The Application Value of Lipoprotein Particle Numbers in the Diagnosis of HBV-Related Hepatocellular Carcinoma with BCLC Stage 0-A
Abstract
:1. Introduction
2. Results
2.1. Clinical Characteristics
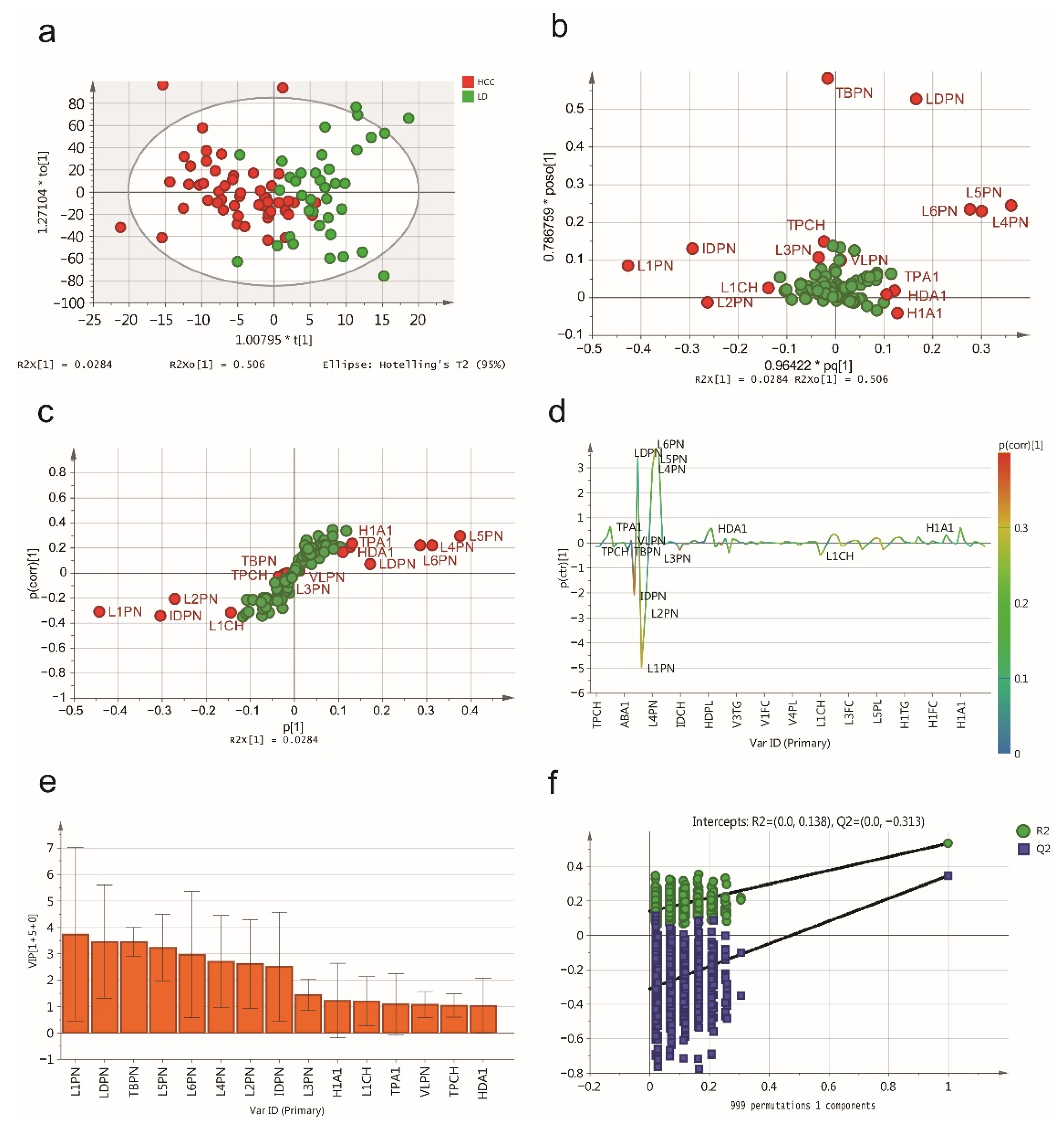
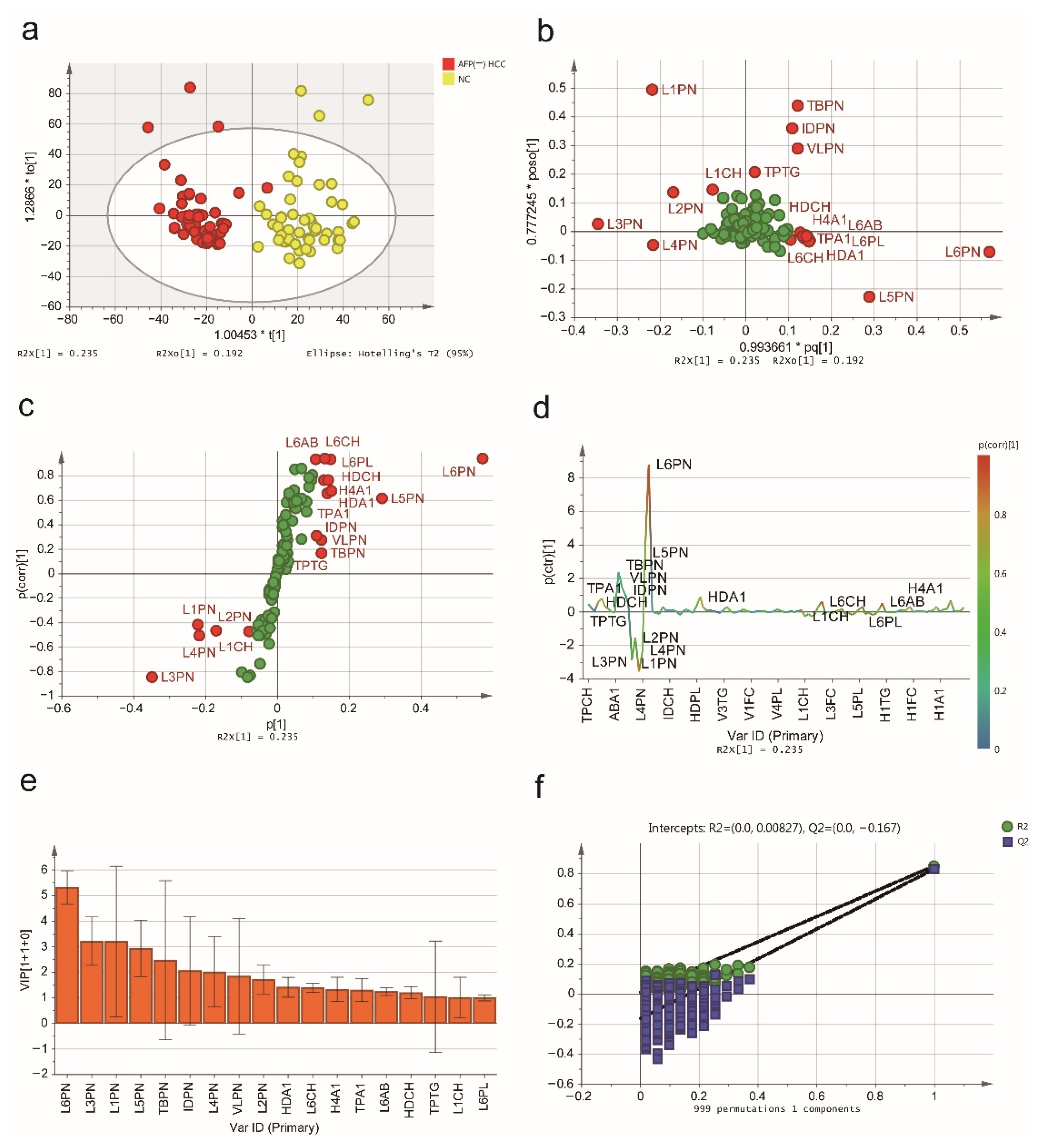
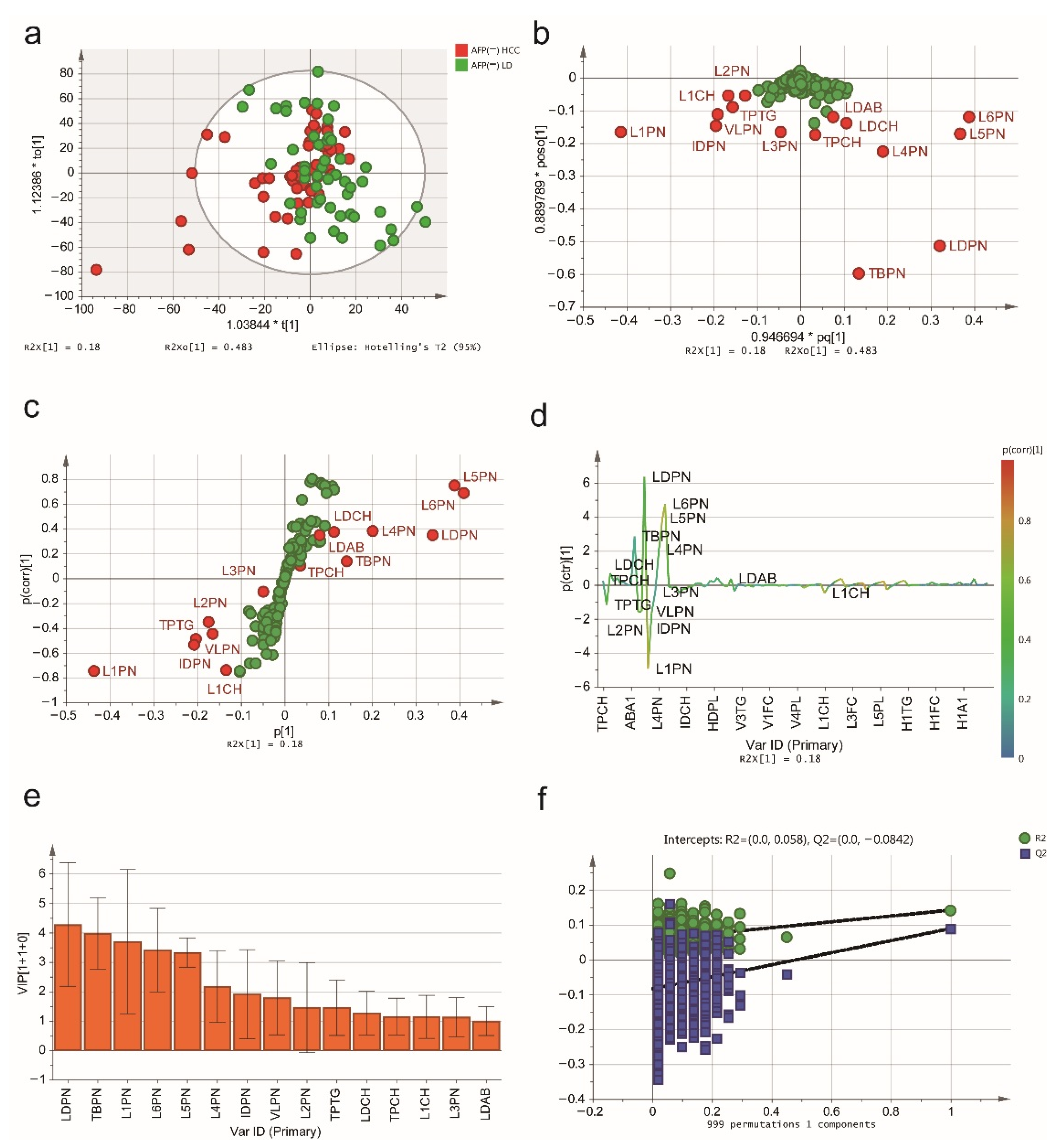
2.2. NMR Spectroscopic Multivariable Analysis
2.3. Biomarker Selection and Validation of the Diagnostic Model
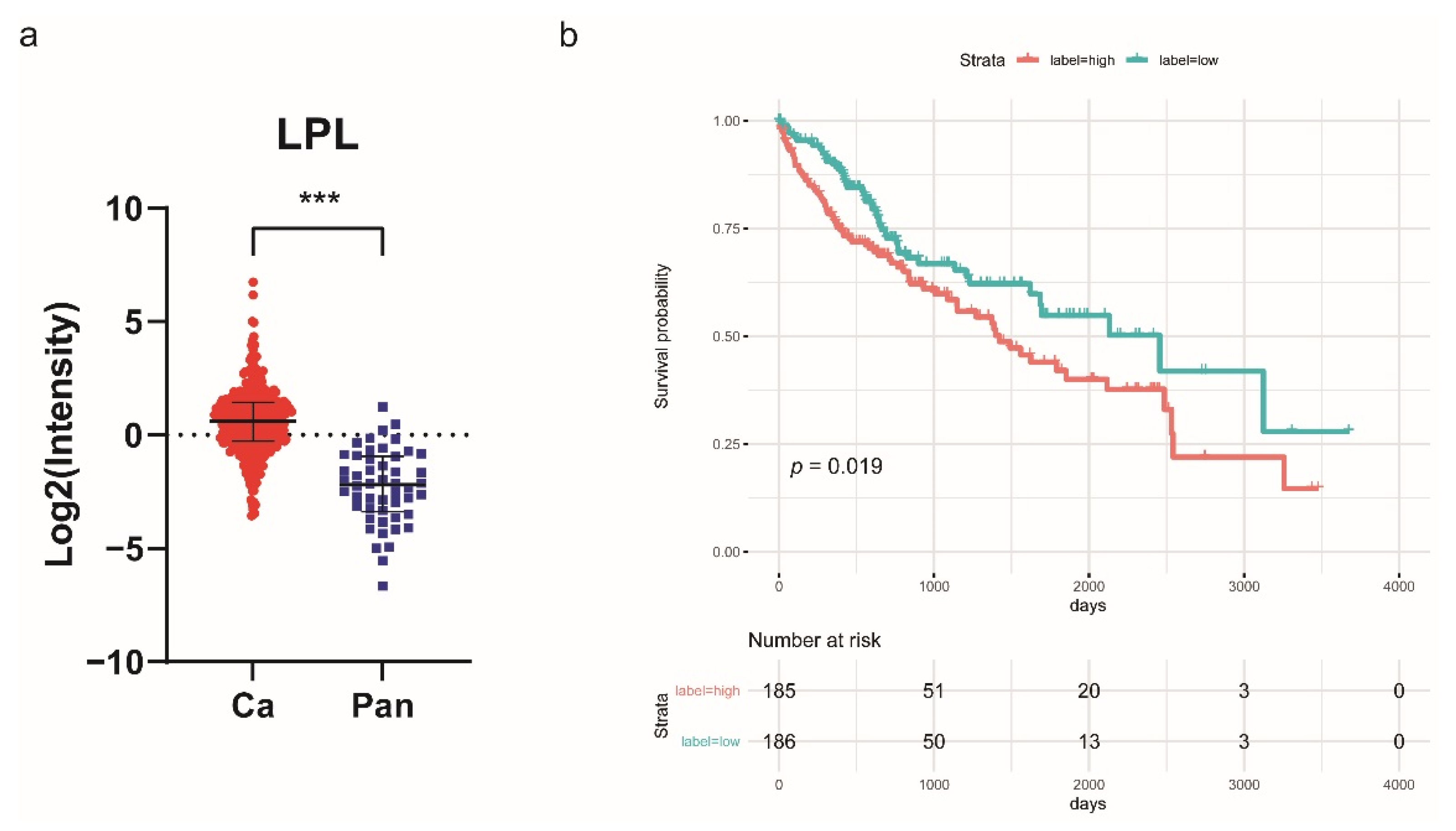
2.4. Lipoprotein Lipase (LPL) Is Upregulated in HCC and Associated with Poor Prognosis
2.5. Identification of Differentially Expressed Protein and Lipid Metabolism-Related Pathways
3. Discussion
4. Materials and Methods
4.1. Ethical Statement
4.2. Patients and Sample Collection
4.3. Inclusion Criteria and Exclusion Criteria
- Primary HCC diagnosed by histological or cellular examination.
- Single tumor (regardless of size) or the number of tumors is less than 3 and the maximum diameter is ≤ 3 cm, and no history of portal invasion or extrahepatic spread.
- HCC, cirrhosis and hepatitis with a history of HBV infection confirmed by virological assay.
- Age > 18 years.
- No previous treatment for HCC.
- Knowledge of the study and agreement to follow-up.
- History of other diagnosed malignancies.
- History of anticancer treatment for HCC.
- History of hepatitis virus infection without HBV.
- Factors can cause abnormal elevation of serum AFP in normal controls, including pregnancy and any type of liver disease.
- Participants with severe illnesses, including cardiovascular disease, endocrine disease and renal impairment.
- Participants with lactation, current smoking and drug dependence.
- Participants were taking lipid-lowering, hyperglycemic, anti-inflammatory, antithrombotic medications, dietary supplements, or antihypertensive treatment.
