Generating a Precision Endoxifen Prediction Algorithm to Advance Personalized Tamoxifen Treatment in Patients with Breast Cancer
Abstract
1. Introduction and Objective
2. Tamoxifen Clinical Use, Metabolism and Metabolic Resistance
2.1. Tamoxifen Treatment and Mechanism of Action
2.2. Tamoxifen Metabolism
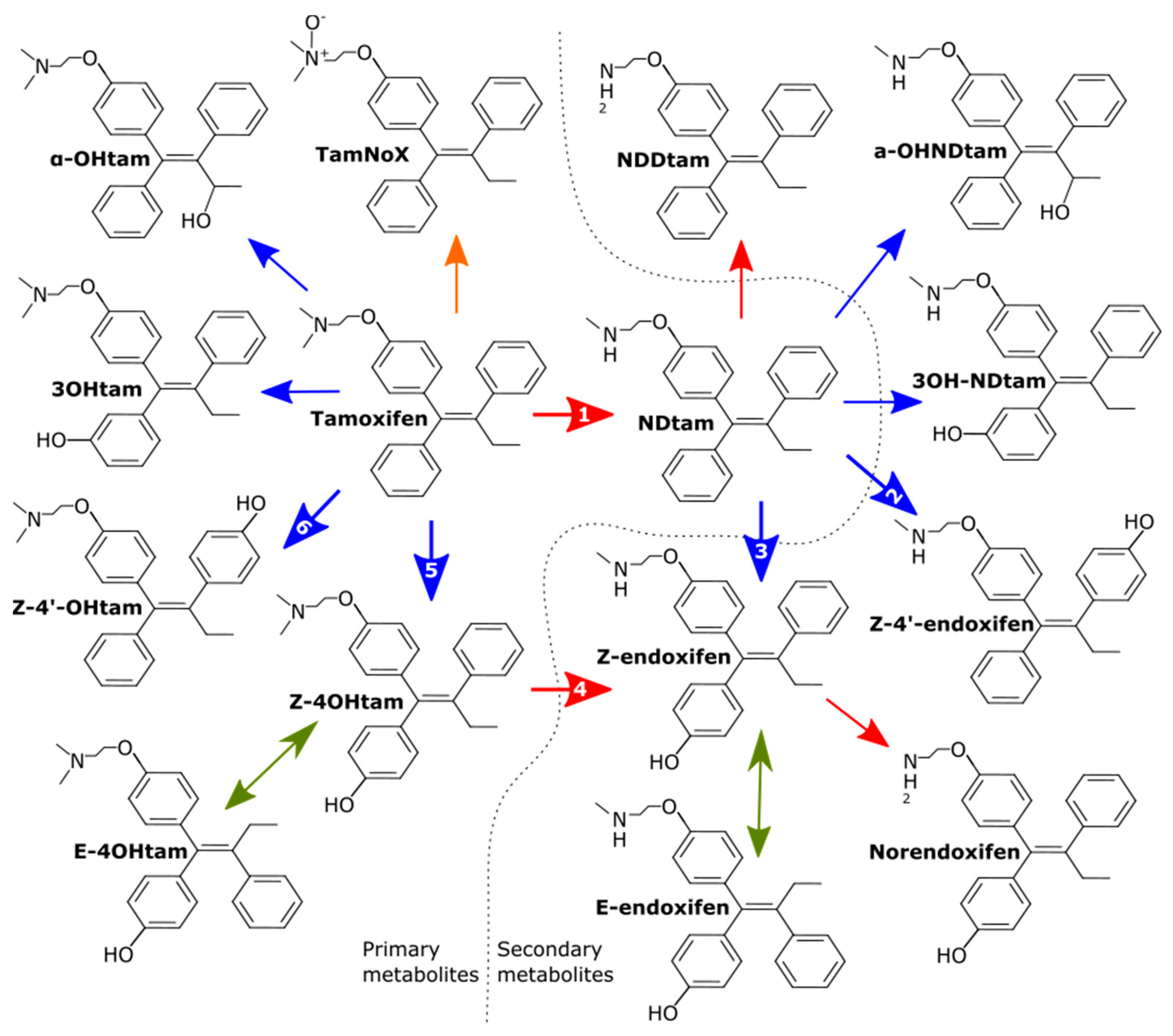
2.3. Active Metabolites: 4OHtam and Endoxifen
| Metabolite | Relative ER Affinity | Relative Abundance |
|---|---|---|
| Tamoxifen | 1 | 1 |
| Tam-NoX | n.d | 0.07 |
| 3OHtam | 10 | <0.01 |
| Norendoxifen | n.d | |
| α-OHtam | n.d | <0.01 |
| NDtam | 0.85 | 1.78–2.01 |
| NNDDtam | 0.46 | 0.29–0.22 |
| 3OHNDtam | n.d | <0.01 |
| 4OHtam Isoforms | ||
| Z-4OHtam | 100 | 0.01–0.02 |
| Z-4′-Ohtam | 10 | 0.02–0.03 |
| E-4OHtam | <0.03 | <0.01 |
| Endoxifen Isoforms | ||
| Z-endoxifen | 100 | 0.07–0.09 |
| Z-4′-endoxifen | 10 | 0.05–0.08 |
| E-endoxifen | <0.03 | <0.01 |
2.4. Associations of Active Metabolite Concentrations with Tamoxifen Efficacy
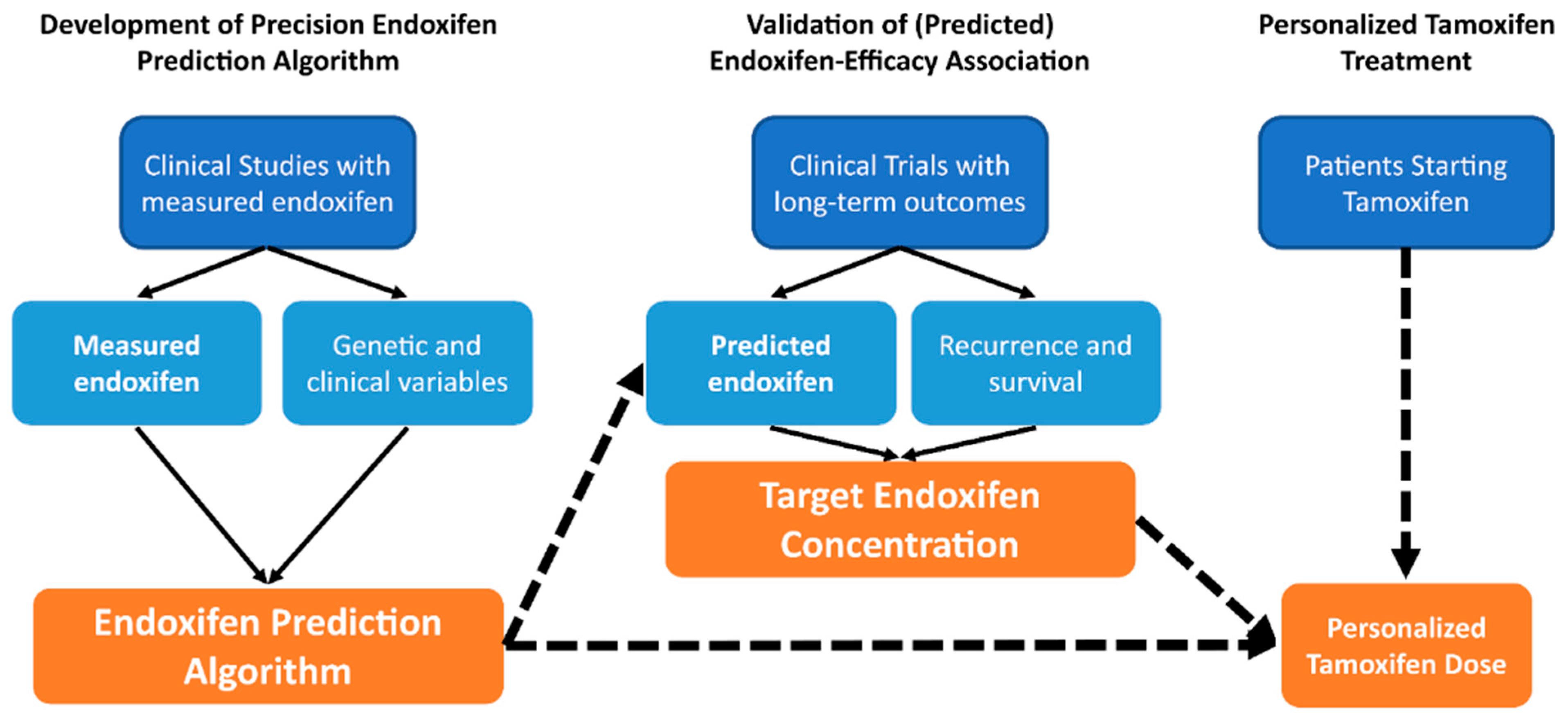
3. Prediction of Active Tamoxifen Metabolite Concentrations
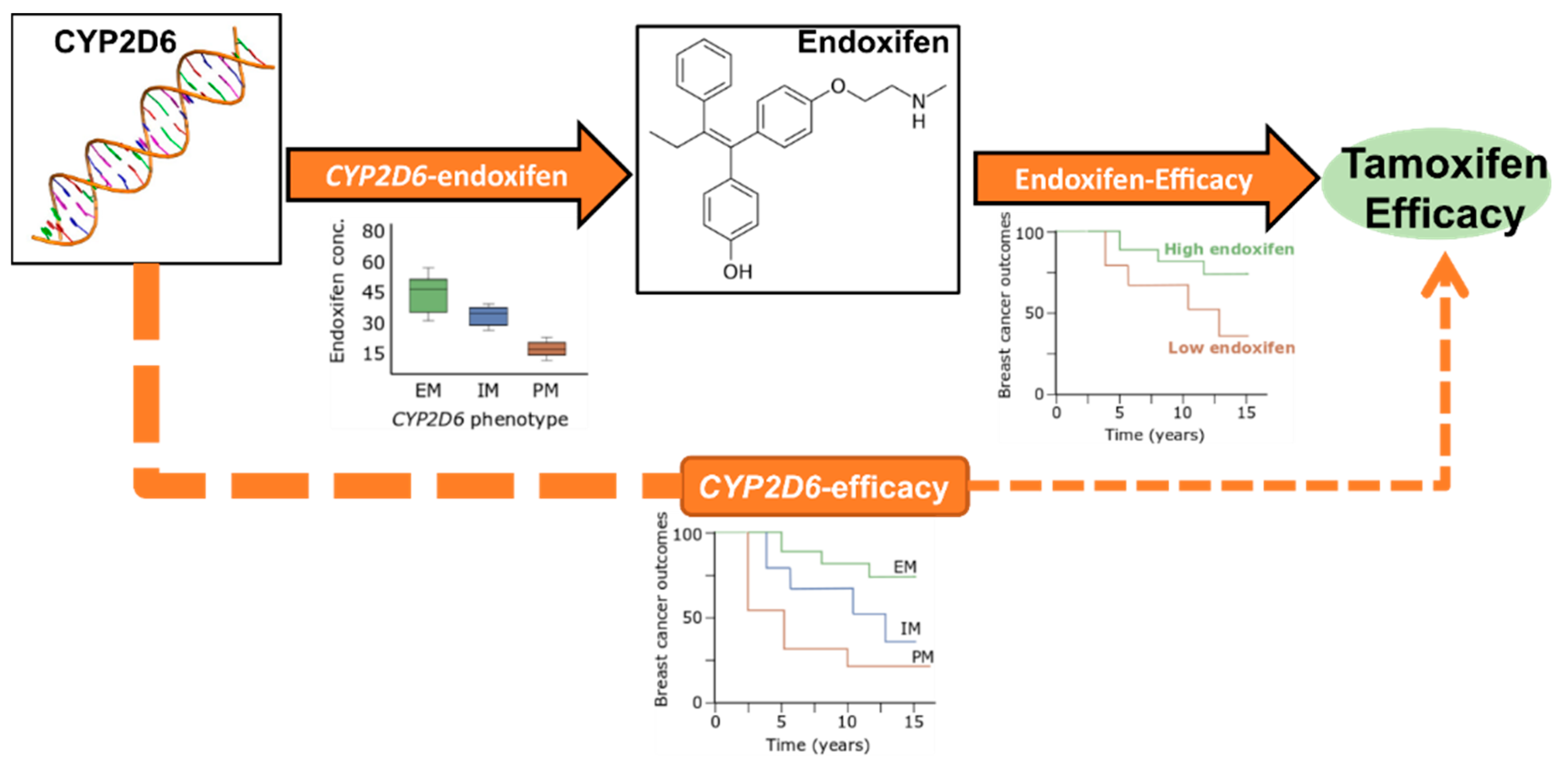
3.1. Genotype-Predicted Endoxifen Concentrations and Tamoxifen Treatment Efficacy
3.2. Effect of CYP2D6 on Endoxifen Formation
3.2.1. CYP2D6 Genetic Variation and Activity Phenotype Translation
3.2.2. Effect of CYP2D6 Genetic Variation on Endoxifen Concentrations
3.2.3. CYP2D6 Genotyping Comprehensiveness
3.2.4. CYP2D6 Activity Phenotype Scoring
3.2.5. Racial Differences in CYP2D6 Allele Frequencies
3.3. Concomitant Use of CYP2D6 Inhibitors
3.4. Tamoxifen Adherence
3.5. Endoxifen Measurement
3.6. Other Clinical Variables Associated with Endoxifen Concentrations
3.7. Effect of CYP2D6 Genetic Variation on Other Active Metabolites
4. Contribution of Other Enzymes to Tamoxifen Metabolite Concentrations
4.1. CYP2Cs
4.2. CYP3As
4.3. SULTs
4.4. UGTs
| Gene | Variants | Metabolites Measured | Reported Association | Ref. |
|---|---|---|---|---|
| CYP2C9 | *2 and *3 | Tam, 4OHtam, endoxifen, glucuronides, E and Z | Lower 4OHtam and endoxifen | [28] |
| *2 and *3 | Tam, NDtam, 4OHtam, endoxifen, N,N-DDtam, norendoxifen | Lower 4OHtam/tam ratio | [5] | |
| *1–*11 | Endoxifen | Lower endoxifen and end/4OHtam ratio | [73] | |
| *2 and *3 | Endoxifen | Lower endoxifen | [72] | |
| CYP2C19 | *2 | Tamoxifen, NDtam, tamNoX, Z-4′-OH-Tam, Z-4OH-tam, Z-endoxifen, Z-4′-endoxifen | Higher Tam/4OHtam ratio | [70] |
| *2, *3, *17 | Endoxifen | Higher 4OHtam | [73] | |
| *2, *3, *17 | Tam, NDtam, 4OHtam, endoxifen, N,N-DDtam, norendoxifen | Lower norendoxifen/N,N-DD-tam ratio | [5] | |
| *2, *3, *17 | Tam, 4OHtam, NDtam | Higher 4OHtam/tam ratio | [99] | |
| CYP3A4 | *22 | Endoxifen | Higher endoxifen | [67] |
| *22 | Tamoxifen, NDtam, tamNoX, Z-4′-OHtam, Z-4-OH-tam, Z-endoxifen, Z-4′-endoxifen | Higher endoxifen | [70] | |
| SULT1A1 | rs6839, rs1042157 | Tamoxifen, endoxifen, 4OHtam and NDtam | Higher endoxifen and 4OHtam | [119] |
| SULT1A2 | *2 and *3 | Tam, 4OHtam, NDtam, endoxifen, TamNoX | Higher endoxifen and 4OH-tam | [82] |
| UGT1A4 | Leu48Val | Tam, 4OHtam, endoxifen, glucuronides, E and Z | Lower tam/Tam-N-glucoronide ratio | [28] |
| Leu48Val | Glucuronidated metabolites of endoxifen and 4OHtam | Lower 4OHtam-N-Gluc and endoxifen-gluc | [125] | |
| Leu48Val | Tamoxifen, E and Z-endoxifen, E- and Z-4-OHtam and the corresponding glucuronides | Higher Tam-N-gluc | [123] | |
| UGT2B7 | *2 (His268Tyr) | Tam and endoxifen | Higher endoxifen | [81] |
| *2 (His268Tyr) | Glucuronidated metabolites of endoxifen and 4OHtam | Higher Tam, 4OHtam, NDtam. Lower 4OHtam-O-gluc and 4-OH-Tam-N-gluc | [125] | |
| UGT2B15 | Lys523Thr | Glucuronidated metabolites of endoxifen and 4OHtam | Higher 4-OH-Tam-O-gluc and endoxifen-gluc | [125] |
| UGT2B17 | Deletion | Glucuronidated metabolites of endoxifen and 4OHtam | Higher 4OHtam-N-gluc | [125] |
5. Conclusions and Directions for Future Research
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Early Breast Cancer Trialists’ Collaborative Group (EBCTCG). Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: An overview of the randomised trials. Lancet 2005, 365, 1687–1717. [Google Scholar] [CrossRef]
- Stearns, V.; Johnson, M.D.; Rae, J.M.; Morocho, A.; Novielli, A.; Bhargava, P.; Hayes, D.F.; Desta, Z.; Flockhart, D.A. Active Tamoxifen Metabolite Plasma Concentrations After Coadministration of Tamoxifen and the Selective Serotonin Reuptake Inhibitor Paroxetine. J. Natl. Cancer Inst. 2003, 95, 1758–1764. [Google Scholar] [CrossRef]
- Jin, Y.; Desta, Z.; Stearns, V.; Ward, B.; Ho, H.; Lee, K.H.; Skaar, T.; Storniolo, A.M.; Li, L.; Araba, A.; et al. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J. Natl. Cancer Inst. 2005, 97, 30–39. [Google Scholar] [CrossRef]
- Madlensky, L.; Natarajan, L.; Tchu, S.; Pu, M.; Mortimer, J.; Flatt, S.W.; Nikoloff, D.M.; Hillman, G.; Fontecha, M.R.; Lawrence, H.J.; et al. Tamoxifen Metabolite Concentrations, CYP2D6 Genotype, and Breast Cancer Outcomes. Clin. Pharmacol. Ther. 2011, 89, 718–725. [Google Scholar] [CrossRef]
