.F.; Shastri, A.J.; Ye, C.; Weldon, C.H.; Filshtein-Sonmez, T.; Coker, D.; Symons, A.; Esparza-Gordillo, J.; 23andMe COVID-19 Team; Aslibekyan, S.; et al. Trans-ethnic analysis reveals genetic and non-genetic associations with COVID-19 susceptibility and severity. Nat. Genet. 2021. [Google Scholar] [CrossRef]
- Roberts, G.H.L.; Park, D.S.; Coignet, M.V.; McCurdy, S.R.; Knight, S.C.; Partha, R.; Rhead, B.; Zhang, M.; Berkowitz, N.; Ancestry DNA Science Team; et al. Ancestry DNA COVID-19 Host Genetic Study Identifies Three Novel Loci. Available online: https://www.medrxiv.org/content/10.1101/2020.10.06.20205864v1 (accessed on 19 May 2021).
- Pairo-Castineira, E.; Clohisey, S.; Klaric, L.; Bretherick, A.D.; Rawlik, K.; Pasko, D.; Walker, S.; Parkinson, N.; Fourman, M.H.; Russell, C.D.; et al. Genetic mechanisms of critical illness in Covid-19. Nature 2020. [Google Scholar] [CrossRef]
- Horowitz, J.E.; Kosmicki, J.A.; Damask, A.; Sharma, D.; Roberts, G.H.L.; Justice, A.E.; Banerjee, N.; Coignet, M.V.; Yadav, A.; Leader, J.B. Common genetic variants identify therapeutic targets for COVID-19 and individuals at high risk of severe disease. MedRxiv 2020. [Google Scholar] [CrossRef]
- The COVID-19 Host Genetics Initiative; Ganna, A. Mapping the human genetic architecture of COVID-19 by worldwide meta-analysis. medRxiv 2021. [Google Scholar] [CrossRef]
- Zeberg, H.; Pääbo, S. The major genetic risk factor for severe COVID-19 is inherited from Neanderthals. Nature 2020, 587, 610–612. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, T.; Pigazzini, S.; Degenhardt, F.; Cordioli, M.; Butler-Laporte, G.; Maya-Miles, D.; Nafría-Jiménez, B.; Bouysran, Y.; Niemi, M.; Palom, A.; et al. Age-dependent impact of the major common genetic risk factor for COVID-19 on severity and mortality. medRxiv 2021. [Google Scholar] [CrossRef]
- Bianco, C.; Baselli, G.; Malvestiti, F.; Santoro, L.; Pelusi, S.; Manunta, M. Genetic insight into COVID-19-related liver injury. Liver Int. 2020. [Google Scholar] [CrossRef]
- Valenti, L.; Griffini, S.; Lamorte, G.; Grovetti, E.; Uceda Renteria, S.C.; Malvestiti, F.; Scudeller, L.; Bandera, A.; Peyvandi, F.; Prati, D.; et al. Chromosome 3 cluster rs11385942 variant links complement activation with severe COVID-19. J. Autoimmun. 2021, 117, 102595. [Google Scholar] [CrossRef]
- Schmiedel, B.J.; Chandra, V.; Rocha, J.; Gonzalez-Colin, C.; Bhattacharyya, S.; Madrigal, A.; Ottensmeier, C.H.; Ay, F.; Vijayanand, P. COVID-19 Genetic Risk Variants Are Associated with Expression of Multiple Genes in Diverse IMMUNE cell Types. bioRxiv 2020. [Google Scholar] [CrossRef]
- Szabo, P.A.; Dogra, P.; Gray, J.I.; Wells, S.B.; Connors, T.J.; Weisberg, S.P.; Krupska, I.; Matsumoto, R.; Poon, M.M.L.; Idzikowski, E.; et al. Analysis of respiratory and systemic immune responses in COVID-19 reveals mechanisms of disease pathogenesis. medRxiv 2020. [Google Scholar] [CrossRef]