4.4. Magnetic Resonance Experiments
4.5. Nanoscale Liquid Chromatography-Tandem Mass Spectrometry (Nano-LC-MS/MS) Analysis
4.6. Statistical Analysis
4.6.1. Multivariable and Univariable Statistical Analysis of NMR Data
4.6.2. Quantification and Statistical Analysis of LC-MS/MS Data
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, B.; Zhang, B.; Zhang, Z.; Huang, Z.; Chen, Y.; Chen, M.; Bie, P.; Peng, B.; Wu, L.; Wang, Z.; et al. 42,573 cases of hepatectomy in China: A multicenter retrospective investigation. Sci. China Life Sci. 2018, 61, 660–670. [Google Scholar] [CrossRef]
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 589–604. [Google Scholar] [CrossRef]
- Bruix, J.; Reig, M.; Sherman, M. Evidence-Based Diagnosis, Staging, and Treatment of Patients with Hepatocellular Carcinoma. Gastroenterology 2016, 150, 835–853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, M.A.; Hegab, B.; Hyde, C.; Guo, B.; Buckels, J.A.C.; Mirza, D.F. Needle track seeding following biopsy of liver lesions in the diagnosis of hepatocellular cancer: A systematic review and meta-analysis. Gut 2008, 57, 1592–1596. [Google Scholar] [CrossRef] [PubMed]
- Wong, R.J.; Ahmed, A.; Gish, R.G. Elevated Alpha-Fetoprotein. Clin. Liver Dis. 2015, 19, 309–323. [Google Scholar] [CrossRef]
- Pavlova, N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef] [Green Version]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Markley, J.L.; Brüschweiler, R.; Edison, A.S.; Eghbalnia, H.R.; Powers, R.; Raftery, D.; Wishart, D.S. The future of NMR-based metabolomics. Curr. Opin. Biotechnol. 2016, 43, 34–40. [Google Scholar] [CrossRef] [Green Version]
- Han, X. Lipidomics for studying metabolism. Nat. Rev. Endocrinol. 2016, 12, 668–679. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Han, X. Lipidomics: Techniques, Applications, and Outcomes Related to Biomedical Sciences. Trends Biochem. Sci. 2016, 41, 954–969. [Google Scholar] [CrossRef] [Green Version]
- Jiménez, B.; Holmes, E.; Heude, C.; Tolson, R.F.; Harvey, N.; Lodge, S.L.; Chetwynd, A.J.; Cannet, C.; Fang, F.; Pearce, J.T.M.; et al. Quantitative Lipoprotein Subclass and Low Molecular Weight Metabolite Analysis in Human Serum and Plasma by 1H NMR Spectroscopy in a Multilaboratory Trial. Anal. Chem. 2018, 90, 11962–11971. [Google Scholar] [CrossRef]
- Jiang, J.; Nilsson-Ehle, P.; Xu, N. Influence of liver cancer on lipid and lipoprotein metabolism. Lipids Health Dis. 2006, 5, 4. [Google Scholar] [CrossRef] [Green Version]
- Privitera, G.; Spadaro, L.; Marchisello, S.; Fede, G.; Purrello, F. Abnormalities of Lipoprotein Levels in Liver Cirrhosis: Clinical Relevance. Dig. Dis. Sci. 2017, 63, 16–26. [Google Scholar] [CrossRef]
- Deprince, A.; Haas, J.T.; Staels, B. Dysregulated lipid metabolism links NAFLD to cardiovascular disease. Mol. Metab. 2020, 42, 101092. [Google Scholar] [CrossRef]
- Mihaleva, V.V.; van Schalkwijk, D.B.; de Graaf, A.A.; van Duynhoven, J.; van Dorsten, F.A.; Vervoort, J.; Smilde, A.; Westerhuis, J.A.; Jacobs, D.M. A Systematic Approach to Obtain Validated Partial Least Square Models for Predicting Lipoprotein Subclasses from Serum NMR Spectra. Anal. Chem. 2013, 86, 543–550. [Google Scholar] [CrossRef]
- El Harchaoui, K.; van der Steeg, W.A.; Stroes, E.S.; Kuivenhoven, J.A.; Otvos, J.D.; Wareham, N.J.; Hutten, B.A.; Kastelein, J.J.; Khaw, K.-T.; Boekholdt, M. Value of Low-Density Lipoprotein Particle Number and Size as Predictors of Coronary Artery Disease in Apparently Healthy Men and Women: The EPIC-Norfolk Prospective Population Study. J. Am. Coll. Cardiol. 2007, 49, 547–553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kostara, C.E.; Papathanasiou, A.; Psychogios, N.; Cung, M.T.; Elisaf, M.S.; Goudevenos, J.; Bairaktari, E.T. NMR-Based Lipidomic Analysis of Blood Lipoproteins Differentiates the Progression of Coronary Heart Disease. J. Proteome Res. 2014, 13, 2585–2598. [Google Scholar] [CrossRef] [PubMed]
