Precision Autism: Genomic Stratification of Disorders Making Up the Broad Spectrum May Demystify Its “Epidemic Rates”
Abstract
1. Introduction
2. Materials and Methods
- CATEGORY 1
- CATEGORY 2
- CATEGORY 3
- SYNDROMIC (former category 4)
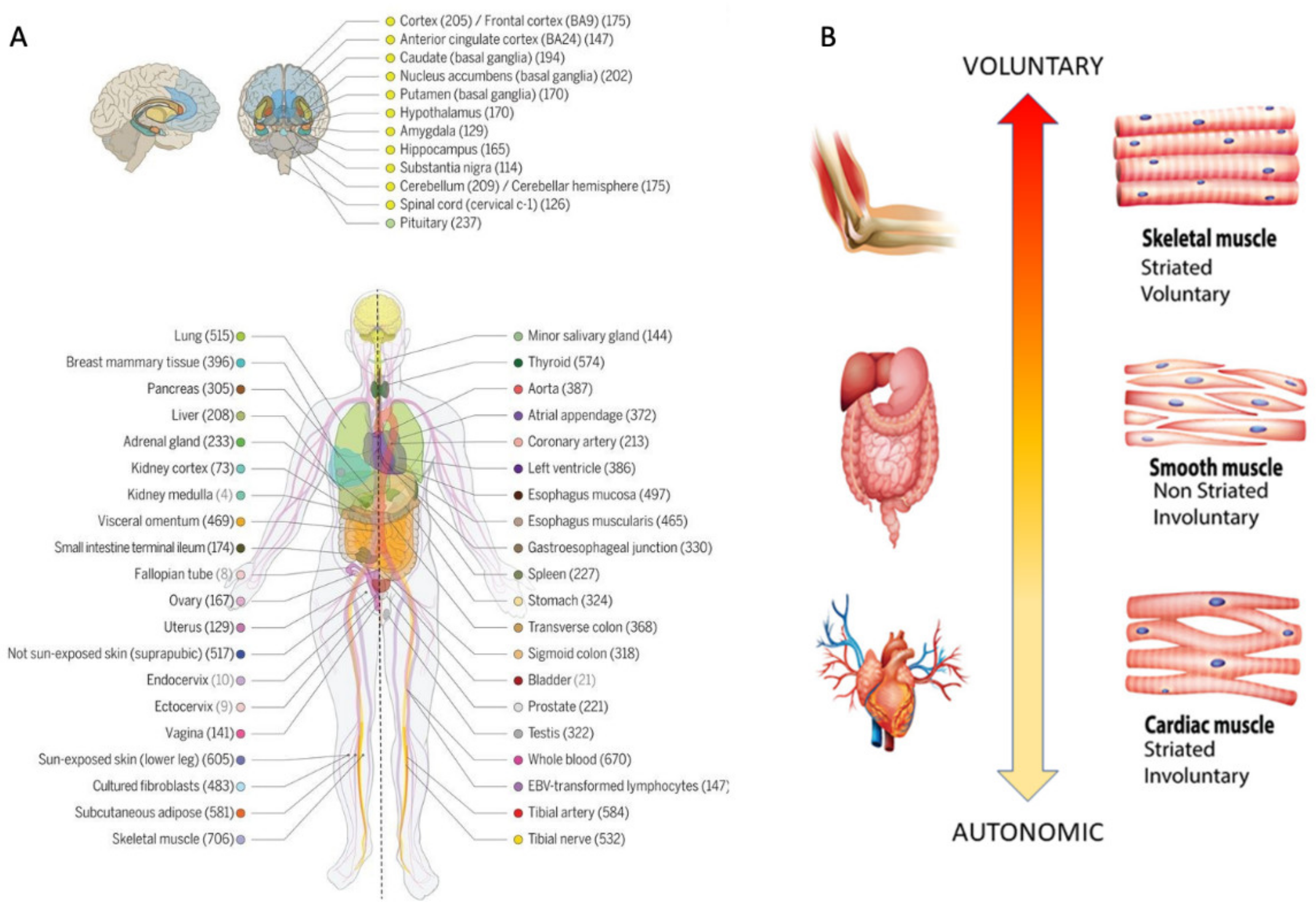
Methods to Obtain Pairwise Comparisons of Genes’ Expression in Autism and Various Disorders