- Saladores, P.; Murdter, T.; Eccles, D.; Chowbay, B.; Zgheib, N.K.; Winter, S.; Ganchev, B.; Eccles, B.; Gerty, S.; Tfayli, A.; et al. Tamoxifen metabolism predicts drug concentrations and outcome in premenopausal patients with early breast cancer. Pharm. J. 2015, 15, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Helland, T.; Naume, B.; Hustad, S.; Bifulco, E.; Kvaløy, J.T.; Saetersdal, A.B.; Synnestvedt, M.; Lende, T.H.; Gilje, B.; Mjaaland, I.; et al. Low Z-4OHtam concentrations are associated with adverse clinical outcome among early stage premenopausal breast cancer patients treated with adjuvant tamoxifen. Mol. Oncol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Drögemöller, B.I.; Wright, G.E.B.; Shih, J.; Monzon, J.G.; Gelmon, K.A.; Ross, C.J.D.; Amstutz, U.; Carleton, B.C. CYP2D6 as a treatment decision aid for ER-positive non-metastatic breast cancer patients: A systematic review with accompanying clinical practice guidelines. Breast Cancer Res. Treat. 2019, 173, 521–532. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Spitman, A.B.; Swen, J.J.; Dezentje, V.O.; Moes, D.; Gelderblom, H.; Guchelaar, H.J. Clinical pharmacokinetics and pharmacogenetics of tamoxifen and endoxifen. Expert Rev. Clin. Pharm. 2019, 12, 523–536. [Google Scholar] [CrossRef] [PubMed]
- Davies, C.; Pan, H.; Godwin, J.; Gray, R.; Arriagada, R.; Raina, V.; Abraham, M.; Alencar, V.H.M.; Badran, A.; Bonfill, X. Long-term effects of continuing adjuvant tamoxifen to 10 years versus stopping at 5 years after diagnosis of oestrogen receptor-positive breast cancer: ATLAS, a randomised trial. Lancet 2013, 381, 805–816. [Google Scholar] [CrossRef]
- Cuzick, J.; Sestak, I.; Bonanni, B.; Costantino, J.P.; Cummings, S.; DeCensi, A.; Dowsett, M.; Forbes, J.F.; Ford, L.; LaCroix, A.Z.; et al. Selective oestrogen receptor modulators in prevention of breast cancer: An updated meta-analysis of individual participant data. Lancet 2013, 381, 1827–1834. [Google Scholar] [CrossRef]
- DeCensi, A.; Puntoni, M.; Guerrieri-Gonzaga, A.; Caviglia, S.; Avino, F.; Cortesi, L.; Taverniti, C.; Pacquola, M.G.; Falcini, F.; Gulisano, M.; et al. Randomized Placebo Controlled Trial of Low-Dose Tamoxifen to Prevent Local and Contralateral Recurrence in Breast Intraepithelial Neoplasia. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2019, 37, 1629–1637. [Google Scholar] [CrossRef]
- Manavathi, B.; Dey, O.; Gajulapalli, V.N.R.; Bhatia, R.S.; Bugide, S.; Kumar, R. Derailed Estrogen Signaling and Breast Cancer: An Authentic Couple. Endocr. Rev. 2013, 34, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Shiau, A.K.; Barstad, D.; Loria, P.M.; Cheng, L.; Kushner, P.J.; Agard, D.A.; Greene, G.L. The Structural Basis of Estrogen Receptor/Coactivator Recognition and the Antagonism of This Interaction by Tamoxifen. Cell 1998, 95, 927–937. [Google Scholar] [CrossRef]
- Brzozowski, A.M.; Pike, A.C.; Dauter, Z.; Hubbard, R.E.; Bonn, T.; Engström, O.; Öhman, L.; Greene, G.L.; Gustafsson, J.-Å.; Carlquist, M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature 1997, 389, 753. [Google Scholar] [CrossRef] [PubMed]
- Jordan, V.C. New insights into the metabolism of tamoxifen and its role in the treatment and prevention of breast cancer. Steroids 2007, 72, 829–842. [Google Scholar] [CrossRef]
- Patterson, J.S.; Settatree, R.S.; Adam, H.K.; Kemp, J.V. Serum concentrations of tamoxifen and major metabolite during long-term nolvadex therapy, correlated with clinical response. Eur. J. Cancer 1980, 1 (Suppl. 1), 89–92. [Google Scholar] [PubMed]
- Desta, Z.; Ward, B.A.; Soukhova, N.V.; Flockhart, D.A. Comprehensive Evaluation of Tamoxifen Sequential Biotransformation by the Human Cytochrome P450 System in Vitro: Prominent Roles for CYP3A and CYP2D6. J. Pharmacol. Exp. Ther. 2004, 310, 1062–1075. [Google Scholar] [CrossRef] [PubMed]
- Klein, D.J.; Thorn, C.F.; Desta, Z.; Flockhart, D.A.; Altman, R.B.; Klein, T.E. PharmGKB summary: Tamoxifen pathway, pharmacokinetics. Pharm. Genom. 2013, 23, 643–647. [Google Scholar] [CrossRef] [PubMed]
- Borges, S.; Desta, Z.; Li, L.; Skaar, T.C.; Ward, B.A.; Nguyen, A.; Jin, Y.; Storniolo, A.M.; Nikoloff, D.M.; Wu, L.; et al. Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen metabolism: Implication for optimization of breast cancer treatment. Clin. Pharmacol. Ther. 2006, 80, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Brauch, H.; Jordan, V.C. Targeting of tamoxifen to enhance antitumour action for the treatment and prevention of breast cancer: The ‘personalised’ approach? Eur. J. Cancer 2009, 45, 2274–2283. [Google Scholar] [CrossRef]
- Poon, G.K.; Chui, Y.C.; McCague, R.; Lłnning, P.E.; Feng, R.; Rowlands, M.G.; Jarman, M. Analysis of phase I and phase II metabolites of tamoxifen in breast cancer patients. Drug Metab. Dispos. 1993, 21, 1119–1124. [Google Scholar]
- Lu, W.J.; Xu, C.; Pei, Z.; Mayhoub, A.S.; Cushman, M.; Flockhart, D.A. The tamoxifen metabolite norendoxifen is a potent and selective inhibitor of aromatase (CYP19) and a potential lead compound for novel therapeutic agents. Breast Cancer Res. Treat. 2012, 133, 99–109. [Google Scholar] [CrossRef]