- Casadei-Gardini, A.; Del Coco, L.; Marisi, G.; Conti, F.; Rovesti, G.; Ulivi, P.; Canale, M.; Frassineti, G.L.; Foschi, F.G.; Longo, S.; et al. 1H-NMR Based Serum Metabolomics Highlights Different Specific Biomarkers between Early and Advanced Hepatocellular Carcinoma Stages. Cancers 2020, 12, 241. [Google Scholar] [CrossRef] [Green Version]
- Fages, A.; Duarte-Salles, T.; Stepien, M.; Ferrari, P.; Fedirko, V.; Pontoizeau, C.; Trichopoulou, A.; Aleksandrova, K.; Tjønneland, A.; Olsen, A.; et al. Metabolomic profiles of hepatocellular carcinoma in a European prospective cohort. BMC Med. 2015, 13, 242. [Google Scholar] [CrossRef]
- Nahon, P.; Amathieu, R.; Triba, M.N.; Bouchemal, N.; Nault, J.C.; Ziol, M.; Seror, O.; Dhonneur, G.; Trinchet, J.-C.; Beaugrand, M.; et al. Identification of Serum Proton NMR Metabolomic Fingerprints Associated with Hepatocellular Carcinoma in Patients with Alcoholic Cirrhosis. Clin. Cancer Res. 2012, 18, 6714–6722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cox, I.J.; Aliev, A.E.; Crossey, M.M.; Dawood, M.; Al-Mahtab, M.; Akbar, S.M.; Rahman, S.; Riva, A.; Williams, R.; Taylor-Robinson, S.D. Urinary nuclear magnetic resonance spectroscopy of a Bangladeshi cohort with hepatitis-B hepatocellular carcinoma: A biomarker corroboration study. World J. Gastroenterol. 2016, 22, 4191–4200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ranjan, R.; Sinha, N. Nuclear magnetic resonance (NMR)-based metabolomics for cancer research. NMR Biomed. 2018, 32, e3916. [Google Scholar] [CrossRef]
- Alves-Bezerra, M.; Cohen, D.E. Triglyceride Metabolism in the Liver. Compr. Physiol. 2017, 8, 1–22. [Google Scholar] [CrossRef]
- Ramasamy, I. Recent advances in physiological lipoprotein metabolism. Clin. Chem. Lab. Med. 2014, 52, 1695–1727. [Google Scholar] [CrossRef]
- Perez-Matos, M.C.; Sandhu, B.; Bonder, A.; Jiang, Z.G. Lipoprotein metabolism in liver diseases. Curr. Opin. Lipidol. 2019, 30, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Lewis, G.F.; Rader, D.J. New Insights Into the Regulation of HDL Metabolism and Reverse Cholesterol Transport. Circ. Res. 2005, 96, 1221–1232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaidi, N.; Lupien, L.; Kuemmerle, N.B.; Kinlaw, W.B.; Swinnen, J.V.; Smans, K. Lipogenesis and lipolysis: The pathways exploited by the cancer cells to acquire fatty acids. Prog. Lipid Res. 2013, 52, 585–589. [Google Scholar] [CrossRef] [Green Version]
- Satriano, L.; Lewinska, M.; Rodrigues, P.M.; Banales, J.M.; Andersen, J.B. Metabolic rearrangements in primary liver cancers: Cause and consequences. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 748–766. [Google Scholar] [CrossRef]
- Sangineto, M.; Villani, R.; Cavallone, F.; Romano, A.; Loizzi, D.; Serviddio, G. Lipid Metabolism in Development and Progression of Hepatocellular Carcinoma. Cancers 2020, 12, 1419. [Google Scholar] [CrossRef]
- Bianco, C.; Jamialahmadi, O.; Pelusi, S.; Baselli, G.; Dongiovanni, P.; Zanoni, I.; Santoro, L.; Maier, S.; Liguori, A.; Meroni, M.; et al. Non-invasive stratification of hepatocellular carcinoma risk in non-alcoholic fatty liver using polygenic risk scores. J. Hepatol. 2020, 74, 775–782. [Google Scholar] [CrossRef]