- Wei, Q.; Gu, Y.-F.; Zhang, Q.-J.; Yu, H.; Peng, Y.; Williams, K.W.; Wang, R.; Yu, K.; Liu, T.; Liu, Z.-P. Lztfl1/BBS17 controls energy homeostasis by regulating the leptin signaling in the hypothalamic neurons. J. Mol. Cell Biol. 2018, 10, 402–410. [Google Scholar] [CrossRef] [PubMed]
- Xiao, F.; Tang, M.; Zheng, X.; Liu, Y.; Li, X.; Shan, H. Evidence for Gastrointestinal Infection of SARS-CoV-2. Gastroenterology 2020, 158, 1831–1833.e3. [Google Scholar] [CrossRef] [PubMed]
- Lamers, M.M.; Beumer, J.; Van Der Vaart, J.; Knoops, K.; Puschhof, J.; Breugem, T.I.; Ravelli, R.B.G.; Van Schayck, J.P.; Mykytyn, A.Z.; Duimel, H.Q.; et al. SARS-CoV-2 productively infects human gut enterocytes. Science 2020, 369, 50–54. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Li, C.; Liu, X.; Chiu, M.C.; Zhao, X.; Wang, D.; Wei, Y.; Lee, A.; Zhang, A.J.; Chu, H.; et al. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat. Med. 2020, 26, 1077–1083. [Google Scholar] [CrossRef]
- Olaussen, R.W.; Karlsson, M.R.; Lundin, K.E.; Jahnsen, J.; Brandtzaeg, P.; Farstad, I.N. Reduced chemokine receptor 9 on intraepithelial lymphocytes in celiac disease suggests persistent epithelial activation. Gastroenterology 2007, 132, 2371–2382. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Jangani, M.; Parmar, A.; Wang, G.; Coe, D.; Spear, S.; Sandrock, I.; Capasso, M.; Coles, M.; Cornish, G.; et al. A Subset of CCL25-Induced Gut-Homing T Cells Affects Intestinal Immunity to Infection and Cancer. Front. Immunol. 2019, 10, 271. [Google Scholar] [CrossRef]
- López-Pacheco, C.; Soldevila, G.; Du Pont, G.; Hernández-Pando, R.; García-Zepeda, E.A. CCR9 Is a Key Regulator of Early Phases of Allergic Airway Inflammation. Mediat. Inflamm. 2016, 2016, 3635809. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Imam, H.; Siddiqui, A. Subversion of cellular autophagy during virus infection: Insights from hepatitis B and hepatitis C viruses. Liver Res. 2018, 2, 146–156. [Google Scholar] [CrossRef]
- Wozniak, A.L.; Long, A.; Jones-Jamtgaard, K.N.; Weinman, S.A. Hepatitis C virus promotes virion secretion through cleavage of the Rab7 adaptor protein RILP. Proc. Natl. Acad. Sci. USA 2016, 113, 12484–12489. [Google Scholar] [CrossRef]
- Boyaka, P.N.; McGhee, J.R.; Czerkinsky, C.; Mestecky, J. Mucosal Vaccines: An Overview. Mucosal Immunol. 2005, 855–874. [Google Scholar] [CrossRef]
- Lillard, J.W., Jr.; Boyaka, P.N.; Hedrick, J.A.; Zlotnik, A.; McGhee, J.R. Lymphotactin acts as an innate mucosal adjuvant. J. Immunol. 1999, 162, 1959–1965. [Google Scholar] [PubMed]
- Shan, L.; Qiao, X.; Oldham, E.; Catron, D.; Kaminski, H.; Lundell, D.; Zlotnik, A.; Gustafson, E.; Hedrick, J.A. Identification of viral macrophage inflammatory protein (vMIP)-II as a ligand for GPR5/XCR1. Biochem. Biophys. Res. Commun. 2000, 268, 938–941. [Google Scholar] [CrossRef]
- Kim, B.O.; Liu, Y.; Zhou, B.Y.; He, J.J. Induction of C chemokine XCL1 (lymphotactin/single C motif-1 alpha/activation-induced, T cell-derived and chemokine-related cytokine) expression by HIV-1 Tat protein. J. Immunol. 2004, 172, 1888–1895. [Google Scholar] [CrossRef]