- Lien, E.A.; Solheim, E.; Kvinnsland, S.; Ueland, P.M. Identification of 4-Hydroxy-N-desmethyltamoxifen as a Metabolite of Tamoxifen in Human Bile. Cancer Res. 1988, 48, 2304. [Google Scholar] [PubMed]
- Falany, C.N. Enzymology of human cytosolic sulfotransferases. FASEB J. 1997, 11, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, T.; Ogura, K.; Nakano, H.; Ohnuma, T.; Kaku, T.; Hiratsuka, A.; Muro, K.; Watabe, T. Reverse geometrical selectivity in glucuronidation and sulfation of cis- and trans-4-hydroxytamoxifens by human liver UDP-glucuronosyltransferases and sulfotransferases. Biochem. Pharmacol. 2002, 63, 1817–1830. [Google Scholar] [CrossRef]
- Fromson, J.M.; Pearson, S.; Bramah, S. The metabolism of tamoxifen (I.C.I. 46,474). I: In laboratory animals. Xenobiotica 1973, 3, 693–709. [Google Scholar] [CrossRef] [PubMed]
- Lien, E.A.; Solheim, E.; Lea, O.A.; Lundgren, S.; Kvinnsland, S.; Ueland, P.M. Distribution of 4-hydroxy-N-desmethyltamoxifen and other tamoxifen metabolites in human biological fluids during tamoxifen treatment. Cancer Res. 1989, 49, 2175–2183. [Google Scholar] [PubMed]
- Mürdter, T.E.; Schroth, W.; Bacchus-Gerybadze, L.; Winter, S.; Heinkele, G.; Simon, W.; Fasching, P.A.; Fehm, T.; The German, T.; Group, A.I.C.; et al. Activity Levels of Tamoxifen Metabolites at the Estrogen Receptor and the Impact of Genetic Polymorphisms of Phase I and II Enzymes on Their Concentration Levels in Plasma. Clin. Pharmacol. Ther. 2011, 89, 708–717. [Google Scholar] [CrossRef]
- Wakeling, A.E.; Slater, S.R. Estrogen-receptor binding and biologic activity of tamoxifen and its metabolites. Cancer Treat. Rep. 1980, 64, 741–744. [Google Scholar]
- Johnson, M.D.; Zuo, H.; Lee, K.H.; Trebley, J.P.; Rae, J.M.; Weatherman, R.V.; Desta, Z.; Flockhart, D.A.; Skaar, T.C. Pharmacological characterization of 4-hydroxy-N-desmethyl tamoxifen, a novel active metabolite of tamoxifen. Breast Cancer Res. Treat. 2004, 85, 151–159. [Google Scholar] [CrossRef]
- Katzenellenbogen, B.S.; Norman, M.J.; Eckert, R.L.; Peltz, S.W.; Mangel, W.F. Bioactivities, estrogen receptor interactions, and plasminogen activator-inducing activities of tamoxifen and hydroxy-tamoxifen isomers in MCF-7 human breast cancer cells. Cancer Res. 1984, 44, 112–119. [Google Scholar]
- Barginear, M.F.; Jaremko, M.; Peter, I.; Yu, C.; Kasai, Y.; Kemeny, M.; Raptis, G.; Desnick, R.J. Increasing Tamoxifen Dose in Breast Cancer Patients Based on CYP2D6 Genotypes and Endoxifen Levels: Effect on Active Metabolite Isomers and the Antiestrogenic Activity Score. Clin. Pharmacol. Ther. 2011, 90, 605–611. [Google Scholar] [CrossRef] [PubMed]
- Murphy, C.S.; Parker, C.J.; McCague, R.; Jordan, V.C. Structure-activity relationships of nonisomerizable derivatives of tamoxifen: Importance of hydroxyl group and side chain positioning for biological activity. Mol. Pharm. 1991, 39, 421–428. [Google Scholar]
- Lim, Y.C.; Desta, Z.; Flockhart, D.A.; Skaar, T.C. Endoxifen (4-hydroxy-N-desmethyl-tamoxifen) has anti-estrogenic effects in breast cancer cells with potency similar to 4-hydroxy-tamoxifen. Cancer Chemother. Pharmacol. 2005, 55, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Hawse, J.R.; Subramaniam, M.; Goetz, M.P.; Ingle, J.N.; Spelsberg, T.C. The Tamoxifen Metabolite, Endoxifen, Is a Potent Antiestrogen that Targets Estrogen Receptor α for Degradation in Breast Cancer Cells. Cancer Res. 2009, 69, 1722–1727. [Google Scholar] [CrossRef]
- Gong, I.Y.; Teft, W.A.; Ly, J.; Chen, Y.-H.; Alicke, B.; Kim, R.B.; Choo, E.F. Determination of clinically therapeutic endoxifen concentrations based on efficacy from human MCF7 breast cancer xenografts. Breast Cancer Res. Treat. 2013, 139, 61–69. [Google Scholar] [CrossRef] [PubMed]
- De Vries Schultink, A.H.M.; Alexi, X.; van Werkhoven, E.; Madlensky, L.; Natarajan, L.; Flatt, S.W.; Zwart, W.; Linn, S.C.; Parker, B.A.; Wu, A.H.B.; et al. An Antiestrogenic Activity Score for tamoxifen and its metabolites is associated with breast cancer outcome. Breast Cancer Res. Treat. 2017, 161, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Löser, R.; Seibel, K.; Eppenberger, U. No loss of estrogenic or anti-estrogenic activity after demethylation of droloxifene (3-OH-tamoxifen). Int. J. Cancer 1985, 36, 701–703. [Google Scholar] [CrossRef]
- Helland, T.; Hagen, K.B.; Haugstoyl, M.E.; Kvaloy, J.T.; Lunde, S.; Lode, K.; Lind, R.A.; Gripsrud, B.H.; Jonsdottir, K.; Gjerde, J.; et al. Drug monitoring of tamoxifen metabolites predicts vaginal dryness and verifies a low discontinuation rate from the Norwegian Prescription Database. Breast Cancer Res. Treat. 2019, 177, 185–195. [Google Scholar] [CrossRef]
- Helland, T.; Henne, N.; Bifulco, E.; Naume, B.; Borgen, E.; Kristensen, V.N.; Kvaløy, J.T.; Lash, T.L.; Alnæs, G.I.G.; van Schaik, R.H.; et al. Serum concentrations of active tamoxifen metabolites predict long-term survival in adjuvantly treated breast cancer patients. Breast Cancer Res. 2017, 19, 125. [Google Scholar] [CrossRef]