- Tang, Y.; Zhou, J.; Hooi, S.C.; Jiang, Y.M.; Lu, G.-D. Fatty acid activation in carcinogenesis and cancer development: Essential roles of long-chain acyl-CoA synthetases (Review). Oncol. Lett. 2018, 16, 1390–1396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Ding, C.; Chen, Y.; Hu, W.; Lu, Y.; Wu, W.; Zhang, Y.; Yang, B.; Wu, H.; Peng, C.; et al. ACSL4 promotes hepatocellular carcinoma progression via c-Myc stability mediated by ERK/FBW7/c-Myc axis. Oncogenesis 2020, 9, 42. [Google Scholar] [CrossRef]
- Dash, S.; Xiao, C.; Morgantini, C.; Lewis, G.F. New Insights into the Regulation of Chylomicron Production. Annu. Rev. Nutr. 2015, 35, 265–294. [Google Scholar] [CrossRef]
- Heeren, J.; Scheja, L. Metabolic-associated fatty liver disease and lipoprotein metabolism. Mol. Metab. 2021, 50, 101238. [Google Scholar] [CrossRef]
- Kumari, A.; Kristensen, K.K.; Ploug, M.; Winther, A.L.-M. The Importance of Lipoprotein Lipase Regulation in Atherosclerosis. Biomedicines 2021, 9, 782. [Google Scholar] [CrossRef] [PubMed]
- Young, S.G.; Fong, L.G.; Beigneux, A.P.; Allan, C.M.; He, C.; Jiang, H.; Nakajima, K.; Meiyappan, M.; Birrane, G.; Ploug, M. GPIHBP1 and Lipoprotein Lipase, Partners in Plasma Triglyceride Metabolism. Cell Metab. 2019, 30, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, K.K.; Leth-Espensen, K.Z.; Kumari, A.; Grønnemose, A.L.; Lund-Winther, A.-M.; Young, S.G.; Ploug, M. GPIHBP1 and ANGPTL4 Utilize Protein Disorder to Orchestrate Order in Plasma Triglyceride Metabolism and Regulate Compartmentalization of LPL Activity. Front. Cell Dev. Biol. 2021, 9, 1900. [Google Scholar] [CrossRef]
- Cao, D.; Song, X.; Che, L.; Li, X.; Pilo, M.G.; Vidili, G.; Porcu, A.; Solinas, A.; Cigliano, A.; Pes, G.M.; et al. Bothde novosynthetized and exogenous fatty acids support the growth of hepatocellular carcinoma cells. Liver Int. 2016, 37, 80–89. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Ma, H.; Wang, L.; Song, X.; Zhang, J.; Liu, W.; Ge, Y.; Sun, Y.; Yu, X.; Wang, Z.; et al. Tumor suppressor ZHX2 inhibits NAFLD-HCC progression via blocking LPL-mediated lipid uptake. Cell Death Differ. 2019, 27, 1693–1708. [Google Scholar] [CrossRef]
- Manupati, K.; Yeeravalli, R.; Kaushik, K.; Singh, D.; Mehra, B.; Gangane, N.; Gupta, A.; Goswami, K.; Das, A. Activation of CD44-Lipoprotein lipase axis in breast cancer stem cells promotes tumorigenesis. Biochim. Biophys. Acta (BBA)—Mol. Basis Dis. 2021, 1867, 166228. [Google Scholar] [CrossRef]
- Lu, C.-W.; Lo, Y.-H.; Chen, C.-H.; Lin, C.-Y.; Tsai, C.-H.; Chen, P.-J.; Yang, Y.-F.; Wang, C.-H.; Tan, C.-H.; Hou, M.-F.; et al. VLDL and LDL, but not HDL, promote breast cancer cell proliferation, metastasis and angiogenesis. Cancer Lett. 2016, 388, 130–138. [Google Scholar] [CrossRef] [PubMed]

| Training Set | Validation Set | |||||
|---|---|---|---|---|---|---|
| Characteristics | HCC | Liver Disease | p-Value | HCC | Liver Disease | p-Value |
| n | 51 | 37 | 24 | 17 | ||
| Age (years) | 58 (33.00 to 71.00) | 59 (43.00 to 66.00) | 0.889 | 60.50 (51.75 to 66.00) | 49.00 (45.50 to 58.00) | 0.010 |
| Sex (male/female) | 37/14 | 21/16 | 0.123 | 16/8 | 8/9 | 0.209 |
| AFP (ng/mL) | 23.88 (4.80 to 126.20) | 2.69 (1.68 to 4.64) | <0.001 | 60.50 (3.42 to 481.00) | 2.66 (1.98 to 3.55) | <0.001 |
| ALT (IU/L) | 36.00 (22.00 to 48.00) | 25.00 (16.00 to 36.50) | 0.018 | 24.00 (15.25 to 31.75) | 21.00 (14.00 to 35.00) | 0.937 |
| AST (IU/L) | 37.00 (26.00 to 59.00) | 28.00 (19.00 to 36.50) | 0.002 | 29.00 (21.50 to 38.50) | 21.00 (16.00 to 29.00) | 0.095 |
| ALB (g/L) | 41.60 (39.40 to 45.50) | 43.50 (37.50 to 46.95) | 0.422 | 42.55 (36.50 to 45.08) | 44.90 (41.45 to 47.60) | 0.058 |