- Uddin, M.; Mustafa, F.; Rizvi, T.A.; Loney, T.; Suwaidi, H.A.; Al-Marzouqi, A.H.H.; Eldin, A.K.; Alsabeeha, N.; Adrian, T.E.; Stefanini, C.; et al. SARS-CoV-2/COVID-19: Viral Genomics, Epidemiology, Vaccines, and Therapeutic Interventions. Viruses 2020, 12, 526. [Google Scholar] [CrossRef]
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics 2018, 34, 4121–4123. [Google Scholar] [CrossRef] [PubMed]
- Duchene, S.; Featherstone, L.; Haritopoulou-Sinanidou, M.; Rambaut, A.; Lemey, P.; Baele, G. Temporal signal and the phylodynamic threshold of SARS-CoV-2. Virus Evol. 2020, 6, veaa061. [Google Scholar] [CrossRef]
- Toyoshima, Y.; Nemoto, K.; Matsumoto, S.; Nakamura, Y.; Kiyotani, K. SARS-CoV-2 genomic variations associated with mortality rate of COVID-19. J. Hum. Genet. 2020, 65, 1075–1082. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wu, J.; Nie, J.; Zhang, L.; Hao, H.; Liu, S.; Zhao, C.; Zhang, Q.; Liu, H.; Nie, L.; et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell 2020, 182, 1284–1294.e9. [Google Scholar] [CrossRef] [PubMed]
- Pachetti, M.; Marini, B.; Benedetti, F.; Giudici, F.; Mauro, E.; Storici, P.; Masciovecchio, C.; Angeletti, S.; Ciccozzi, M.; Gallo, R.C.; et al. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020, 18, 1–9. [Google Scholar] [CrossRef]
- Young, B.E.; Fong, S.W.; Chan, Y.H.; Mak, T.M.; Ang, L.W.; Anderson, D.E.; Lee, C.Y.; Amrun, S.N.; Lee, B.; Goh, Y.S.; et al. Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: An observational cohort study. Lancet 2020, 396, 603–611. [Google Scholar] [CrossRef]
- Popa, A.; Genger, J.W.; Nicholson, M.D.; Penz, T.; Schmid, D.; Aberle, S.W.; Agerer, B.; Lercher, A.; Endler, L.; Colaço, H.; et al. Genomic epidemiology of superspreading events in Austria reveals mutational dynamics and transmission properties of SARS-CoV-2. Sci. Transl. Med. 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Oude Munnink, B.B.; Sikkema, R.S.; Nieuwenhuijse, D.F.; Molenaar, R.J.; Munger, E.; Molenkamp, R.; van der Spek, A.; Tolsma, P.; Rietveld, A.; Brouwer, M.; et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science 2021, 371, 172–177. [Google Scholar] [CrossRef]
- McCarthy, K.R.; Rennick, L.J.; Nambulli, S.; Robinson-McCarthy, L.R.; Bain, W.G.; Haidar, G.; Duprex, W.P. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science 2021, 371, 1139–1142. [Google Scholar] [CrossRef]
- Davies, N.G.; Jarvis, C.I.; CMMID COVID-19 Working Group; Edmunds, W.J.; Jewell, N.P.; Diaz-Ordaz, K.; Keogh, R.H. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature 2021. [Google Scholar] [CrossRef]
- Van Dorp, L.; Richard, D.; Tan, C.; Shaw, L.; Acman, M.; Balloux, F. No evidence for increased transmissibility from recurrent mutations in SARS-CoV-2. Nat. Commun. 2020, 11, 1–8. [Google Scholar] [CrossRef]
