Circulating MicroRNA: Incident Asthma Prediction and Vitamin D Effect Modification
Abstract
1. Introduction
2. Materials and Methods
2.1. Participant Selection
2.2. Small RNA Sequencing
2.3. Statistical Analysis
2.4. Validation
2.5. Meta-Analysis
2.6. MiRNA-Target Gene Network and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Enrichment Analysis
2.7. Prediction
3. Results
3.1. Baseline Characteristics
3.2. Main Analysis
3.3. Significant miRNAs in Treatment Interactions Analysis
3.4. Significant miRNAs in the Stratified Analysis
3.5. Validation in Project Viva
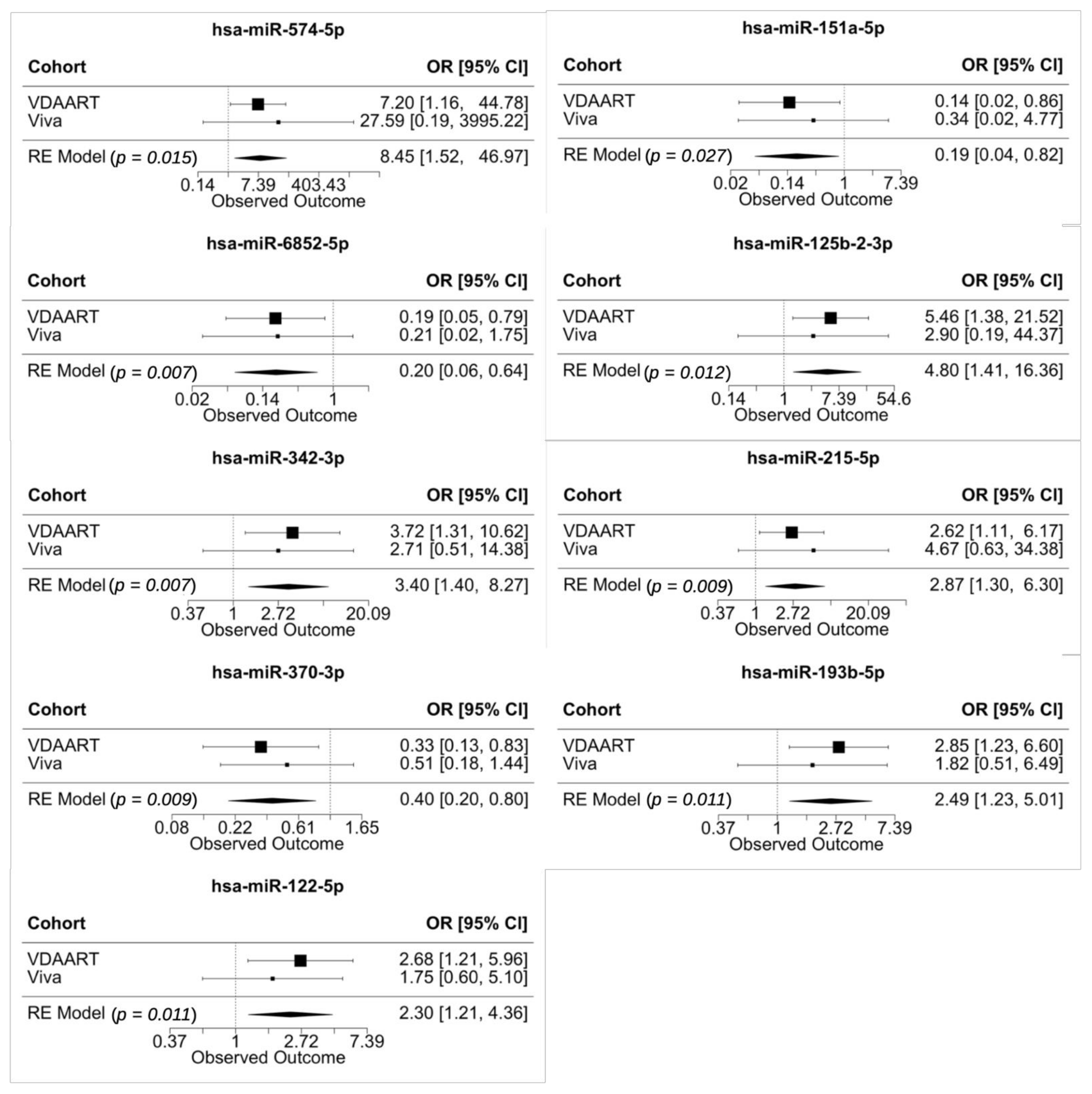
3.6. Meta-Analysis of Validated miRNAs in Vitamin D Antenatal Asthma Reduction Trial (VDAART) and Project Viva
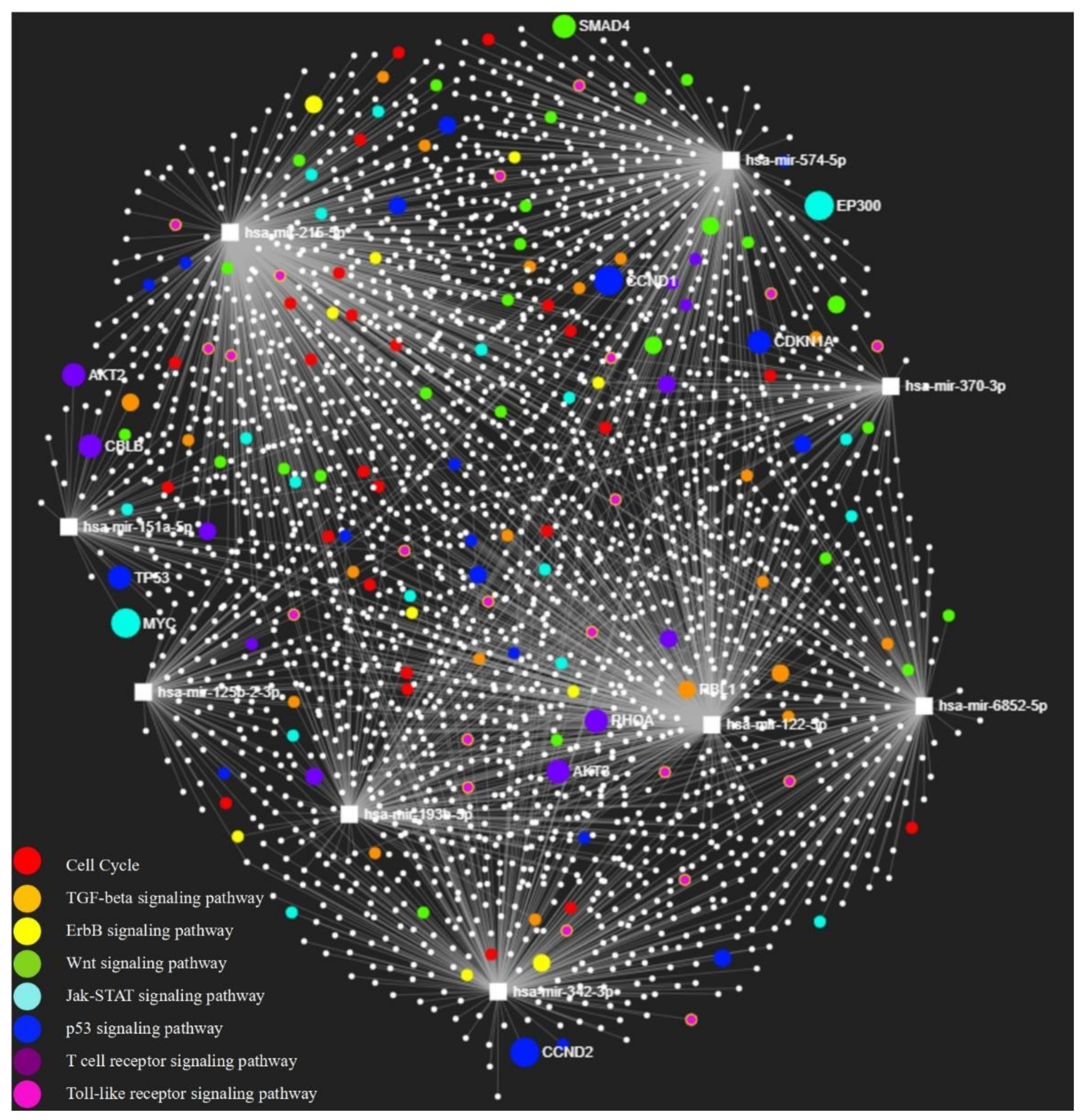
3.7. MiRNA-Target Gene Network and KEGG Pathway Enrichment Analysis
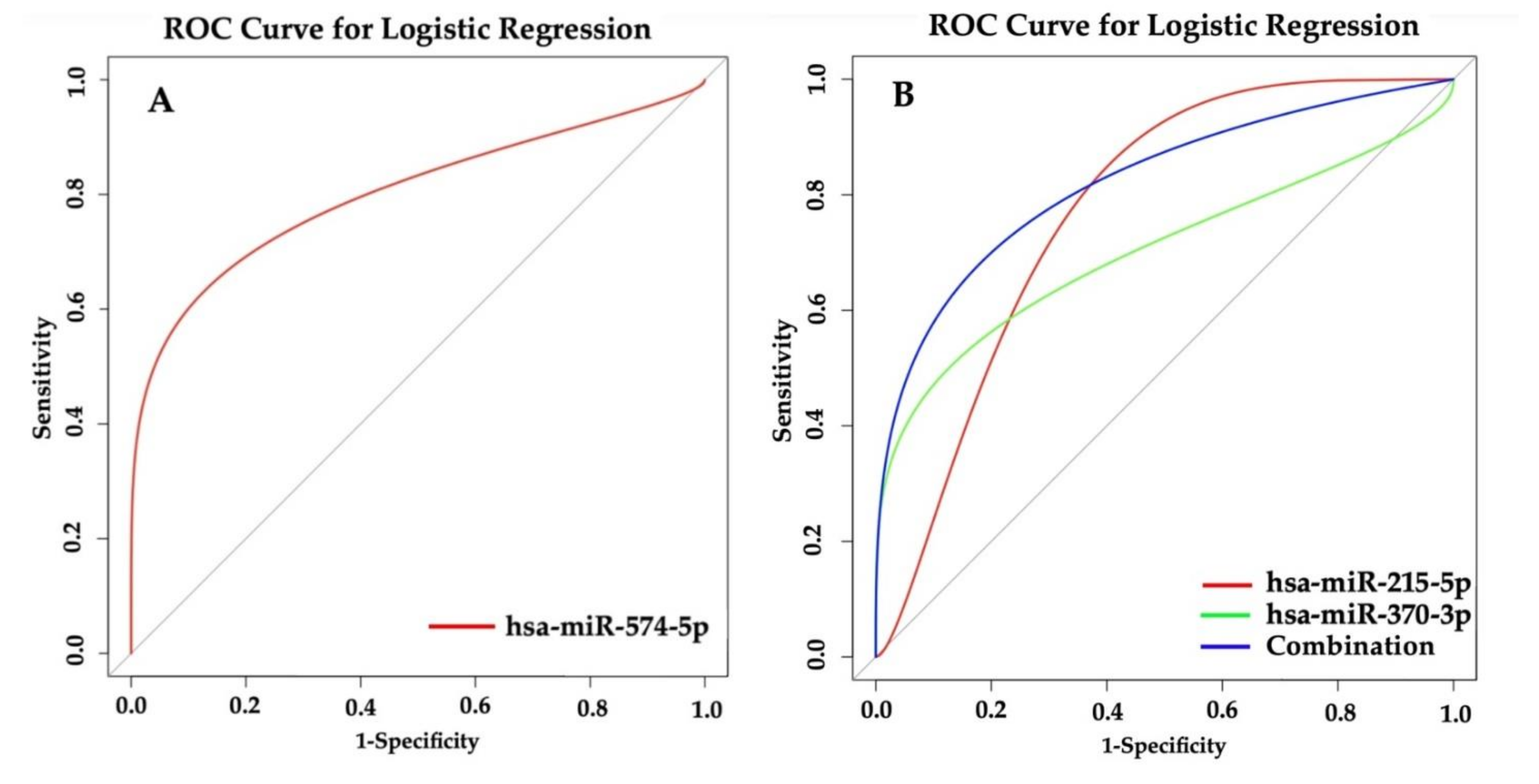
3.8. Prediction of Asthma Based on miRNAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Martinez, F.D.; Vercelli, D. Asthma. Lancet 2013, 382, 1360–1372. [Google Scholar] [CrossRef]
- CDC.gov. National Current Asthma Prevalence. 2018. Available online: https://www.cdc.gov/asthma/asthmadata.htm (accessed on 10 January 2021).
- Lambrecht, B.N.; Hammad, H. The immunology of asthma. Nat. Immunol. 2015, 16, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Martinez, F.D. Development of wheezing disorders and asthma in preschool children. Pediatrics 2002, 109, 362–367. [Google Scholar]
- Spanier, A.J.; Lasso-Pirot, A.; Delgado-Villalta, S. Early childhood wheezers: Identifying asthma in later life. J. Asthma Allergy 2015, 8, 63–73. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Martinez, F.D. What have we learned from the Tucson Children’s Respiratory Study? Paediatr. Respir. Rev. 2002, 3, 193–197. [Google Scholar] [CrossRef]
- Nair, R.; Maseeh, A. Vitamin D: The “sunshine” vitamin. J. Pharm. Pharm. 2012, 3, 118–126. [Google Scholar]
- Kulie, T.; Groff, A.; Redmer, J.; Hounshell, J.; Schrager, S. Vitamin D: An Evidence-Based Review. J. Am. Board Fam. Med. 2009, 22, 698–706. [Google Scholar] [CrossRef]
- Ali, N.S.; Nanji, K. A Review on the Role of Vitamin D in Asthma. Cureus 2017, 9, 1288. [Google Scholar] [CrossRef]
- Litonjua, A.A.; Lange, N.E.; Carey, V.J.; Brown, S.; Laranjo, N.; Harshfield, B.J.; O’Connor, G.T.; Sandel, M.; Strunk, R.C.; Bacharier, L.B.; et al. The Vitamin D Antenatal Asthma Reduction Trial (VDAART): Rationale, design, and methods of a randomized, controlled trial of vitamin D supplementation in pregnancy for the primary prevention of asthma and allergies in children. Contemp. Clin. Trials 2014, 38, 37–50. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Zhao, C.; Sun, X.; Li, L. Biogenesis and function of extracellular miRNAs. ExRNA 2019, 1, 38. [Google Scholar] [CrossRef]