| TP (g/L) | 74.50 (70.90 to 77.20) | 68.40 (53.50 to 77.65) | 0.016 | 70.45 (64.60 to 75.93) | 74.40 (66.25 to 76.25) | 0.404 |
| TBIL (μmmol/L) | 16.50 (13.90 to 23.60) | 14.50 (11.05 to 18.63) | 0.154 | 14.70 (11.08 to 20.98) | 12.40 (9.80 to 18.25) | 0.375 |
| CRE (μmol/L) | 61.00 (53.00 to 69.00) | 62.00 (52.00 to 79.50) | 0.244 | 61.50 (56.00 to 78.00) | 60.00 (51.50 to 68.00) | 0.255 |
| BCLC stage | ||||||
| stage 0 | 9 | / | 1 | / | ||
| stage A | 42 | / | 23 | / | ||
| Child-Pugh class | ||||||
| A | 46 | 33 | 22 | 16 | ||
| B-C | 5 | 4 | 2 | 1 | ||
| Tumor diameter (cm) | ||||||
| ≤3 | 25 | / | 10 | / | ||
| >3 | 26 | / | 14 | / | ||
| HCC vs. LD | HCC vs. NCs | ||||||
|---|---|---|---|---|---|---|---|
| Index | Description | Unit | VIP | p (corr) | VIP | p (corr) | p-Value |
| TPCH | Total Cholesterol | mg/dL | 1.045 | −0.026 | 1.030 | 0.165 | 0.596 |
| HDCH | HDL-C | mg/dL | 0.414 | 0.120 | 1.032 | 0.703 | 2.931 × 10−14 |
| TPA1 | Apo-A1 | mg/dL | 1.089 | 0.202 | 1.146 | 0.631 | 3.742 × 10−10 |
| TBPN | Total Particle Number | nmol/L | 3.451 | −0.006 | 3.512 | 0.107 | 0.775 |
| VLPN | VLDL Particle Number | nmol/L | 1.083 | 0.014 | 1.329 | 0.310 | 0.003 |
| IDPN | IDL Particle Number | nmol/L | 2.514 | −0.347 | 1.379 | 0.277 | 6.318 × 10−5 |
| LDPN | LDL Particle Number | nmol/L | 3.465 | 0.069 | 2.772 | −0.016 | 0.339 |
| L1PN | LDL-1 Particle Number | nmol/L | 3.743 | −0.310 | 3.024 | −0.512 | 1.399 × 10−9 |
| L2PN | LDL-2 Particle Number | nmol/L | 2.612 | −0.209 | 2.118 | −0.511 | 1.712 × 10−4 |
| L3PN | LDL-3 Particle Number | nmol/L | 1.451 | −0.032 | 3.144 | −0.838 | <0.001 |
| L4PN | LDL-4 Particle Number | nmol/L | 2.708 | 0.221 | 1.645 | −0.443 | 2.148 × 10−10 |
| L5PN | LDL-5 Particle Number | nmol/L | 3.232 | 0.296 | 2.737 | 0.643 | 5.121 × 10−8 |
| L6PN | LDL-6 Particle Number | nmol/L | 2.971 | 0.218 | 5.050 | 0.934 | <0.001 |
| HDA1 | HDL Apo-A1 | mg/dL | 1.037 | 0.167 | 1.227 | 0.644 | 1.538 × 10−11 |
| L1CH | LDL-1 Cholesterol | mg/dL | 1.201 | −0.316 | 0.993 | −0.563 | 2.456 × 10−9 |
| L6CH | LDL-6 Cholesterol | mg/dL | 0.942 | 0.227 | 1.324 | 0.936 | <0.001 |
| L6AB | LDL-6 Apo-B | mg/dL | 0.802 | 0.218 | 1.184 | 0.934 | <0.001 |
| H1A1 | HDL-1 Apo-A1 | mg/dL | 1.229 | 0.230 | 0.146 | −0.271 | 0.011 |
| H4A1 | HDL-4 Apo-A1 | mg/dL | 0.811 | −0.083 | 1.306 | 0.773 | <0.001 |
| AFP (-) HCC vs. AFP (-) LD | AFP (-) HCC vs. NCs | ||||||
|---|---|---|---|---|---|---|---|
| Index | Description | Unit | VIP | p (corr) | VIP | p (corr) | p-Value |
| TPCH | Total Cholesterol | mg/dL | 1.160 | 0.105 | 0.836 | 0.281 | 0.463 |
| TPTG | Total Triglycerides | mg/dL | 1.456 | −0.450 | 1.047 | 0.060 | 0.075 |
| LDCH | LDL-C | mg/dL | 1.274 | 0.373 | 0.204 | −0.013 | 0.214 |
| HDCH | HDL-C | mg/dL | 0.414 | 0.261 | 1.207 | 0.763 | <0.001 |
| TPA1 | Apo-A1 | mg/dL | 0.690 | 0.294 | 1.307 | 0.655 | 5.091 × 10−14 |
| TBPN | Total Particle Number | nmol/L | 3.986 | 0.138 | 2.477 | 0.163 | 0.594 |
| VLPN | VLDL Particle Number | nmol/L | 1.794 | −0.490 | 1.850 | 0.271 | 0.005 |
| IDPN | IDL Particle Number | nmol/L | 1.919 | −0.537 | 2.066 | 0.309 | 1.076 × 10−5 |
| LDPN | LDL Particle Number | nmol/L | 4.282 | 0.350 | 0.463 | 0.035 | 0.084 |
| L1PN | LDL-1 Particle Number | nmol/L | 3.702 | −0.747 | 3.206 | −0.422 | 9.240 × 10−8 |
| L2PN | LDL-2 Particle Number | nmol/L | 1.467 | −0.354 | 1.715 | −0.465 | 1.573 × 10−4 |
| L3PN | LDL-3 Particle Number | nmol/L | 1.137 | −0.110 | 3.224 | −0.849 | <0.001 |
| L4PN | LDL-4 Particle Number | nmol/L | 2.181 | 0.382 | 2.018 | −0.511 | 1.325 × 10−11 |