- Sheahan, T.P.; Sims, A.C.; Leist, S.R.; Schäfer, A.; Won, J.; Brown, A.J.; Montgomery, S.A.; Hogg, A.; Babusis, D.; Clarke, M.O.; et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Agostini, M.L.; Andres, E.L.; Sims, A.C.; Graham, R.L.; Sheahan, T.P.; Lu, X.; Smith, E.C.; Case, J.B.; Feng, J.Y.; Jordan, R.; et al. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. MBio 2018, 9, e00221-18. [Google Scholar] [CrossRef] [PubMed]
- Martinot, M.; Jary, A.; Fafi-Kremer, S.; Leducq, V.; Delagreverie, H.; Garnier, M.; Pacanowski, J.; Mékinian, A.; Pirenne, F.; Tiberghien, P.; et al. Remdesivir failure with SARS-CoV-2 RNA-dependent RNA-polymerase mutation in a B-cell immunodeficient patient with protracted Covid-19. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control. ECDC Strategic Framework for the Integration of Molecular and Genomic Typing into European Surveillance and Multi-Country Outbreak Investigations. Available online: https://www.ecdc.europa.eu/en/publications-data/ecdc-strategic-framework-integration-molecular-and-genomic-typing-european (accessed on 31 December 2020).
- Expert Opinion on Whole Genome Sequencing for Public Health Surveillance. Available online: https://www.ecdc.europa.eu/en/publications-data/expert-opinion-whole-genome-sequencing-public-health-surveillance (accessed on 31 December 2020).
- Report 42—Transmission of SARS-CoV-2 Lineage B.1.1.7 in England: Insights from Linking Epidemiological and Genetic Data. 2020. Available online: https://www.imperial.ac.uk/mrc-global-infectious-disease-analysis/covid-19/report-42-sars-cov-2-variant/ (accessed on 2 January 2021).
- The PIRASOA Programme. 2014. Available online: http://pirasoa.iavante.es/ (accessed on 4 January 2021).
- SIEGA (Integrated System for Genomic Epidemiology in Andalusia). 2020. Available online: http://clinbioinfosspa.es/projects/siega/ (accessed on 4 January 2021).
- The Andalusian SARS-CoV-2 Genomic Surveillance Project. 2020. Available online: http://clinbioinfosspa.es/projects/covseq/ (accessed on 4 January 2021).
- Mas, V.; Nair, H.; Campbell, H.; Melero, J.A.; Williams, T.C. Antigenic and sequence variability of the human respiratory syncytial virus F glycoprotein compared to related viruses in a comprehensive dataset. Vaccine 2018, 36, 6660–6673. [Google Scholar] [CrossRef] [PubMed]
- Simões, E.A.F.; Forleo-Neto, E.; Geba, G.P.; Kamal, M.; Yang, F.; Cicirello, H.; Houghton, M.R.; Rideman, R.; Zhao, Q.; Benvin, S.L.; et al. Suptavumab for the prevention of medically attended respiratory syncytial virus infection in preterm infants. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Romanò, L.; Paladini, S.; Galli, C.; Raimondo, G.; Pollicino, T.; Zanetti, A.R. Hepatitis B vaccination: Are escape mutant viruses a matter of concern? Human Vaccines Immunother. 2015, 1, 53–57. [Google Scholar] [CrossRef]
- Ali, H.; Donovan, B.; Wand, H.; Read, T.R.; Regan, D.G.; Grulich, A.E.; Fairley, C.K.; Guy, R.J. Genital warts in young Australians five years into national human papillomavirus vaccination programme: National surveillance data. Br. Med. J. 2013, 346, f2032. [Google Scholar] [CrossRef]