- Li, J.; Panganiban, R.; Kho, A.T.; McGeachie, M.J.; Farnam, L.; Chase, R.P.; Weiss, S.T.; Lu, Q.; Tantisira, K.G. Circulating MicroRNAs and Treatment Response in Childhood Asthma. Am. J. Respir. Crit. Care Med. 2020, 202, 65–72. [Google Scholar] [CrossRef]
- Li, J.; Kho, A.T.; Chase, R.P.; Pantano, L.; Farnam, L.; Amr, S.S.; Tantisira, K.G. COMPSRA: A COMprehensive Platform for Small RNA-Seq data Analysis. Sci. Rep. 2020, 10, 4552–4557. [Google Scholar] [CrossRef]
- Kishikawa, T.; Otsuka, M.; Ohno, M.; Yoshikawa, T.; Takata, A.; Koike, K. Circulating RNAs as new biomarkers for detecting pancreatic cancer. World J. Gastroenterol. 2015, 21, 8527–8540. [Google Scholar] [CrossRef]
- Viereck, J.; Thum, T. Circulating Noncoding RNAs as Biomarkers of Cardiovascular Disease and Injury. Circ. Res. 2017, 120, 381–399. [Google Scholar] [CrossRef]
- Kho, A.T.; Sharma, S.; Davis, J.S.; Spina, J.; Howard, D.; McEnroy, K.; Moore, K.; Sylvia, J.; Qiu, W.; Weiss, S.T.; et al. Circulating MicroRNAs: Association with Lung Function in Asthma. PLoS ONE 2016, 11, e0157998. [Google Scholar] [CrossRef]
- Tiwari, A.; Li, J.; Kho, A.T.; Sun, M.; Lu, Q.; Weiss, S.T.; Tantisira, K.G.; McGeachie, M.J. COPD-associated miR-145-5p is downregulated in early-decline FEV1 trajectories in childhood asthma. J. Allergy Clin. Immunol. 2020. [Google Scholar] [CrossRef]
- Kho, A.T.; McGeachie, M.J.; Moore, K.G.; Sylvia, J.M.; Weiss, S.T.; Tantisira, K.G. Circulating microRNAs and prediction of asthma exacerbation in childhood asthma. Respir. Res. 2018, 19, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.L.; Li, J.; Kho, A.T.; McGeachie, M.J.; Tantisira, K.G. Enhancing the prediction of childhood asthma remission: Integrating clinical factors with microRNAs. J. Allergy Clin. Immunol. 2021, 147, 1093–1095. [Google Scholar] [CrossRef]
- Davis, J.S.; Sun, M.; Kho, A.T.; Moore, K.G.; Sylvia, J.M.; Weiss, S.T.; Lü, Q.; Tantisira, K.G. Circulating microRNAs and association with methacholine PC20 in the Childhood Asthma Management Program (CAMP) cohort. PLoS ONE 2017, 12, e0180329. [Google Scholar] [CrossRef] [PubMed]
- Karam, R.A.; Elrahman, D.M.A. Differential expression of miR-155 and Let-7a in the plasma of childhood asthma: Potential biomarkers for diagnosis and severity. Clin. Biochem. 2019, 68, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Rozowsky, J.; Kitchen, R.R.; Park, J.J.; Galeev, T.R.; Diao, J.; Warrell, J.; Thistlethwaite, W.; Subramanian, S.L.; Milosavljevic, A.; Gerstein, M. ExceRpt: A Comprehensive Analytic Platform for Extracellular RNA Profiling. Cell Syst. 2019, 8, 352–357. [Google Scholar] [CrossRef] [PubMed]