- Love, R.R.; Desta, Z.; Flockhart, D.; Skaar, T.; Ogburn, E.T.; Ramamoorthy, A.; Uy, G.B.; Laudico, A.V.; Van Dinh, N.; Quang, L.H.; et al. CYP2D6 genotypes, endoxifen levels, and disease recurrence in 224 Filipino and Vietnamese women receiving adjuvant tamoxifen for operable breast cancer. SpringerPlus 2013, 2, 52. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Spitman, A.; Dezentjé, V.; Swen, J.; Moes, D.J.A.R.; Böhringer, S.; Batman, E.; van Druten, E.; Smorenburg, C.; van Bochove, A.; Zeillemaker, A.; et al. Tamoxifen Pharmacogenetics and Metabolism: Results From the Prospective CYPTAM Study. J. Clin. Oncol. 2019, 37, 636–646. [Google Scholar] [CrossRef]
- Goetz, M.P.; Suman, V.J.; Nakamura, Y.; Kiyotani, K.; Jordan, V.C.; Ingle, J.N. Tamoxifen Metabolism and Breast Cancer Recurrence: A Question Unanswered by CYPTAM. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2019, 37, 1982–1983. [Google Scholar] [CrossRef] [PubMed]
- Brauch, H.; Schroth, W.; Murdter, T.; Schwab, M. Tamoxifen Pharmacogenetics and Metabolism: The Same Is Not the Same. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2019, 37, 1981–1982. [Google Scholar] [CrossRef] [PubMed]
- Neven, P.; Jongen, L.; Lintermans, A.; Van Asten, K.; Blomme, C.; Lambrechts, D.; Poppe, A.; Wildiers, H.; Dieudonne, A.S.; Brouckaert, O.; et al. Tamoxifen Metabolism and Efficacy in Breast Cancer: A Prospective Multicenter Trial. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2018, 24, 2312–2318. [Google Scholar] [CrossRef]
- Tamura, K.; Imamura, C.K.; Takano, T.; Saji, S.; Yamanaka, T.; Yonemori, K.; Takahashi, M.; Tsurutani, J.; Nishimura, R.; Sato, K.; et al. CYP2D6 Genotype–Guided Tamoxifen Dosing in Hormone Receptor–Positive Metastatic Breast Cancer (TARGET-1): A Randomized, Open-Label, Phase II Study. J. Clin. Oncol. 2020, 38, 558–566. [Google Scholar] [CrossRef] [PubMed]
- Johansson, H.; Gandini, S.; Serrano, D.; Gjerde, J.; Lattanzi, M.; Macis, D.; Guerrieri-Gonzaga, A.; Aristarco, V.; Mellgren, G.; Lien, E.; et al. A pooled analysis of CYP2D6 genotype in breast cancer prevention trials of low-dose tamoxifen. Breast Cancer Res. Treat. 2016, 159, 97–108. [Google Scholar] [CrossRef]
- Engstrøm, M.J.; Opdahl, S.; Hagen, A.I.; Romundstad, P.R.; Akslen, L.A.; Haugen, O.A.; Vatten, L.J.; Bofin, A.M. Molecular subtypes, histopathological grade and survival in a historic cohort of breast cancer patients. Breast Cancer Res. Treat. 2013, 140, 463–473. [Google Scholar] [CrossRef]
- Sanchez-Spitman, A.B.; Moes, D.A.R.; Swen, J.J.; Dezentjé, V.O.; Lambrechts, D.; Neven, P.; Gelderblom, H.; Guchelaar, H.J. Exposure-response analysis of endoxifen serum concentrations in early-breast cancer. Cancer Chemother. Pharmacol. 2020, 85, 1141–1152. [Google Scholar] [CrossRef]
- Ribelles, N.; Perez-Villa, L.; Jerez, J.M.; Pajares, B.; Vicioso, L.; Jimenez, B.; de Luque, V.; Franco, L.; Gallego, E.; Marquez, A.; et al. Pattern of recurrence of early breast cancer is different according to intrinsic subtype and proliferation index. Breast Cancer Res. 2013, 15, R98. [Google Scholar] [CrossRef]
- Robinson, D.R.; Wu, Y.M.; Vats, P.; Su, F.; Lonigro, R.J.; Cao, X.; Kalyana-Sundaram, S.; Wang, R.; Ning, Y.; Hodges, L.; et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat. Genet. 2013, 45, 1446–1451. [Google Scholar] [CrossRef]
- Kudela, E.; Samec, M.; Koklesova, L.; Liskova, A.; Kubatka, P.; Kozubik, E.; Rokos, T.; Pribulova, T.; Gabonova, E.; Smolar, M.; et al. miRNA Expression Profiles in Luminal A Breast Cancer—Implications in Biology, Prognosis, and Prediction of Response to Hormonal Treatment. Int. J. Mol. Sci. 2020, 21, 7691. [Google Scholar] [CrossRef]
- Johnston, S.R.; Saccani-Jotti, G.; Smith, I.E.; Salter, J.; Newby, J.; Coppen, M.; Ebbs, S.R.; Dowsett, M. Changes in estrogen receptor, progesterone receptor, and pS2 expression in tamoxifen-resistant human breast cancer. Cancer Res. 1995, 55, 3331–3338. [Google Scholar] [PubMed]
- Regan, M.M.; Leyland-Jones, B.; Bouzyk, M.; Pagani, O.; Tang, W.; Kammler, R.; Dell’Orto, P.; Biasi, M.O.; Thürlimann, B.; Lyng, M.B.; et al. CYP2D6 Genotype and Tamoxifen Response in Postmenopausal Women with Endocrine-Responsive Breast Cancer: The Breast International Group 1-98 Trial. J. Natl. Cancer Inst. 2012. [Google Scholar] [CrossRef]
- Rae, J.M.; Drury, S.; Hayes, D.F.; Stearns, V.; Thibert, J.N.; Haynes, B.P.; Salter, J.; Sestak, I.; Cuzick, J.; Dowsett, M. CYP2D6 and UGT2B7 genotype and risk of recurrence in tamoxifen-treated breast cancer patients. J. Natl. Cancer Inst. 2012, 104, 452–460. [Google Scholar] [CrossRef]
- Referenced from the NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) for Breast Cancer V.1.2019.© National Comprehensive Cancer Network, Inc. 2019. National Comprehensive Cancer Network®, NCCN®, NCCN Guidelines®, and all other NCCN Content are trademarks owned by the National Comprehensive Cancer Network, Inc. Available online: www.nccn.org (accessed on 27 June 2019).