- Weisblum, Y.; Schmidt, F.; Zhang, F.; DaSilva, J.; Poston, D.; Lorenzi, J.C.; Muecksch, F.; Rutkowska, M.; Hoffmann, H.H.; Michailidis, E. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife 2020, 9, e61312. [Google Scholar] [CrossRef] [PubMed]
- Andreano, E.; Piccini, G.; Licastro, D.; Casalino, L.; Johnson, N.V.; Paciello, I.; Monego, S.D.; Pantano, E.; Manganaro, N.; Manenti, A. SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. bioRxiv 2020. (Preprint). [Google Scholar] [CrossRef]
- Poland, G.A.; Ovsyannikova, I.G.; Kennedy, R.B. Personalized vaccinology: A review. Vaccine 2018, 36, 5350–5357. [Google Scholar] [CrossRef] [PubMed]
- Grifoni, A.; Sidney, J.; Zhang, Y.; Scheuermann, R.H.; Peters, B.; Sette, A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe 2020, 27, 671–680.e2. [Google Scholar] [CrossRef]
- Kiyotani, K.; Toyoshima, Y.; Nemoto, K.; Nakamura, Y. Bioinformatic prediction of potential T cell epitopes for SARS-Cov-2. J. Human Genet. 2020, 65, 569–575. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.; David, J.K.; Maden, S.K.; Wood, M.A.; Weeder, B.R.; Nellore, A.; Thompson, R.F. Human leukocyte antigen susceptibility map for SARS-CoV-2. J. Virol. 2020, 94, e00510–e00520. [Google Scholar] [CrossRef]
- Barquera, R.; Collen, E.; Di, D.; Buhler, S.; Teixeira, J.; Llamas, B.; Nunes, J.M.; Sanchez-Mazas, A. Binding affinities of 438 HLA proteins to complete proteomes of seven pandemic viruses and distributions of strongest and weakest HLA peptide binders in populations worldwide. HLA 2020, 96, 277–298. [Google Scholar] [CrossRef] [PubMed]
- Omersel, J.; Karas Kuželički, N. Vaccinomics and Adversomics in the Era of Precision Medicine: A Review Based on HBV, MMR, HPV, and COVID-19 Vaccines. J. Clin. Med. 2020, 9, 3561. [Google Scholar] [CrossRef]
- Densen, P. Challenges and opportunities facing medical education. Trans. Am. Clin. Climatol. Assoc. 2011, 122, 48–58. [Google Scholar] [PubMed]
- Sherman, R.E.; Anderson, S.A.; Dal Pan, G.J.; Gray, G.W.; Gross, T.; Hunter, N.L.; LaVange, L.; Marinac-Dabic, D.; Marks, P.W.; Robb, M.A.; et al. Real-World Evidence—What Is It and What Can It Tell Us? N. Engl. J. Med. 2016, 375, 2293–2297. [Google Scholar] [CrossRef] [PubMed]
- Topol, E.J. High-performance medicine: The convergence of human and artificial intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef]
- Rajkomar, A.; Dean, J.; Kohane, I. Machine learning in medicine. N. Engl. J. Med. 2019, 380, 1347–1358. [Google Scholar] [CrossRef]
- van der Schaar, M.; Alaa, A.M.; Floto, A.; Gimson, A.; Scholtes, S.; Wood, A.; McKinney, E.; Jarrett, D.; Lio, P.; Ercole, A. How artificial intelligence and machine learning can help healthcare systems respond to COVID-19. Mach Learn 2020, 110, 1–14. [Google Scholar] [CrossRef]
- Martini, K.; Blüthgen, C.; Walter, J.E.; Messerli, M.; Nguyen-Kim, T.D.L.; Frauenfelder, T. Accuracy of Conventional and Machine Learning Enhanced Chest Radiography for the Assessment of COVID-19 Pneumonia: Intra-Individual Comparison with CT. J. Clin. Med. 2020, 9, 3576. [Google Scholar] [CrossRef]