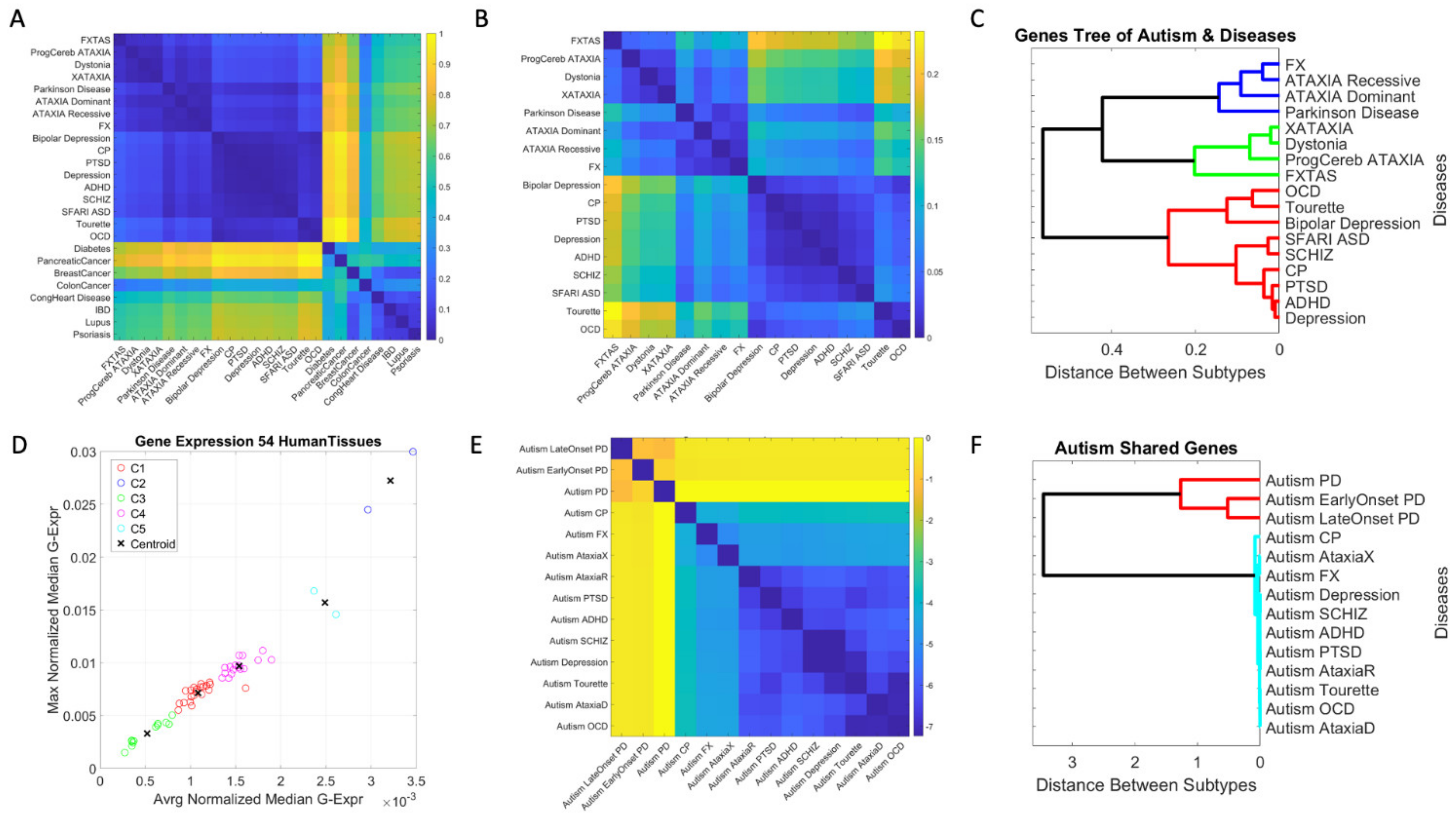
3. Results
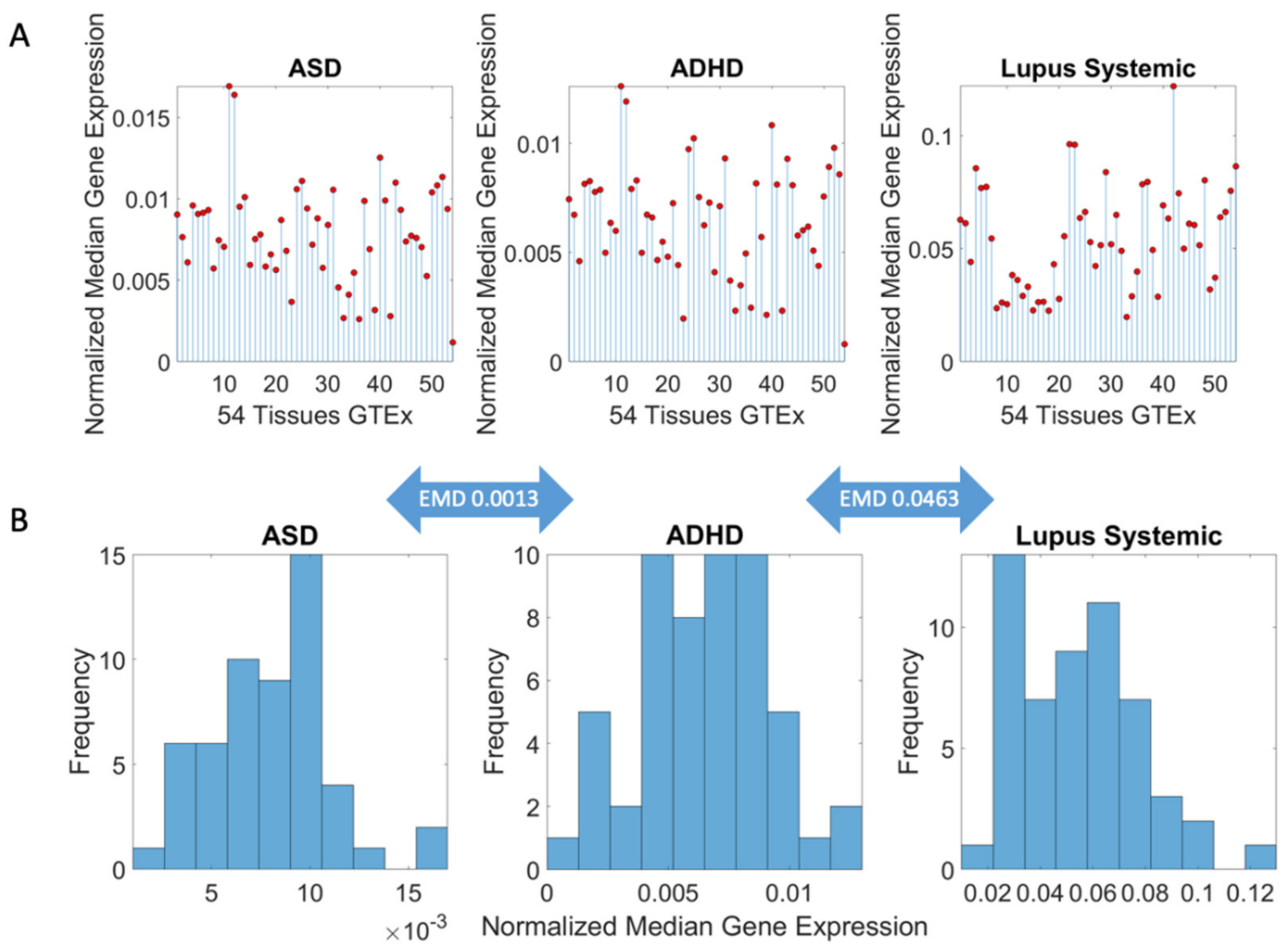
3.1. Neuropsychiatric Disorders and Autism Share Common Genes Expressed in Brain Tissues for Motor, Emotional and Self-Regulatory Control