- Goetz, M.P.; Sangkuhl, K.; Guchelaar, H.-J.; Schwab, M.; Province, M.; Whirl-Carrillo, M.; Symmans, W.F.; McLeod, H.L.; Ratain, M.J.; Zembutsu, H.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and Tamoxifen Therapy. Clin. Pharmacol. Ther. 2018, 103, 770–777. [Google Scholar] [CrossRef] [PubMed]
- Harris, L.; Ismaila, N.; McShane, L.; Andre, F.; Collyar, D.; Gonzalez-Angulo, A.; Hammond, E.; Kuderer, N.; Liu, M.; Mennel, R.; et al. Use of Biomarkers to Guide Decisions on Adjuvant Systemic Therapy for Women With Early-Stage Invasive Breast Cancer: American Society of Clinical Oncology Clinical Practice Guideline. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2016, 34, 1134–1150. [Google Scholar] [CrossRef]
- Schroth, W.; Antoniadou, L.; Fritz, P.; Schwab, M.; Muerdter, T.; Zanger, U.M.; Simon, W.; Eichelbaum, M.; Brauch, H. Breast Cancer Treatment Outcome With Adjuvant Tamoxifen Relative to Patient CYP2D6 and CYP2C19 Genotypes. J. Clin. Oncol. 2007, 25, 5187–5193. [Google Scholar] [CrossRef]
- Wegman, P.; Elingarami, S.; Carstensen, J.; Stal, O.; Nordenskjold, B.; Wingren, S. Genetic variants of CYP3A5, CYP2D6, SULT1A1, UGT2B15 and tamoxifen response in postmenopausal patients with breast cancer. Breast Cancer Res. 2007, 9, R7. [Google Scholar] [CrossRef] [PubMed]
- Andrea, G. Pharmacogene Variation Consortium (PharmVar). Available online: https://www.pharmvar.org/gene/CYP2D6 (accessed on 13 July 2020).
- (PharmVar), P.V.C. Human Cytochrome P450 (CYP) Allele Nomenclature Database. 2020. Available online: www.PharmVar.org (accessed on 15 January 2021).
- Caudle, K.E.; Sangkuhl, K.; Whirl-Carrillo, M.; Swen, J.J.; Haidar, C.E.; Klein, T.E.; Gammal, R.S.; Relling, M.V.; Scott, S.A.; Hertz, D.L.; et al. Standardizing CYP2D6 Genotype to Phenotype Translation: Consensus Recommendations from the Clinical Pharmacogenetics Implementation Consortium and Dutch Pharmacogenetics Working Group. Clin. Transl. Sci. 2020, 13, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Hicks, J.K.; Swen, J.J.; Gaedigk, A. Challenges in CYP2D6 phenotype assignment from genotype data: A critical assessment and call for standardization. Curr. Drug Metab. 2014, 15, 218–232. [Google Scholar] [CrossRef]
- Gjerde, J.; Hauglid, M.; Breilid, H.; Lundgren, S.; Varhaug, J.E.; Kisanga, E.R.; Mellgren, G.; Steen, V.M.; Lien, E.A. Effects of CYP2D6 and SULT1A1 genotypes including SULT1A1 gene copy number on tamoxifen metabolism. Ann. Oncol. 2007, 19, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Hertz, D.L.; McLeod, H.L.; Irvin, W.J. Tamoxifen and CYP2D6: A contradiction of data. Oncologist 2012, 17, 620–630. [Google Scholar] [CrossRef] [PubMed]
- Teft, W.A.; Gong, I.Y.; Dingle, B.; Potvin, K.; Younus, J.; Vandenberg, T.A.; Brackstone, M.; Perera, F.E.; Choi, Y.-H.; Zou, G.; et al. CYP3A4 and seasonal variation in vitamin D status in addition to CYP2D6 contribute to therapeutic endoxifen level during tamoxifen therapy. Breast Cancer Res. Treat. 2013, 139, 95–105. [Google Scholar] [CrossRef]
- Hennig, E.E.; Piatkowska, M.; Karczmarski, J.; Goryca, K.; Brewczynska, E.; Jazwiec, R.; Kluska, A.; Omiotek, R.; Paziewska, A.; Dadlez, M.; et al. Limited predictive value of achieving beneficial plasma (Z)-endoxifen threshold level by CYP2D6 genotyping in tamoxifen-treated Polish women with breast cancer. BMC Cancer 2015, 15, 570. [Google Scholar] [CrossRef]
- Antunes, M.V.; Linden, R.; Santos, T.V.; Wallemacq, P.; Haufroid, V.; Classen, J.F.; Andreolla, H.; Costa, N.; Fontanive, T.O.; Rosa, D.D. Endoxifen levels and its association with CYP2D6 genotype and phenotype: Evaluation of a southern Brazilian population under tamoxifen pharmacotherapy. Ther. Drug Monit. 2012, 34, 422–431. [Google Scholar] [CrossRef]
- Puszkiel, A.; Arellano, C.; Vachoux, C.; Evrard, A.; Le Morvan, V.; Boyer, J.C.; Robert, J.; Delmas, C.; Dalenc, F.; Debled, M.; et al. Factors Affecting Tamoxifen Metabolism in Patients With Breast Cancer: Preliminary Results of the French PHACS Study. Clin. Pharmacol. Ther. 2019, 106, 585–595. [Google Scholar] [CrossRef]
- Antunes, M.V.; de Oliveira, V.; Raymundo, S.; Staudt, D.E.; Gössling, G.; Biazús, J.V.; Cavalheiro, J.A.; Rosa, D.D.; Mathy, G.; Wallemacq, P.; et al. CYP3A4*22 is related to increased plasma levels of 4-hydroxytamoxifen and partially compensates for reduced CYP2D6 activation of tamoxifen. Pharmacogenomics 2015, 16, 601–617. [Google Scholar] [CrossRef] [PubMed]
- Marcath, L.A.; Deal, A.M.; Van Wieren, E.; Danko, W.; Walko, C.M.; Ibrahim, J.G.; Weck, K.E.; Jones, D.R.; Desta, Z.; McLeod, H.L.; et al. Comprehensive assessment of cytochromes P450 and transporter genetics with endoxifen concentration during tamoxifen treatment. Pharm. Genom. 2017, 27, 402–409. [Google Scholar] [CrossRef]
- Powers, J.L.; Buys, S.S.; Fletcher, D.; Melis, R.; Johnson-Davis, K.L.; Lyon, E.; Malmberg, E.M.; McMillin, G.A. Multigene and Drug Interaction Approach for Tamoxifen Metabolite Patterns Reveals Possible Involvement of CYP2C9, CYP2C19, and ABCB1. J. Clin. Pharmacol. 2016, 56, 1570–1581. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.S.; Ju Lee, H.; Seok Lee, K.; Sook Lee, E.; Jang, I.J.; Ro, J. Clinical implications of CYP2D6 genotypes predictive of tamoxifen pharmacokinetics in metastatic breast cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2007, 25, 3837–3845. [Google Scholar] [CrossRef]
- Fox, P.; Balleine, R.L.; Lee, C.; Gao, B.; Balakrishnar, B.; Menzies, A.M.; Yeap, S.H.; Ali, S.S.; Gebski, V.; Provan, P.; et al. Dose Escalation of Tamoxifen in Patients with Low Endoxifen Level: Evidence for Therapeutic Drug Monitoring-The TADE Study. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016, 22, 3164–3171. [Google Scholar] [CrossRef] [PubMed]
- Safgren, S.L.; Suman, V.J.; Kosel, M.L.; Gilbert, J.A.; Buhrow, S.A.; Black, J.L.; Northfelt, D.W.; Modak, A.S.; Rosen, D.; Ingle, J.N.; et al. Evaluation of CYP2D6 enzyme activity using a 13C-dextromethorphan breath test in women receiving adjuvant tamoxifen. Pharm. Genom. 2015, 25, 157–163. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kiyotani, K.; Mushiroda, T.; Imamura, C.K.; Tanigawara, Y.; Hosono, N.; Kubo, M.; Sasa, M.; Nakamura, Y.; Zembutsu, H. Dose-adjustment study of tamoxifen based on CYP2D6 genotypes in Japanese breast cancer patients. Breast Cancer Res. Treat. 2012, 131, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Jr, W.J.I.; Walko, C.M.; Weck, K.E.; Ibrahim, J.G.; Chiu, W.K.; Dees, E.C.; Moore, S.G.; Olajide, O.A.; Graham, M.L.; Canale, S.T.; et al. Genotype-Guided Tamoxifen Dosing Increases Active Metabolite Exposure in Women With Reduced CYP2D6 Metabolism: A Multicenter Study. J. Clin. Oncol. 2011, 29, 3232–3239. [Google Scholar] [CrossRef]