| L5PN | LDL-5 Particle Number | nmol/L | 3.338 | 0.746 | 2.940 | 0.610 | 7.372 × 10−9 |
| L6PN | LDL-6 Particle Number | nmol/L | 3.419 | 0.685 | 5.325 | 0.939 | <0.001 |
| HDA1 | HDL Apo-A1 | mg/dL | 0.703 | 0.298 | 1.418 | 0.675 | 4.663 × 10−14 |
| LDAB | LDL Apo-B | mg/dL | 1.004 | 0.350 | 0.109 | 0.035 | 0.084 |
| L1CH | LDL-1 Cholesterol | mg/dL | 1.149 | −0.738 | 1.013 | −0.475 | 1.382 × 10−8 |
| L6CH | LDL-6 Cholesterol | mg/dL | 0.942 | 0.717 | 1.398 | 0.930 | <0.001 |
| L6PL | LDL-6 Phospholipids | mg/dL | 0.709 | 0.757 | 1.006 | 0.932 | <0.001 |
| L6AB | LDL-6 Apo-B | mg/dL | 0.802 | 0.685 | 1.249 | 0.939 | <0.001 |
| H4A1 | HDL-4 Apo-A1 | mg/dL | 0.811 | 0.421 | 1.335 | 0.760 | <0.001 |
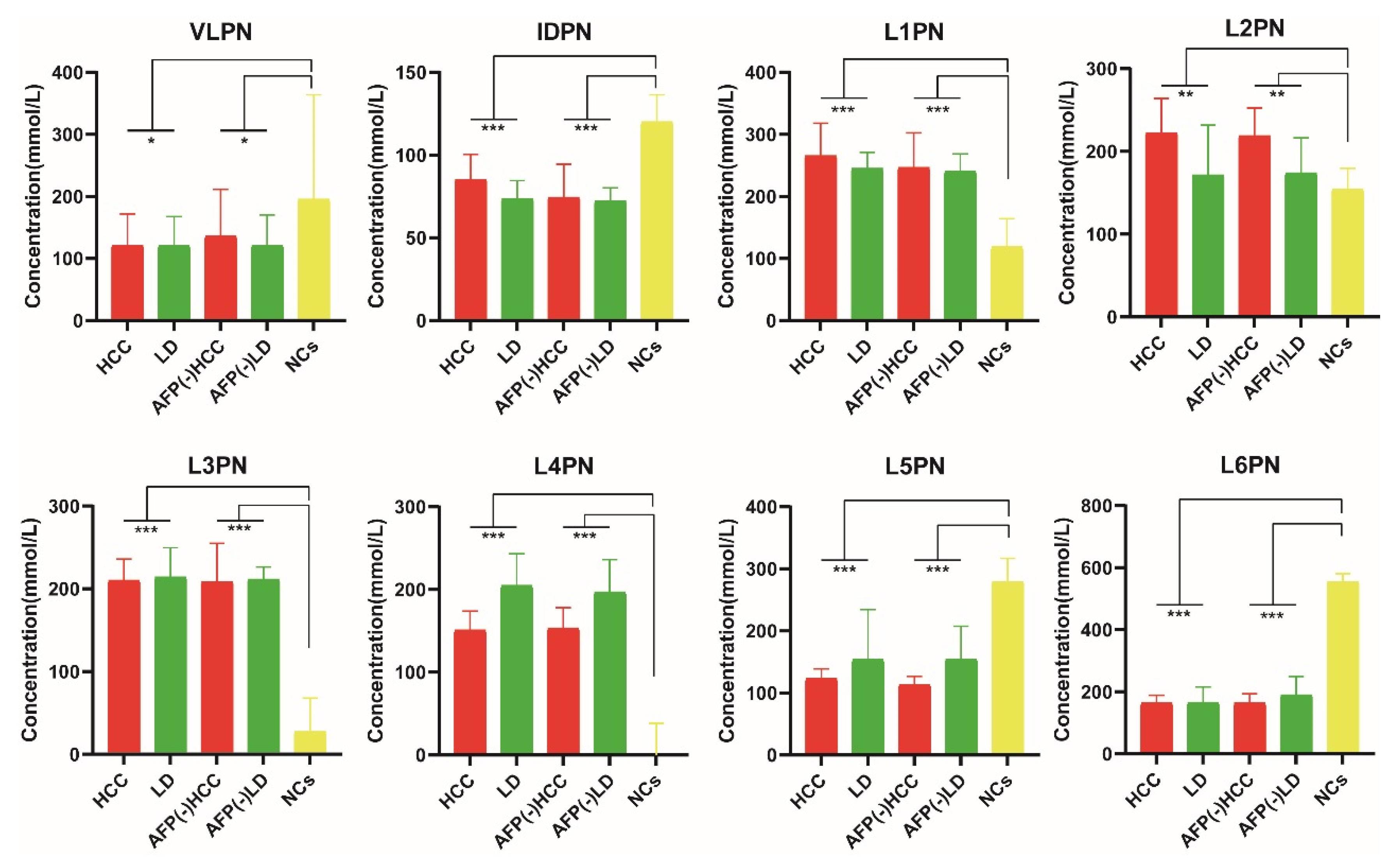
| HCC vs. LD | HCC vs. NCs | AFP (-) HCC vs. AFP (-) LD | AFP (-) HCC vs. NCs | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Index | VIP | Trend | VIP | Trend | p-Value | VIP | Trend | VIP | Trend | p-Value |
| VLPN | 1.083 | up | 1.329 | down | 3.023 × 10−3 | 1.794 | up | 1.850 | down | 4.733 × 10−3 |
| IDPN | 2.514 | up | 1.379 | down | 6.318 × 10−5 | 1.919 | up | 2.066 | down | 1.076 × 10−5 |
| L1PN | 3.743 | up | 3.024 | up | 1.398 × 10−8 | 3.702 | up | 3.206 | up | 9.240 × 10−8 |
| L2PN | 2.613 | up | 2.118 | up | 1.712 × 10−4 | 1.467 | up | 1.715 | up | 1.573 × 10−4 |
| L3PN | 1.451 | down | 3.144 | up | <0.001 | 1.137 | down | 3.224 | up | <0.001 |
| L4PN | 2.708 | down | 1.645 | up | 2.148 × 10−10 | 2.181 | down | 2.018 | up | 1.325 × 10−11 |
| L5PN | 3.232 | down | 2.737 | down | 5.121 × 10−8 | 3.338 | down | 2.940 | down | 7.372 × 10−9 |
| L6PN | 2.971 | down | 5.050 | down | <0.001 | 3.419 | down | 5.325 | down | <0.001 |
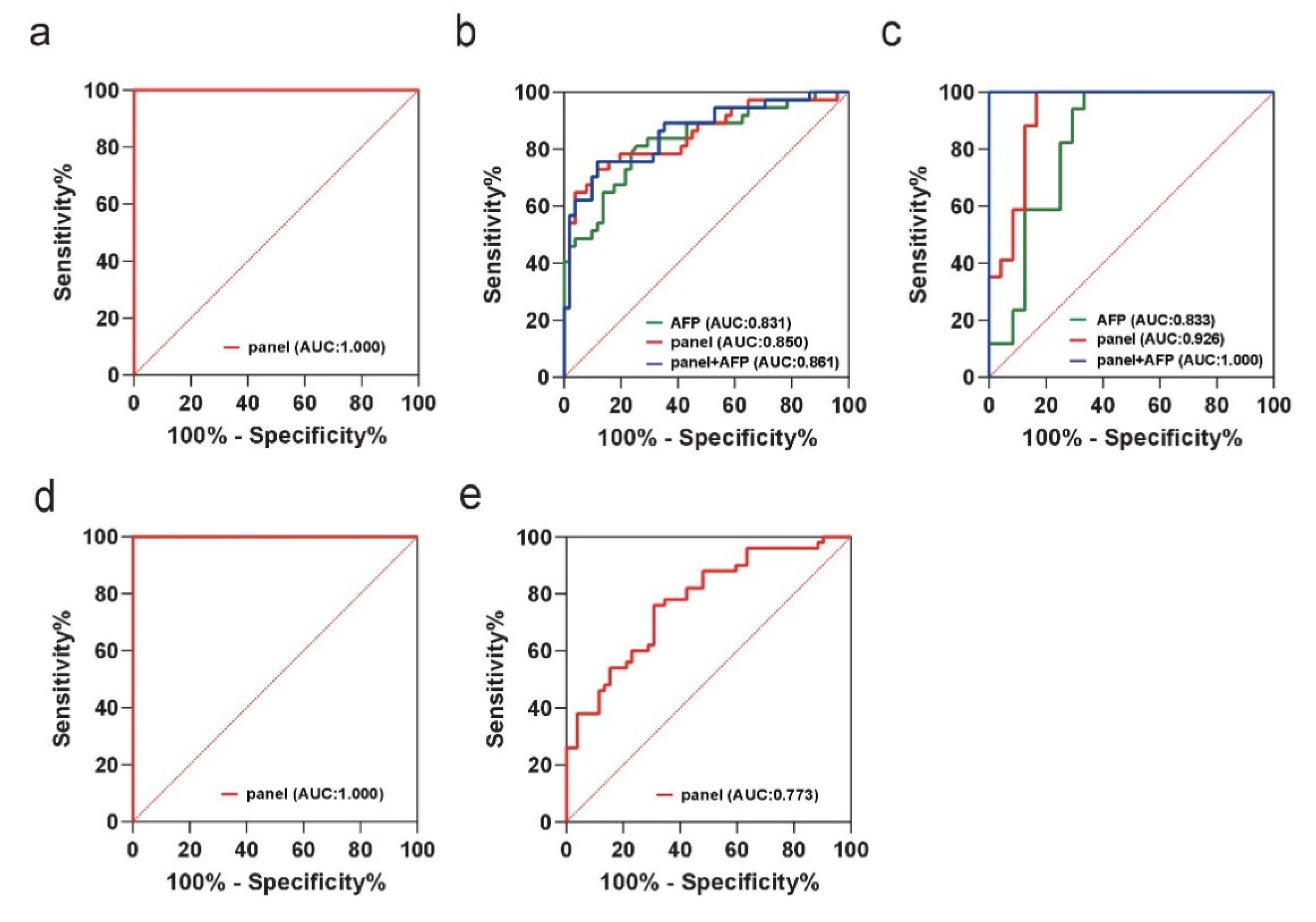
| Experiment Set | Group | Dataset | AUC (95% CI) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|
| Training set | HCC vs. LD | AFP | 0.831 (0.736 to 0.902) | 74.51 | 81.08 |
| panel | 0.850 (0.758 to 0.917) | 88.24 | 72.97 | ||
| Panel + AFP | 0.861 (0.771 to 0.926) | 88.24 | 75.68 | ||
| HCC vs. NCs | panel | 1.000 (0.964 to 1.000) | 100.00 | 100.00 | |
| Validation set | HCC vs. LD | AFP | 0.833 (0.684 to 0.931) | 66.67 | 100.00 |
| panel | 0.926 (0.800 to 0.984) | 83.33 | 100.00 | ||
| Panel + AFP | 1.000 (0.914 to 1.000) | 100.00 | 100.00 | ||
| AFP-negative | HCC vs. LD | panel | 0.773 (0.680 to 0.850) | 69.23 | 76.00 |
| HCC vs. NCs | panel | 1.000 (0.964 to 1.000) | 100.00 | 100.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zuo, D.; An, H.; Li, J.; Xiao, J.; Ren, L. The Application Value of Lipoprotein Particle Numbers in the Diagnosis of HBV-Related Hepatocellular Carcinoma with BCLC Stage 0-A. J. Pers. Med. 2021, 11, 1143. https://doi.org/10.3390/jpm11111143
Zuo D, An H, Li J, Xiao J, Ren L. The Application Value of Lipoprotein Particle Numbers in the Diagnosis of HBV-Related Hepatocellular Carcinoma with BCLC Stage 0-A. Journal of Personalized Medicine. 2021; 11(11):1143. https://doi.org/10.3390/jpm11111143
Chicago/Turabian StyleZuo, Duo, Haohua An, Jianhua Li, Jiawei Xiao, and Li Ren. 2021. "The Application Value of Lipoprotein Particle Numbers in the Diagnosis of HBV-Related Hepatocellular Carcinoma with BCLC Stage 0-A" Journal of Personalized Medicine 11, no. 11: 1143. https://doi.org/10.3390/jpm11111143
APA StyleZuo, D., An, H., Li, J., Xiao, J., & Ren, L. (2021). The Application Value of Lipoprotein Particle Numbers in the Diagnosis of HBV-Related Hepatocellular Carcinoma with BCLC Stage 0-A. Journal of Personalized Medicine, 11(11), 1143. https://doi.org/10.3390/jpm11111143