- Oken, E.; Baccarelli, A.A.; Gold, D.R.; Kleinman, K.P.; Litonjua, A.A.; De Meo, D.; Rich-Edwards, J.W.; Rifas-Shiman, S.L.; Sagiv, S.; Taveras, E.M.; et al. Cohort Profile: Project Viva. Int. J. Epidemiol. 2015, 44, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Camargo, C.A.; Rifas-Shiman, S.L.; Litonjua, A.A.; Rich-Edwards, J.W.; Weiss, S.T.; Gold, D.R.; Kleinman, K.; Gillman, M.W. Maternal intake of vitamin D during pregnancy and risk of recurrent wheeze in children at 3 y of age. Am. J. Clin. Nutr. 2007, 85, 788–795. [Google Scholar] [CrossRef]
- Ross, A.C.; Manson, J.E.; Abrams, S.A.; Aloia, J.F.; Brannon, P.M.; Clinton, S.K.; Durazo-Arvizu, R.A.; Gallagher, J.C.; Gallo, R.L.; Jones, G.; et al. The 2011 report on dietary reference intakes for calcium and vitamin D from the Institute of Medicine: What clinicians need to know. J. Clin. Endocrinol. Metab 2011, 96, 53–58. [Google Scholar] [CrossRef]
- Viechtbauer, W. Conducting meta-analyses in R with the metafor package. J. Stat. Softw. 2010, 36, 1–48. [Google Scholar] [CrossRef]
- Chang, L.; Zhou, G.; Soufan, O.; Xia, J. miRNet 2.0: Network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020, 48, W244–W251. [Google Scholar] [CrossRef]
- Chou, C.H.; Shrestha, S.; Yang, C.D.; Chang, N.W.; Lin, Y.L.; Liao, K.W.; Huang, W.C.; Sun, T.H.; Tu, S.J.; Lee, W.H.; et al. miRTarBase update 2018: A resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018, 46, D296–D302. [Google Scholar] [CrossRef]
- Johnson, P.R.A.; Roth, M.; Tamm, M.; Hughes, M.; Ge, Q.; King, G.; Burgess, J.K.; Black, J.L. Airway Smooth Muscle Cell Proliferation Is Increased in Asthma. Am. J. Respir. Crit. Care Med. 2001, 164, 474–477. [Google Scholar] [CrossRef]
- Halwani, R.; Al-Muhsen, S.; Al-Jahdali, H.; Hamid, Q. Role of transforming growth factor-beta in airway remodeling in asthma. Am. J. Respir Cell Mol. Biol. 2011, 44, 127–133. [Google Scholar] [CrossRef]
- Boxall, C.; Holgate, S.T.; Davies, D.E. The contribution of transforming growth factor-beta and epidermal growth factor signalling to airway remodelling in chronic asthma. Eur. Respir. J. 2006, 27, 208–229. [Google Scholar] [CrossRef]
- Polosa, R.; Puddicombe, S.M.; Krishna, M.T.; Tuck, A.B.; Howarth, P.H.; Holgate, S.T.; Davies, D.E. Expression of c-erbB receptors and ligands in the bronchial epithelium of asthmatic subjects. J. Allergy Clin. Immunol. 2002, 109, 75–81. [Google Scholar] [CrossRef]