- Ximenez, J.P.B.; de Andrade, J.M.; Marques, M.P.; Coelho, E.B.; Suarez-Kurtz, G.; Lanchote, V.L. Hormonal status affects plasma exposure of tamoxifen and its main metabolites in tamoxifen-treated breast cancer patients. BMC Pharm. Toxicol 2019, 20, 81. [Google Scholar] [CrossRef] [PubMed]
- Woo, H.I.; Lee, S.K.; Kim, J.; Kim, S.W.; Yu, J.; Bae, S.Y.; Lee, J.E.; Nam, S.J.; Lee, S.-Y. Variations in plasma concentrations of tamoxifen metabolites and the effects of genetic polymorphisms on tamoxifen metabolism in Korean patients with breast cancer. Oncotarget 2017, 8, 100296–100311. [Google Scholar] [CrossRef]
- Areepium, N.; Panomvana, D.; Rungwanonchai, P.; Sathaporn, S.; Voravud, N. Effects of CYP2D6 and UGT2B7 polymorphisms on pharmacokinetics of tamoxifen in Thai breast cancer patients. Breast Cancer 2013, 5, 73–78. [Google Scholar] [CrossRef]
- Fernandez-Santander, A.; Gaibar, M.; Novillo, A.; Romero-Lorca, A.; Rubio, M.; Chicharro, L.M.; Tejerina, A.; Bandres, F. Relationship between genotypes Sult1a2 and Cyp2d6 and tamoxifen metabolism in breast cancer patients. PLoS ONE 2013, 8, e70183. [Google Scholar] [CrossRef]
- Thorén, L.; Lindh, J.D.; Ackehed, G.; Kringen, M.K.; Hall, P.; Bergh, J.; Molden, E.; Margolin, S.; Eliasson, E. Impairment of endoxifen formation in tamoxifen-treated premenopausal breast cancer patients carrying reduced-function CYP2D6 alleles. Br. J. Clin. Pharm. 2020. [Google Scholar] [CrossRef]
- Khan, B.A.; Robinson, R.; Fohner, A.E.; Muzquiz, L.I.; Schilling, B.D.; Beans, J.A.; Olnes, M.J.; Trawicki, L.; Frydenlund, H.; Laukes, C.; et al. Cytochrome P450 Genetic Variation Associated with Tamoxifen Biotransformation in American Indian and Alaska Native People. Clin. Transl. Sci. 2018, 11, 312–321. [Google Scholar] [CrossRef] [PubMed]
- Schroth, W.; Hamann, U.; Fasching, P.A.; Dauser, S.; Winter, S.; Eichelbaum, M.; Schwab, M.; Brauch, H. CYP2D6 polymorphisms as predictors of outcome in breast cancer patients treated with tamoxifen: Expanded polymorphism coverage improves risk stratification. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2010, 16, 4468–4477. [Google Scholar] [CrossRef]
- He, Y.; Hoskins, J.M.; McLeod, H.L. Copy number variants in pharmacogenetic genes. Trends Mol. Med. 2011, 17, 244–251. [Google Scholar] [CrossRef]
- Schroth, W.; Winter, S.; Mürdter, T.; Schaeffeler, E.; Eccles, D.; Eccles, B.; Chowbay, B.; Khor, C.C.; Tfayli, A.; Zgheib, N.K.; et al. Improved Prediction of Endoxifen Metabolism by CYP2D6 Genotype in Breast Cancer Patients Treated with Tamoxifen. Front Pharm. 2017, 8, 582. [Google Scholar] [CrossRef] [PubMed]
- Gaedigk, A.; Simon, S.D.; Pearce, R.E.; Bradford, L.D.; Kennedy, M.J.; Leeder, J.S. The CYP2D6 activity score: Translating genotype information into a qualitative measure of phenotype. Clin. Pharmacol. Ther. 2008, 83, 234–242. [Google Scholar] [CrossRef]
- Kiyotani, K.; Mushiroda, T.; Imamura, C.K.; Hosono, N.; Tsunoda, T.; Kubo, M.; Tanigawara, Y.; Flockhart, D.A.; Desta, Z.; Skaar, T.C.; et al. Significant effect of polymorphisms in CYP2D6 and ABCC2 on clinical outcomes of adjuvant tamoxifen therapy for breast cancer patients. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2010, 28, 1287–1293. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.S.; Chen, X.A.; Singh, O.; Yap, Y.S.; Ng, R.C.; Wong, N.S.; Wong, M.; Lee, E.J.; Chowbay, B. Impact of CYP2D6, CYP3A5, CYP2C9 and CYP2C19 polymorphisms on tamoxifen pharmacokinetics in Asian breast cancer patients. Br. J. Clin. Pharm. 2011, 71, 737–750. [Google Scholar] [CrossRef]
- Helland, T.; Baurley, J.; Thomas-Jean, F.; Mellgren, G.; Swenn, J.; Brauch, H.; Gaedigk, A.; Hertz, D. Consensus CYP2D6 translation system improves explanation of tamoxifen bioactivation in analysis of 15 multi-racial cohorts. In Proceedings of the American Society of Clinical Pharmacology and Therapeutics Annual Meeting, Houston, TX, USA, 17–21 March 2020. [Google Scholar]
- Haque, R.; Shi, J.; Schottinger, J.E.; Ahmed, S.A.; Cheetham, T.C.; Chung, J.; Avila, C.; Kleinman, K.; Habel, L.A.; Fletcher, S.W.; et al. Tamoxifen and Antidepressant Drug Interaction in a Cohort of 16,887 Breast Cancer Survivors. J. Natl. Cancer Inst. 2016, 108. [Google Scholar] [CrossRef]
- Pistilli, B.; Paci, A.; Ferreira, A.R.; Meglio, A.D.; Poinsignon, V.; Bardet, A.; Menvielle, G.; Dumas, A.; Pinto, S.; Dauchy, S.; et al. Serum Detection of Nonadherence to Adjuvant Tamoxifen and Breast Cancer Recurrence Risk. J. Clin. Oncol. 2020, 38, 2762–2772. [Google Scholar] [CrossRef]
- Mueller-Schoell, A.; Klopp-Schulze, L.; Michelet, R.; van Dyk, M.; Mürdter, T.E.; Schwab, M.; Joerger, M.; Huisinga, W.; Mikus, G.; Kloft, C. Simulation-Based Assessment of the Impact of Non-Adherence on Endoxifen Target Attainment in Different Tamoxifen Dosing Strategies. Pharmaceuticals 2021, 14, 115. [Google Scholar] [CrossRef]
- Nardin, J.M.; Schroth, W.; Almeida, T.A.; Mürdter, T.; Picolotto, S.; Vendramini, E.C.L.; Hoppe, R.; Kogin, J.P.; Miqueleto, D.; de Moraes, S.D.R.; et al. The Influences of Adherence to Tamoxifen and CYP2D6 Pharmacogenetics on Plasma Concentrations of the Active Metabolite (Z)-Endoxifen in Breast Cancer. Clin. Transl. Sci. 2020, 13, 284–292. [Google Scholar] [CrossRef]
- Jager, N.G.; Rosing, H.; Linn, S.C.; Schellens, J.H.; Beijnen, J.H. Importance of highly selective LC-MS/MS analysis for the accurate quantification of tamoxifen and its metabolites: Focus on endoxifen and 4-hydroxytamoxifen. Breast Cancer Res. Treat. 2012, 133, 793–798. [Google Scholar] [CrossRef][Green Version]
- Mueller-Schoell, A.; Klopp-Schulze, L.; Schroth, W.; Mürdter, T.; Michelet, R.; Brauch, H.; Huisinga, W.; Joerger, M.; Neven, P.; Koolen, S.L.W.; et al. Obesity Alters Endoxifen Plasma Levels in Young Breast Cancer Patients: A Pharmacometric Simulation Approach. Clin. Pharmacol. Ther. 2020, 108, 661–670. [Google Scholar] [CrossRef]
- Lien, E.A.; Søiland, H.; Lundgren, S.; Aas, T.; Steen, V.M.; Mellgren, G.; Gjerde, J. Serum concentrations of tamoxifen and its metabolites increase with age during steady-state treatment. Breast Cancer Res. Treat. 2013, 141, 243–248. [Google Scholar] [CrossRef] [PubMed]
- Gjerde, J.; Geisler, J.; Lundgren, S.; Ekse, D.; Varhaug, J.E.; Mellgren, G.; Steen, V.M.; Lien, E.A. Associations between tamoxifen, estrogens, and FSH serum levels during steady state tamoxifen treatment of postmenopausal women with breast cancer. BMC Cancer 2010, 10, 313. [Google Scholar] [CrossRef] [PubMed]