- Khanday, A.M.U.D.; Rabani, S.T.; Khan, Q.R.; Rouf, N.; Din, M.M.U. Machine learning based approaches for detecting COVID-19 using clinical text data. Int. J. Inf. Technol. 2020, 12, 731–739. [Google Scholar] [CrossRef]
- Yan, L.; Zhang, H.; Goncalves, J.; Xiao, Y.; Wang, M.; Guo, Y.; Sun, C.; Tang, X.; Jin, L.; Zhang, M.; et al. Prediction of survival for severe Covid-19 patients with three clinical features: Development of a machine learning-based prognostic model with clinical data in Wuhan. medRxiv 2020. [Google Scholar] [CrossRef]
- Alaa, A.M.; van der Schaar, M. Autoprognosis: Automated clinical prognostic modeling via bayesian optimization with structured kernel learning. arXiv 2018, arXiv:180207207. [Google Scholar]
- Ostaszewski, M.; Mazein, A.; Gillespie, M.E.; Kuperstein, I.; Niarakis, A.; Hermjakob, H.; Pico, A.R.; Willighagen, E.L.; Evelo, C.T.; Hasenauer, J.; et al. COVID-19 Disease Map, building a computational repository of SARS-CoV-2 virus-host interaction mechanisms. Sci. Data 2020, 7, 136. [Google Scholar] [CrossRef] [PubMed]
- Harrison, C. Coronavirus puts drug repurposing on the fast track. Nat. Biotechnol. 2020, 38, 379–381. [Google Scholar] [CrossRef]
- Fragkou, P.C.; Belhadi, D.; Peiffer-Smadja, N.; Moschopoulos, C.D.; Lescure, F.X.; Janocha, H.; Karofylakis, E.; Yazdanpanah, Y.; Mentré, F.; Skevaki, C.; et al. Review of trials currently testing treatment and prevention of COVID-19. Clin. Microbiol. Infect. 2020, 26, 988–998. [Google Scholar] [CrossRef]
- The 1000 Genomes Project. Available online: http://www.internationalgenome.org/ (accessed on 10 May 2021).
- dbGaP. Available online: https://www.ncbi.nlm.nih.gov/gap (accessed on 10 May 2021).
- The European Genome-Phenome Archive EGA. Available online: https://www.ebi.ac.uk/ega/home (accessed on 10 May 2021).
- NHGRI AnVIL. Available online: https://anvilproject.org/ (accessed on 10 May 2021).
- COVID-19 HGI: How to Share Data. Available online: https://www.covid19hg.org/data-sharing/ (accessed on 12 May 2021).
- Muñoyerro-Muñiz, D.; Goicoechea-Salazar, J.; García-León, F.; Laguna-Tellez, A.; Larrocha-Mata, D.; Cardero-Rivas, M. Health record linkage: Andalusian health population database. Gaceta Sanitaria 2019, 34, 105–113. [Google Scholar] [CrossRef]
- BPS and Research. Andalusian Health Population Database (Base Poblacional de Salud), 2020. Available online: https://www.sspa.juntadeandalucia.es/servicioandaluzdesalud/sites/default/files/sincfiles/wsas-media mediafile_sasdocumento/2019/BPS_Investigaci%C3%B3n.pdf (accessed on 3 January 2021).
- García-León, F.; Villegas-Portero, R.; Goicoechea-Salazar, J.; Muñoyerro-Muñiz, D.; Dopazo, J. Impact assessment on data protection in research projects. Gaceta Sanitaria 2020, 34, 521–523. [Google Scholar] [CrossRef]
- Clinical Bioinformatics Area. Progress and Health Foundation, 2017. Available online: http://clinbioinfosspa.es/projects/covseq/indexEng.html (accessed on 3 April 2021).