- Le Cras, T.D.; Acciani, T.H.; Mushaben, E.M.; Kramer, E.L.; Pastura, P.A.; Hardie, W.D.; Korfhagen, T.R.; Sivaprasad, U.; Ericksen, M.; Gibson, A.M.; et al. Epithelial EGF receptor signaling mediates airway hyperreactivity and remodeling in a mouse model of chronic asthma. Am. J. Physiol. Cell. Mol. Physiol. 2011, 300, L414–L421. [Google Scholar] [CrossRef]
- Sharma, S.; Tantisira, K.; Carey, V.; Murphy, A.J.; Lasky-Su, J.; Celedón, J.C.; Lazarus, R.; Klanderman, B.; Rogers, A.; Soto-Quirós, M.; et al. A Role for Wnt Signaling Genes in the Pathogenesis of Impaired Lung Function in Asthma. Am. J. Respir. Crit. Care Med. 2010, 181, 328–336. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, L.; Jia, X.X.; Lin, X.X.; Zhang, W.X. Vitamin D alleviates airway remodeling in asthma by down-regulating the activity of Wnt/beta-catenin signaling pathway. Int. Immunopharmacol. 2019, 68, 88–94. [Google Scholar] [CrossRef]
- Pernis, A.B.; Rothman, P.B. JAK-STAT signaling in asthma. J. Clin. Investig. 2002, 109, 1279–1283. [Google Scholar] [CrossRef]
- Salvi, S.S.; Babu, K.S.; Holgate, S.T. Is Asthma Really Due to a Polarized T Cell Response Toward a Helper T Cell Type 2 Phenotype? Am. J. Respir. Crit. Care Med. 2001, 164, 1343–1346. [Google Scholar] [CrossRef]
- Trian, T.; Allard, B.; Ozier, A.; Maurat, E.; Dupin, I.; Thumerel, M.; Ousova, O.; Gillibert-Duplantier, J.; Le Morvan, V.; Begueret, H.; et al. Selective dysfunction of p53 for mitochondrial biogenesis induces cellular proliferation in bronchial smooth muscle from asthmatic patients. J. Allergy Clin. Immunol. 2016, 137, 1717–1726. [Google Scholar] [CrossRef]
- Colgan, J.D.; Hankel, I.L. Signaling pathways critical for allergic airway inflammation. Curr. Opin. Allergy Clin. Immunol. 2010, 10, 42–47. [Google Scholar] [CrossRef]
- Athari, S.S.; Athari, S.M.; Beyzay, F.; Movassaghi, M.; Mortaz, E.; Taghavi, M. Critical role of Toll-like receptors in pathophysiology of allergic asthma. Eur. J. Pharmacol. 2017, 808, 21–27. [Google Scholar] [CrossRef]
- Zuo, L.; Lucas, K.; Fortuna, C.A.; Chuang, C.-C.; Best, T.M. Molecular Regulation of Toll-like Receptors in Asthma and COPD. Front. Physiol. 2015, 6, 312. [Google Scholar] [CrossRef]
- Sinha, A.; Yadav, A.K.; Chakraborty, S.; Kabra, S.; Lodha, R.; Kumar, M.; Kulshreshtha, A.; Sethi, T.; Pandey, R.; Malik, G.; et al. Exosome-enclosed microRNAs in exhaled breath hold potential for biomarker discovery in patients with pulmonary diseases. J. Allergy Clin. Immunol. 2013, 132, 219–222. [Google Scholar] [CrossRef]