- Martinez de Dueñas, E.; Ochoa Aranda, E.; Blancas Lopez-Barajas, I.; Ferrer Magdalena, T.; Bandrés Moya, F.; Chicharro García, L.M.; Gómez Capilla, J.A.; Zafra Ceres, M.; de Haro, T.; Romero Llorens, R.; et al. Adjusting the dose of tamoxifen in patients with early breast cancer and CYP2D6 poor metabolizer phenotype. Breast 2014, 23, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Sun, Y.; Yao, L.; Shi, L.; Wu, Y.; Ouyang, T.; Li, J.; Wang, T.; Fan, Z.; Fan, T.; et al. Association between CYP2D6 *10 genotype and survival of breast cancer patients receiving tamoxifen treatment. Ann. Oncol. 2008, 19, 1423–1429. [Google Scholar] [CrossRef]
- Lei, L.; Wang, X.; Wu, X.D.; Wang, Z.; Chen, Z.H.; Zheng, Y.B.; Wang, X.J. Association of CYP2D6*10 (c.100C>T) polymorphisms with clinical outcome of breast cancer after tamoxifen adjuvant endocrine therapy in Chinese population. Am. J. Transl. Res. 2016, 8, 3585–3592. [Google Scholar]
- Paine, M.F.; Hart, H.L.; Ludington, S.S.; Haining, R.L.; Rettie, A.E.; Zeldin, D.C. The human intestinal cytochrome P450 “pie”. Drug Metab. Dispos. 2006, 34, 880–886. [Google Scholar] [CrossRef]
- Boocock, D.J.; Brown, K.; Gibbs, A.H.; Sanchez, E.; Turteltaub, K.W.; White, I.N. Identification of human CYP forms involved in the activation of tamoxifen and irreversible binding to DNA. Carcinogenesis 2002, 23, 1897–1901. [Google Scholar] [CrossRef]
- Coller, J.K.; Krebsfaenger, N.; Klein, K.; Endrizzi, K.; Wolbold, R.; Lang, T.; Nüssler, A.; Neuhaus, P.; Zanger, U.M.; Eichelbaum, M.; et al. The influence of CYP2B6, CYP2C9 and CYP2D6 genotypes on the formation of the potent antioestrogen Z-4-hydroxy-tamoxifen in human liver. Br. J. Clin. Pharm. 2002, 54, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Botton, M.R.; Whirl-Carrillo, M.; Del Tredici, A.L.; Sangkuhl, K.; Cavallari, L.H.; Agúndez, J.A.G.; Duconge, J.; Lee, M.T.M.; Woodahl, E.L.; Claudio-Campos, K.; et al. PharmVar GeneFocus: CYP2C19. Clin. Pharmacol. Ther. 2021, 109, 352–366. [Google Scholar] [CrossRef] [PubMed]
- Aquilante, C.L.; Niemi, M.; Gong, L.; Altman, R.B.; Klein, T.E. PharmGKB summary: Very important pharmacogene information for cytochrome P450, family 2, subfamily C, polypeptide 8. Pharm. Genom. 2013, 23, 721–728. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Liu, D.; Wang, H.; Zhu, J.; Chen, C. Functional characterization of five CYP2C8 variants and prediction of CYP2C8 genotype-dependent effects on in vitro and in vivo drug-drug interactions. Xenobiotica 2010, 40, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Elens, L.; van Gelder, T.; Hesselink, D.A.; Haufroid, V.; van Schaik, R.H. CYP3A4*22: Promising newly identified CYP3A4 variant allele for personalizing pharmacotherapy. Pharmacogenomics 2013, 14, 47–62. [Google Scholar] [CrossRef]
- Wang, D.; Guo, Y.; Wrighton, S.A.; Cooke, G.E.; Sadee, W. Intronic polymorphism in CYP3A4 affects hepatic expression and response to statin drugs. Pharm. J. 2011, 11, 274–286. [Google Scholar] [CrossRef]
- Baxter, S.D.; Teft, W.A.; Choi, Y.H.; Winquist, E.; Kim, R.B. Tamoxifen-associated hot flash severity is inversely correlated with endoxifen concentration and CYP3A4*22. Breast Cancer Res. Treat. 2014, 145, 419–428. [Google Scholar] [CrossRef]
- Sanchez Spitman, A.B.; Moes, D.; Gelderblom, H.; Dezentje, V.O.; Swen, J.J.; Guchelaar, H.J. Effect of CYP3A4*22, CYP3A5*3, and CYP3A combined genotypes on tamoxifen metabolism. Eur. J. Clin. Pharm. 2017, 73, 1589–1598. [Google Scholar] [CrossRef]
- Sensorn, I.; Sirachainan, E.; Chamnanphon, M.; Pasomsub, E.; Trachu, N.; Supavilai, P.; Sukasem, C.; Pinthong, D. Association of CYP3A4/5, ABCB1 and ABCC2 polymorphisms and clinical outcomes of Thai breast cancer patients treated with tamoxifen. Pharmgenomics Pers. Med. 2013, 6, 93–98. [Google Scholar] [CrossRef]
- Apellániz-Ruiz, M.; Inglada-Pérez, L.; Naranjo, M.E.; Sánchez, L.; Mancikova, V.; Currás-Freixes, M.; de Cubas, A.A.; Comino-Méndez, I.; Triki, S.; Rebai, A.; et al. High frequency and founder effect of the CYP3A4*20 loss-of-function allele in the Spanish population classifies CYP3A4 as a polymorphic enzyme. Pharm. J. 2015, 15, 288–292. [Google Scholar] [CrossRef]
- Tucker, A.N.; Tkaczuk, K.A.; Lewis, L.M.; Tomic, D.; Lim, C.K.; Flaws, J.A. Polymorphisms in cytochrome P4503A5 (CYP3A5) may be associated with race and tumor characteristics, but not metabolism and side effects of tamoxifen in breast cancer patients. Cancer Lett. 2005, 217, 61–72. [Google Scholar] [CrossRef]
- Charoenchokthavee, W.; Areepium, N.; Panomvana, D.; Sriuranpong, V. Effects of CYP2D6 and CYP3A5 polymorphisms on tamoxifen and its metabolites in Thai breast cancer patients. Breast Cancer (Dove Med. Press) 2017, 9, 249–256. [Google Scholar] [CrossRef]
- Thota, K.; Prasad, K.; Basaveswara Rao, M.V. Detection of Cytochrome P450 Polymorphisms in Breast Cancer Patients May Impact on Tamoxifen Therapy. Asian Pac. J. Cancer Prev. 2018, 19, 343–350. [Google Scholar] [CrossRef]
- Nowell, S.; Falany, C.N. Pharmacogenetics of human cytosolic sulfotransferases. Oncogene 2006, 25, 1673–1678. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Spitman, A.B.; Dezentjé, V.O.; Swen, J.J.; Moes, D.; Gelderblom, H.; Guchelaar, H.J. Genetic polymorphisms of 3’-untranslated region of SULT1A1 and their impact on tamoxifen metabolism and efficacy. Breast Cancer Res. Treat. 2018, 172, 401–411. [Google Scholar] [CrossRef]
- Nowell, S.; Sweeney, C.; Winters, M.; Stone, A.; Lang, N.P.; Hutchins, L.F.; Kadlubar, F.F.; Ambrosone, C.B. Association between sulfotransferase 1A1 genotype and survival of breast cancer patients receiving tamoxifen therapy. J. Natl. Cancer Inst. 2002, 94, 1635–1640. [Google Scholar] [CrossRef]
- Carlini, E.J.; Raftogianis, R.B.; Wood, T.C.; Jin, F.; Zheng, W.; Rebbeck, T.R.; Weinshilboum, R.M. Sulfation pharmacogenetics: SULT1A1 and SULT1A2 allele frequencies in Caucasian, Chinese and African-American subjects. Pharmacogenetics 2001, 11, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Lazarus, P.; Sun, D. Potential role of UGT pharmacogenetics in cancer treatment and prevention: Focus on tamoxifen and aromatase inhibitors. Drug Metab. Rev. 2010, 42, 182–194. [Google Scholar] [CrossRef]
- Sutiman, N.; Lim, J.S.L.; Muerdter, T.E.; Singh, O.; Cheung, Y.B.; Ng, R.C.H.; Yap, Y.S.; Wong, N.S.; Ang, P.C.S.; Dent, R.; et al. Pharmacogenetics of UGT1A4, UGT2B7 and UGT2B15 and Their Influence on Tamoxifen Disposition in Asian Breast Cancer Patients. Clin. Pharm. 2016, 55, 1239–1250. [Google Scholar] [CrossRef] [PubMed]
- Blevins-Primeau, A.S.; Sun, D.; Chen, G.; Sharma, A.K.; Gallagher, C.J.; Amin, S.; Lazarus, P. Functional significance of UDP-glucuronosyltransferase variants in the metabolism of active tamoxifen metabolites. Cancer Res. 2009, 69, 1892–1900. [Google Scholar] [CrossRef]