| Chr | SNPs | Position | Genetic Variation(Effect Allele/Reference Allele) | Genes in LD Region | Associated Phenotype(s) | ß-Coefficient (COVID HGI) or ODDS RATIO (rest) | p-Value | Reference Study, nº of Patients & Phenotype(s) Definition |
|---|---|---|---|---|---|---|---|---|
| 1 | rs67579710 | 155203736 | A/G | THBS3, KRTCAP2, TRIM46, MUC1, MTX1 | Hospitalization | −0.138 | 3.4 × 10−8 | COVID HGI (Data freeze nº5 Jan 2021) 46 studies across 19 countries worldwide Critically Ill (6.179) vs population control (1.483.780) Hospitalized COVID-19 (13.641) vs population control (2.070.709) SARS-CoV-2 infection (49.562) vs population control (1.770.206) PHENOTYPES: Critically ill: Required respiratory support or COVID-19 related death Hospitalized: Required hospitalization due to COVID-19 SARS-CoV2 infection: Laboratory confirmed OR electronic health record, ICD coding OR Physician-confirmed COVID-19 OR self-reported COVID-19 |
| 2 | rs1381109 | 166061783 | T/G | SCN1A | Hospitalization | −0.096 | 4.2 × 10−8 | |
| 3 | rs10490770 rs11919389 | 45823240 101705614 | C/T C/T | LZTFL1 RPL24, ZBTB11, CEP97, NXPE3 | Critical Illness Hospitalization Infection susceptibility Infection susceptibility | 0.634 0.5 0.149 −0.06 | 2.2 × 10−61 1.4 × 10−73 9.7 × 10−30 3.5 × 10−15 | |
| 5 | rs10070196 | 13939721 | C/A | DNAH5 | Infection susceptibility | 0.044 | 9.7 × 10−22 | |
| 6 | rs1886814 | 41534945 | C/A | FOXP4 | Hospitalization Infection susceptibility | 0.233 0.101 | 1.1 × 10−9 2.4 × 10−8 | |
| 8 | rs72711165 | 124324323 | C/T | TMEM65 | Hospitalization | 0.314 | 2.1 × 10−9 | |
| 9 | rs912805253 | 133274084 | T/C | ABO | Hospitalization Infection susceptibility | −0.103 −0.1 | 5.4 × 10−10 1.5 X 10−39 | |
| 12 | rs10774671 | 112919388 | A/G | OAS1, OAS2, OAS3 | Critical Illness Hospitalization Infection susceptibility | 0.231 0.144 0.048 | 4.1 × 10−13 6.1 × 10−10 1.6 X 10−11 | |
| 17 | rs1819040 rs77534576 | 46142465 49863303 | A/T T/C | ARHGAP27, PLEKHM1, LINC02210 CRHR1, SPPL2C, MAPT, STH, KANSL1, LRRC37A, ARL17B, LRRC37AA2, ARL17A, NSF, WNT3 KAT7, TAC4 | Hospitalization Critical illness | −0.129 0.369 | 1.8 × 10−10 4.4 × 10−9 | |
| 19 | rs2109069 rs74956615 rs4801778 | 4719431 10317045 48867352 | A/G A/T T/G | DPP9 TYK2, ICAM1 ICAM3, ICAM4, ICAMS, ZGLP1, FDX2, RAVER1. PLEKHA4, PPP1R115A, TULP2, NUCB1 | Critical Illness Hospitalization Infection susceptibility Hospitalization Critical Illness Infection susceptibility | 0.231 0.144 0.048 0.36 0.236 −0.055 | 9.7 × 10−22 2.8 × 10−17 4.1 × 10−9 5.1 × 10−10 9.7 × 10−22 1.2 × 10−8 | |
| 21 | rs13050728 | 33242905 | C/T | IFNAR2 | Critical Illness Hospitalization | −0.20 −0.15 | 1.1 × 10−16 2.7 × 10−20 | |