- Garbacki, N.; Di Valentin, E.; Huynh-Thu, V.A.; Geurts, P.; Irrthum, A.; Crahay, C.; Arnould, T.; Deroanne, C.; Piette, J.; Cataldo, D.; et al. MicroRNAs Profiling in Murine Models of Acute and Chronic Asthma: A Relationship with mRNAs Targets. PLoS ONE 2011, 6, e16509. [Google Scholar] [CrossRef]
- Gomez, J.L.; Chen, A.; Diaz, M.P.; Zirn, N.; Gupta, A.; Britto, C.; Sauler, M.; Yan, X.; Stewart, E.; Santerian, K.; et al. A Network of Sputum MicroRNAs Is Associated with Neutrophilic Airway Inflammation in Asthma. Am. J. Respir. Crit. Care Med. 2020, 202, 51–64. [Google Scholar] [CrossRef]
- Jorde, R.; Svartberg, J.; Joakimsen, R.M.; Coucheron, D.H. Plasma profile of microRNA after supplementation with high doses of vitamin D3 for 12 months. BMC Res. Notes 2012, 5, 245. [Google Scholar] [CrossRef]
- Giangreco, A.A.; Vaishnav, A.; Wagner, D.; Finelli, A.; Fleshner, N.; Van Der Kwast, T.; Vieth, R.; Nonn, L. Tumor Suppressor microRNAs, miR-100 and -125b, Are Regulated by 1,25-dihydroxyvitamin D in Primary Prostate Cells and in Patient Tissue. Cancer Prev. Res. 2013, 6, 483–494. [Google Scholar] [CrossRef]
- Enquobahrie, D.A.; Williams, M.A.; Qiu, C.; Siscovick, D.S.; Sorensen, T.K. Global maternal early pregnancy peripheral blood mRNA and miRNA expression profiles according to plasma 25-hydroxyvitamin D concentrations. J. Matern. Neonatal Med. 2011, 24, 1002–1012. [Google Scholar] [CrossRef][Green Version]
- Francisco-Garcia, A.S.; Garrido-Martín, E.M.; Rupani, H.; Lau, L.C.K.; Martinez-Nunez, R.T.; Howarth, P.H.; Sanchez-Elsner, T. Small RNA Species and microRNA Profiles are Altered in Severe Asthma Nanovesicles from Broncho Alveolar Lavage and Associate with Impaired Lung Function and Inflammation. Non-Coding RNA 2019, 5, 51. [Google Scholar] [CrossRef]
- Tsuchiya, M.; Kumar, P.; Bhattacharyya, S.; Chattoraj, S.; Srivastava, M.; Pollard, H.B.; Biswas, R. Differential Regulation of Inflammation by Inflammatory Mediators in Cystic Fibrosis Lung Epithelial Cells. J. Interf. Cytokine Res. 2013, 33, 121–129. [Google Scholar] [CrossRef]
- Gupta, R.; Radicioni, G.; Abdelwahab, S.; Dang, H.; Carpenter, J.; Chua, M.; Mieczkowski, P.A.; Sheridan, J.T.; Randell, S.H.; Kesimer, M. Intercellular Communication between Airway Epithelial Cells Is Mediated by Exosome-Like Vesicles. Am. J. Respir. Cell Mol. Biol. 2019, 60, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Mao, F.; Zhao, G.; Wang, H.; Yan, X.; Zhang, Q. Long non-coding RNA SNHG16 promotes lipopolysaccharides-induced acute pneumonia in A549 cells via targeting miR-370-3p/IGF2 axis. Int. Immunopharmacol. 2020, 78, 106065. [Google Scholar] [CrossRef] [PubMed]