- Romero-Lorca, A.; Novillo, A.; Gaibar, M.; Bandres, F.; Fernandez-Santander, A. Impacts of the Glucuronidase Genotypes UGT1A4, UGT2B7, UGT2B15 and UGT2B17 on Tamoxifen Metabolism in Breast Cancer Patients. PLoS ONE 2015, 10, e0132269. [Google Scholar] [CrossRef]
- Klopp-Schulze, L.; Joerger, M.; Wicha, S.G.; ter Heine, R.; Csajka, C.; Parra-Guillen, Z.P.; Kloft, C. Exploiting Pharmacokinetic Models of Tamoxifen and Endoxifen to Identify Factors Causing Subtherapeutic Concentrations in Breast Cancer Patients. Clin. Pharmacokinet. 2018, 57, 229–242. [Google Scholar] [CrossRef]
- Puszkiel, A.; Arellano, C.; Vachoux, C.; Evrard, A.; Le Morvan, V.; Boyer, J.C.; Robert, J.; Delmas, C.; Dalenc, F.; Debled, M.; et al. Model-Based Quantification of Impact of Genetic Polymorphisms and Co-Medications on Pharmacokinetics of Tamoxifen and Six Metabolites in Breast Cancer. Clin. Pharmacol. Ther. 2020. [Google Scholar] [CrossRef] [PubMed]
- Klopp-Schulze, L.; Mueller-Schoell, A.; Neven, P.; Koolen, S.L.W.; Mathijssen, R.H.J.; Joerger, M.; Kloft, C. Integrated Data Analysis of Six Clinical Studies Points Toward Model-Informed Precision Dosing of Tamoxifen. Front. Pharmacol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Van der Lee, M.; Allard, W.G.; Vossen, R.H.A.M.; Baak-Pablo, R.F.; Menafra, R.; Deiman, B.A.L.M.; Deenen, M.J.; Neven, P.; Johansson, I.; Gastaldello, S.; et al. A unifying model to predict variable drug response for personalised medicine. bioRxiv 2003. [Google Scholar] [CrossRef]
| Allelic Activity Scoring | Metabolic Phenotype Scoring | |||
|---|---|---|---|---|
| Activity | Genotype Examples | Activity Score | Activity Score Sum | Phenotype |
| High | *1XN, *2XN, *35XN | 2 | >2.25 | UM |
| Normal | *1, *2, and *35 | 1 | 1.25–2 | NM |
| Reduced | *9, *17, *29, *36, *41 | 0.5 | 0.25–1 | IM |
| Slow | *10 | 0.25 | 0.25–1 | IM |
| None | *3, *4, *5, *6, *7, *8 | 0 | 0 | PM |
| Patients | CYP2D6 Genotype and Phenotype Scoring | Endox Assay | % Endox or Metabolic Ratio (MR) Explained r2, (Adjusted. r2) | Study | |||||
|---|---|---|---|---|---|---|---|---|---|
| n | Race | Exclusion Criteria | CYP2D6 * Alleles | Scoring | Z/Total | Endox | MR | Covariates | Ref |
| 236 | Caucasian | Tam <10% mean | 3, 4, 5, 6, 7, 9, 10, 41, CNV | DT | Sqrt-z | 38.6 | 68 | None | [28] |
| 583 | Mixed | Tam<150 nM, inhibitors | 3, 4, 5, 6, 9, 10, 14, 15, 17, 41, CNV | AS | Log-z | 33–38 | 53 | Age, BMI, non-2D6 genetics | [5] |
| 196 | Caucasian | Unspecified tam cut-off | 3, 4, 5, 9, 10, 41, CNV | DTs | Log-z | 30.1 (46) | - | Age, BMI, genetics, inhibitors, season | [67] |
| 279 | Caucasian | Tam <10% mean | 2, 3, 4, 5, 6, 7, 9, 10, 17, 41, CVN | DTs | Z | 27 | 51 | - | [68] |
| 97 | Caucasian | None | 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 15, 17, 29, 35, 41, CNV | MG # | Total | (26) | (38) | Age, ethnicity | [69] |
| 80 | Caucasian | None | 4, 6, 8 | DTs | Total | 23 | - | None | [3] |
| 1370 | Caucasian | None | CYP450 AmpliChip, CNV | MG | Z | 18 (46) | - | Age, BMI, race, tam conc | [4] |
| 730 | European | Tam<LLQ | 4, 9, 10, 7, 6, 17, 41, CNV | DTs | Z | 16.8 (19.4) | - | Inhibitors, non-2D6 genetics | [70] |
| 178 | Caucasian | Inhibitors, non-adherence | 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 15, 17, 29, 35, 41, CNV | MG # | Z | (16) | - | CYP3A4, demographics | [71] |
| 302 | Caucasian | Inhibitors, non-adherence | CYP450 AmpliChip, CNV | DTs | Log-z | 15.4 (23) | - | Weight, season, CYP2C9 | [72] |
| 224 | Asian | Unspecified tam cut off, inhibitors | 2, 3, 4, 5, 6, 10, 41 | MG | Total | 10 | - | None | [41] |
| 116 | Caucasian | None | 2, 2A, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 14, 17, 29, 35, 41, CNV | AS | Total | - | 29.6 (33.7) | Inhibitors | [73] |
| 83 | Caucasian | Endoxifen < LLQ | 2, 2A, 3, 4, 5, 6, 7, 8, 9, 10, 12, 14, 17, 29, 41, CNV | MG | Z | - | - | None | [40] |
| 202 | Asian | Inhibitors or inducers | 2, 5, 10, CNV | DTs * | Z | - | - | None | [74] |
| 122 | Caucasian | Tam <150 nM | 2, 3, 4, 5, 6, 7, 8, 14, 9, 10, 18, 41, 14B, CNV | MG | Z | None | [75] | ||
| 77 | Caucasian | Inhibitors, non-adherence | 2, 2A, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 17, 41, CNV | AS | Z | - | - | None | [76] |
| 98 | Asian | SSRIs | 5, 10, 21, 41 | DTs | Z | - | - | None | [77] |
| 117 | Caucasian | Inhibitors | CYP450 AmpliChip, CNV | MG | Total | - | - | None | [78] |
| 119 | Mixed | None | 2, 2A, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 17, CNV | AS | Log-z | - | - | Inhibitors | [32] |
| 35 | Mixed | Inhibitors | 2, 3, 4, 9, 10, 17, 29, 35, 41, | AS | Total | - | - | Race, menopausal status | [79] |
| 667 | Caucasian | None | CYP450 AmpliChip, CNV | MG * | Z | - | - | None | [42] |
| 80 | Asian | None | 2, 5, 6, 10, 39, 41, CNV | AS | Total | - | - | Tam conc., BMI, non-2D6 genetics | [80] |
| 59 | Asian | Inhibitors, non-adherence | 10 | DTs | Total | - | - | None | [81] |
| 152 | Caucasian | Unspecified tam cut-off | 2, 3, 4, 5, 6, CNV | MG * | Total | - | - | Age | [65] |
| 120 | Caucasian | None | CYP450 AmpliChip, CNV | DTs/MG | Total | - | - | Inhibitors | [82] |
| 114 | Caucasian | None | 3, 4, 5, 6, 9, 41 | AS/MG | Z | - | - | None | [83] |
| 183 | Asian | Inhibitors | 2, 4, 6, 10, 14, 18, 21, 36, 41, 44, CNV | DT | Z | - | - | None | [46] |
| 42 | Native American | None | 2, 3, 4, 5, 9, 10, 28, 33, 35, 41 | AS/DTs | Total | - | - | Age, site, non-2D6 genetics | [84] |
| 158 | Caucasian | None | CYP450 AmpliChip, CNV | DTs | Total | - | - | Inhibitors | [19] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Helland, T.; Alsomairy, S.; Lin, C.; Søiland, H.; Mellgren, G.; Hertz, D.L. Generating a Precision Endoxifen Prediction Algorithm to Advance Personalized Tamoxifen Treatment in Patients with Breast Cancer. J. Pers. Med. 2021, 11, 201. https://doi.org/10.3390/jpm11030201
Helland T, Alsomairy S, Lin C, Søiland H, Mellgren G, Hertz DL. Generating a Precision Endoxifen Prediction Algorithm to Advance Personalized Tamoxifen Treatment in Patients with Breast Cancer. Journal of Personalized Medicine. 2021; 11(3):201. https://doi.org/10.3390/jpm11030201
Chicago/Turabian StyleHelland, Thomas, Sarah Alsomairy, Chenchia Lin, Håvard Søiland, Gunnar Mellgren, and Daniel Louis Hertz. 2021. "Generating a Precision Endoxifen Prediction Algorithm to Advance Personalized Tamoxifen Treatment in Patients with Breast Cancer" Journal of Personalized Medicine 11, no. 3: 201. https://doi.org/10.3390/jpm11030201
APA StyleHelland, T., Alsomairy, S., Lin, C., Søiland, H., Mellgren, G., & Hertz, D. L. (2021). Generating a Precision Endoxifen Prediction Algorithm to Advance Personalized Tamoxifen Treatment in Patients with Breast Cancer. Journal of Personalized Medicine, 11(3), 201. https://doi.org/10.3390/jpm11030201