| 3 | rs11385942 | 45876460 | insertion–deletion GA or G | LZTFL1, SLC6A20, CCR9, FYCO1, CXCR6, XCR1 | Severe Covid Intubation | 1.77 (1.48–2.11) 1.70; (1.27 to 2.26) | 1.2 × 10−10 3.3 × 10−4 | COVID 19 Host(a)ge (1st release) (Spain+Italy) Severe Covid (1980) vs Population controls (2381) Severe Covid: Hospitalization + respiratory failure + confirmed SARS-CoV-2 |
| 9 | rs657152 rs8176719 rs41302905 rs8176747 | Between 133255928 and 136139265 | A/C 261delG A/G C/G | ABO | Severe Covid | A group 1.32 (1.20–1.47) O group 0.65 (0.53 to 0.79) | 1.48 × 10−4 1.1 × 10−5 | |
| 3 | rs73064425 | 45901089 | T/C | LZTFL1 | Severe Covid | 2.1 (1.88–2.45) | 4.8 × 10−30 | GenOMICC (Genetics Of Mortality In Critical Care) UK Severe Covid (2771) vs Population control (45.875) Severe Covid: Patients in critical care, being profound hypoxaemic respiratory failure the archetypal presentation. |
| 6 | rs9380142 rs143334143 rs3131294 | 29798794 31121426 32180146 | A/G A/G G/A | HLA-G CCHCR1 NOTCH4 | Severe Covid Severe Covid Severe Covid | 1.3 (1.18–1.43) 1.9 (1.61–2.13) 1.5 (1.28–1.66) | 3.2 × 10−8 8.8 × 10−18 2.8 × 10−8 | |
| 12 | rs10735079 | 113380008 | A/G | OAS1/3 | Severe Covid | 1.3 (1.18–1.42) | 1.6 × 10−8 | |
| 19 | rs2109069 rs74956615 | 4719443 10427721 | A/G A/T | DPP9 TYK2 | Severe Covid Severe Covid | 1.4 (1.25–1.48) 1.6 (1.35–1.87) | 4.0 × 10−12 2.3 × 10−8 | |
| 21 | rs2236757 | 33252612 | A/G | IFNAR2 | Severe Covid | 1.3 (1.17–1.41) | 2.3 × 10−8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dopazo, J.; Maya-Miles, D.; García, F.; Lorusso, N.; Calleja, M.Á.; Pareja, M.J.; López-Miranda, J.; Rodríguez-Baño, J.; Padillo, J.; Túnez, I.; et al. Implementing Personalized Medicine in COVID-19 in Andalusia: An Opportunity to Transform the Healthcare System. J. Pers. Med. 2021, 11, 475. https://doi.org/10.3390/jpm11060475
Dopazo J, Maya-Miles D, García F, Lorusso N, Calleja MÁ, Pareja MJ, López-Miranda J, Rodríguez-Baño J, Padillo J, Túnez I, et al. Implementing Personalized Medicine in COVID-19 in Andalusia: An Opportunity to Transform the Healthcare System. Journal of Personalized Medicine. 2021; 11(6):475. https://doi.org/10.3390/jpm11060475
Chicago/Turabian StyleDopazo, Joaquín, Douglas Maya-Miles, Federico García, Nicola Lorusso, Miguel Ángel Calleja, María Jesús Pareja, José López-Miranda, Jesús Rodríguez-Baño, Javier Padillo, Isaac Túnez, and et al. 2021. "Implementing Personalized Medicine in COVID-19 in Andalusia: An Opportunity to Transform the Healthcare System" Journal of Personalized Medicine 11, no. 6: 475. https://doi.org/10.3390/jpm11060475
APA StyleDopazo, J., Maya-Miles, D., García, F., Lorusso, N., Calleja, M. Á., Pareja, M. J., López-Miranda, J., Rodríguez-Baño, J., Padillo, J., Túnez, I., & Romero-Gómez, M. (2021). Implementing Personalized Medicine in COVID-19 in Andalusia: An Opportunity to Transform the Healthcare System. Journal of Personalized Medicine, 11(6), 475. https://doi.org/10.3390/jpm11060475