- HollyRutledge, H.; Baran-Gale, J.; De Villena, F.P.-M.; Chesler, E.J.; Churchill, G.A.; Sethupathy, P.; Kelada, S.N.P. Identification of microRNAs associated with allergic airway disease using a genetically diverse mouse population. BMC Genom. 2015, 16, 1–14. [Google Scholar] [CrossRef]
- Wang, L.; Xia, J.-W.; Ke, Z.-P.; Zhang, B.-H. Blockade of NEAT1 represses inflammation response and lipid uptake via modulating miR-342-3p in human macrophages THP-1 cells. J. Cell. Physiol. 2019, 234, 5319–5326. [Google Scholar] [CrossRef] [PubMed]
- Bahmer, T.; Krauss-Etschmann, S.; Buschmann, D.; Behrends, J.; Watz, H.; Kirsten, A.-M.; Pedersen, F.; Waschki, B.; Fuchs, O.; Pfaffl, M.; et al. miR-122-5p and miR-191-5p are increased in plasma small extracellular vesicles in asthma. In Airway Cell Biology and Immunopathology; European Respiratory Society (ERS): Lausanne, Switzerland, 2019; Volume 54. [Google Scholar]
- Sheu, C.-C.; Tsai, M.-J.; Chen, F.-W.; Chang, K.-F.; Chang, W.-A.; Chong, I.-W.; Kuo, P.-L.; Hsu, Y.-L. Identification of novel genetic regulations associated with airway epithelial homeostasis using next-generation sequencing data and bioinformatics approaches. Oncotarget 2017, 8, 82674–82688. [Google Scholar] [CrossRef]
- Vaswani, C.; Varkouhi, A.; Teixeira, A.; Ektesabi, A.; Kim, M.; Plant, P.; Watts, T.; Moraes, T.; Capelozzi, V.; Cruz, F.; et al. The Trojan, miR193b-5p, in Virus-Induced Acute Respiratory Distress Syndrome (ARDS). In D101. Mechanistic Studies of Lung Injury and Repair; American Thoracic Society (virtual conference), 2020; p. 7705. [Google Scholar]
- Shi, J.; Ye, G.; Zhao, G.; Wang, X.; Ye, C.; Thammavong, K.; Xu, J.; Dong, J. Coordinative control of G2/M phase of the cell cycle by non-coding RNAs in hepatocellular carcinoma. Peer J. 2018, 6, e5787. [Google Scholar] [CrossRef]
| Characteristic | Asthma Status at Age 5 Years | ||
|---|---|---|---|
| Yes (n = 31) | No (n = 42) | p Value | |
| Sex | 1.0 | ||
| Male | 21(68%) | 28(67%) | |
| Female | 10(32%) | 14(33%) | |
| Race | 0.39 * | ||
| White | 8(26%) | 16(38%) | |
| Black/African American | 22(71%) | 22(52%) | |
| Asian | 0(0%) | 1(2%) | |
| Other | 1(3%) | 3(7%) | |
| Treatment | 0.817 | ||
| 4400 IU daily | 15(48%) | 18(43%) | |
| 400 IU daily | 16(52%) | 24(57%) | |
| History of asthma in mother | 1.0 | ||
| Yes | 17(55%) | 23(55%) | |
| No | 14(45%) | 19(45%) | |
| Maternal age at enrollment | 24.51 ± 4.8 | 25.66 ± 5.8 | 0.36 |
| Gestation at delivery weeks | 37.38 ± 3.57 | 38.19 ± 2.85 | 0.3 |
| 95% CI for OR | |||
|---|---|---|---|
| miRNA | OR | Lower Bound | Upper Bound |
| hsa-miR-3942-5p | 178.73 | 4.6 | 6947.95 |
| hsa-miR-151a-5p | 0.05 | 0.01 | 0.42 |
| hsa-miR-574-5p | 19.2 | 2.38 | 155.11 |
| hsa-miR-125b-2-3p | 8.62 | 1.77 | 41.84 |
| hsa-miR-6852-5p | 0.12 | 0.03 | 0.58 |
| hsa-miR-7-1-3p | 7.81 | 1.67 | 36.4 |
| hsa-miR-505-3p | 0.13 | 0.02 | 0.79 |
| hsa-miR-103a-3p | 0.15 | 0.03 | 0.81 |
| hsa-miR-1294 | 5.86 | 1.55 | 22.09 |
| hsa-miR-342-3p | 5.83 | 1.63 | 20.89 |
| hsa-miR-95-3p | 5.8 | 1.93 | 17.41 |
| hsa-miR-193b-5p | 5.55 | 1.99 | 15.45 |
| hsa-miR-29a-3p | 4.77 | 1.61 | 14.11 |
| hsa-miR-331-5p | 4.62 | 1.17 | 18.18 |
| hsa-miR-146b-3p | 4.53 | 1.23 | 16.63 |
| hsa-miR-141-3p | 4.1 | 1.36 | 12.37 |
| hsa-miR-3605-5p | 3.87 | 1.16 | 12.94 |
| hsa-miR-760 | 0.3 | 0.09 | 0.98 |
| hsa-miR-5010-5p | 3.26 | 1.18 | 9.05 |
| hsa-miR-122-5p | 3.05 | 1.2 | 7.74 |
| hsa-miR-215-5p | 3.05 | 1.14 | 8.13 |
| hsa-miR-370-3p | 0.33 | 0.12 | 0.94 |
| hsa-miR-29c-3p | 2.93 | 1.25 | 6.86 |
| hsa-miR-1273h-3p | 0.36 | 0.13 | 0.96 |
| hsa-miR-144-3p | 2.74 | 1.04 | 7.23 |
| hsa-miR-1908-5p | 0.37 | 0.14 | 1 |
| hsa-miR-4732-5p | 2.71 | 1.09 | 6.75 |
| hsa-miR-339-5p | 0.37 | 0.18 | 0.76 |
| hsa-miR-483-5p | 2.67 | 1.13 | 6.28 |
| hsa-miR-214-3p | 2.58 | 1.14 | 5.85 |
| hsa-miR-671-3p | 0.39 | 0.16 | 0.95 |
| hsa-miR-342-5p | 2.51 | 1.3 | 4.85 |
| hsa-miR-150-3p | 2.3 | 1.11 | 4.77 |
| hsa-miR-134-5p | 0.44 | 0.19 | 1 |
| hsa-miR-29b-3p | 2.26 | 1.02 | 4.99 |
| hsa-miR-369-3p | 0.45 | 0.21 | 1 |
| hsa-miR-130b-5p | 0.46 | 0.23 | 0.93 |
| hsa-miR-409-3p | 0.55 | 0.31 | 0.99 |
| 95% CI for OR | |||
|---|---|---|---|
| miRNA | OR | Lower Bound | Upper Bound |
| hsa-miR-3942-5p | 73.05 | 2.23 | 2390.05 |
| hsa-miR-574-5p | 7.2 | 1.16 | 44.78 |
| hsa-miR-151a-5p | 0.14 | 0.02 | 0.86 |
| hsa-miR-125b-2-3p | 5.46 | 1.38 | 21.52 |
| hsa-miR-6852-5p | 0.19 | 0.05 | 0.79 |
| hsa-miR-6842-3p | 0.22 | 0.05 | 0.86 |
| hsa-miR-95-3p | 3.83 | 1.44 | 10.14 |
| hsa-miR-342-3p | 3.72 | 1.31 | 10.62 |
| hsa-miR-6509-5p | 3.66 | 1.02 | 13.15 |
| hsa-miR-1294 | 3.47 | 1.08 | 11.11 |
| hsa-miR-141-3p | 3 | 1.1 | 8.18 |
| hsa-miR-370-3p | 0.33 | 0.13 | 0.83 |
| hsa-miR-331-5p | 2.99 | 1.02 | 8.76 |
| hsa-miR-424-3p | 2.93 | 1.06 | 8.1 |
| hsa-miR-193b-5p | 2.85 | 1.23 | 6.6 |
| hsa-miR-146b-3p | 2.82 | 1.01 | 7.88 |
| hsa-miR-195-5p | 2.71 | 1.09 | 6.76 |
| hsa-miR-122-5p | 2.68 | 1.21 | 5.96 |
| hsa-miR-29a-3p | 2.65 | 1.08 | 6.54 |
| hsa-miR-127-3p | 0.38 | 0.15 | 0.97 |
| hsa-miR-215-5p | 2.62 | 1.11 | 6.17 |
| hsa-miR-30d-3p | 0.42 | 0.18 | 0.98 |
| hsa-miR-136-3p | 0.43 | 0.19 | 0.95 |
| hsa-miR-1224-5p | 2.26 | 1.06 | 4.82 |
| hsa-miR-4732-5p | 2.25 | 1.09 | 4.66 |
| hsa-miR-642a-3p | 2.1 | 1.01 | 4.36 |
| hsa-miR-29c-3p | 2.05 | 1.03 | 4.12 |
| hsa-miR-134-5p | 0.5 | 0.25 | 1 |
| hsa-miR-150-3p | 1.9 | 1.02 | 3.53 |
| hsa-miR-342-5p | 1.89 | 1.09 | 3.29 |
| hsa-miR-130b-5p | 0.53 | 0.3 | 0.95 |
| hsa-miR-339-5p | 0.59 | 0.35 | 1 |
| Name | Hits | p Value | FDR | Function | Reference |
|---|---|---|---|---|---|
| Cell cycle | 41 | 1.44 × 10−8 | 4.8 × 10−7 | Airway smooth muscle (ASM) proliferation. | [30] |
| TGF-beta signaling pathway | 28 | 2.47 × 10−6 | 4.12 × 10−5 | Airway epithelial cells apoptosis, subepithelial fibrosis, airway smooth muscle remodeling, and microvascular changes. | [31,32] |
| ErbB signaling pathway | 27 | 1.69 × 10−5 | 1.88 × 10−4 | Airway hyperreactivity and remodeling. | [33,34] |
| Wnt signaling pathway | 36 | 1.35 × 10−4 | 1.23 × 10−3 | Airway remodeling. | [35,36] |
| Jak-STAT signaling pathway | 27 | 2.03 × 10−4 | 1.45 × 10−3 | Th cell polarization and airway inflammatory response. | [37,38] |
| p53 signaling pathway | 19 | 1.27 × 10−3 | 5.77 × 10−3 | Bronchial smooth muscle (BSM) proliferation and mitochondrial biogenesis. | [39] |
| T cell receptor signaling pathway | 22 | 1.04 × 10−2 | 3.18 × 10−2 | T cell development and immune system. | [40] |
| Toll-like receptor signaling pathway | 21 | 1.81 × 10−2 | 4.55 × 10−2 | Airway inflammation. | [41,42] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Tiwari, A.; Mirzakhani, H.; Wang, A.L.; Kho, A.T.; McGeachie, M.J.; Litonjua, A.A.; Weiss, S.T.; Tantisira, K.G. Circulating MicroRNA: Incident Asthma Prediction and Vitamin D Effect Modification. J. Pers. Med. 2021, 11, 307. https://doi.org/10.3390/jpm11040307
Li J, Tiwari A, Mirzakhani H, Wang AL, Kho AT, McGeachie MJ, Litonjua AA, Weiss ST, Tantisira KG. Circulating MicroRNA: Incident Asthma Prediction and Vitamin D Effect Modification. Journal of Personalized Medicine. 2021; 11(4):307. https://doi.org/10.3390/jpm11040307
Chicago/Turabian StyleLi, Jiang, Anshul Tiwari, Hooman Mirzakhani, Alberta L. Wang, Alvin T. Kho, Michael J. McGeachie, Augusto A. Litonjua, Scott T. Weiss, and Kelan G. Tantisira. 2021. "Circulating MicroRNA: Incident Asthma Prediction and Vitamin D Effect Modification" Journal of Personalized Medicine 11, no. 4: 307. https://doi.org/10.3390/jpm11040307
APA StyleLi, J., Tiwari, A., Mirzakhani, H., Wang, A. L., Kho, A. T., McGeachie, M. J., Litonjua, A. A., Weiss, S. T., & Tantisira, K. G. (2021). Circulating MicroRNA: Incident Asthma Prediction and Vitamin D Effect Modification. Journal of Personalized Medicine, 11(4), 307. https://doi.org/10.3390/jpm11040307