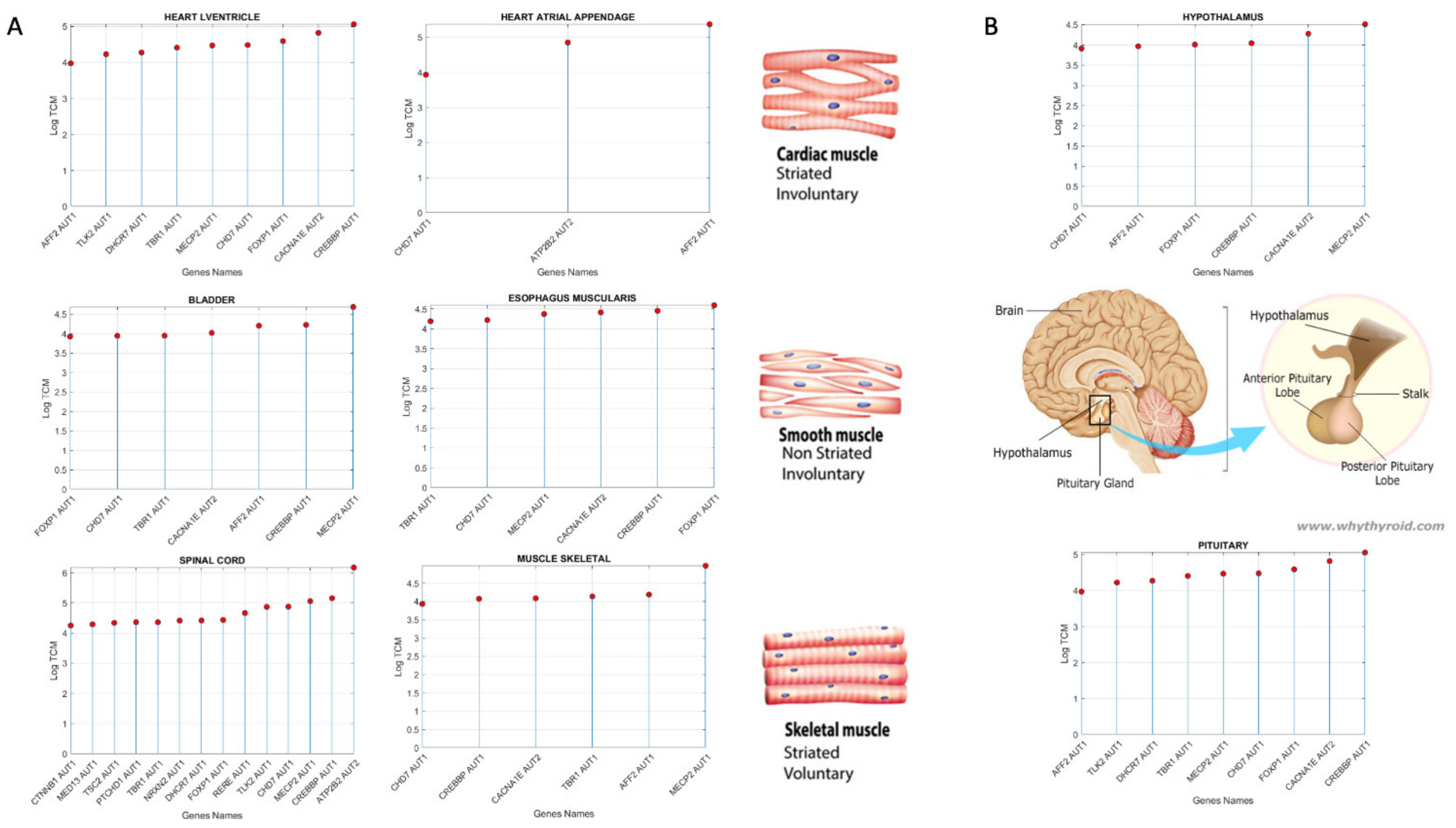
3.3. Examination of the Maximal Gene Expression on the Tissues for Genes Common to Autism and These Disorders Revealed a Compact Gene Pool
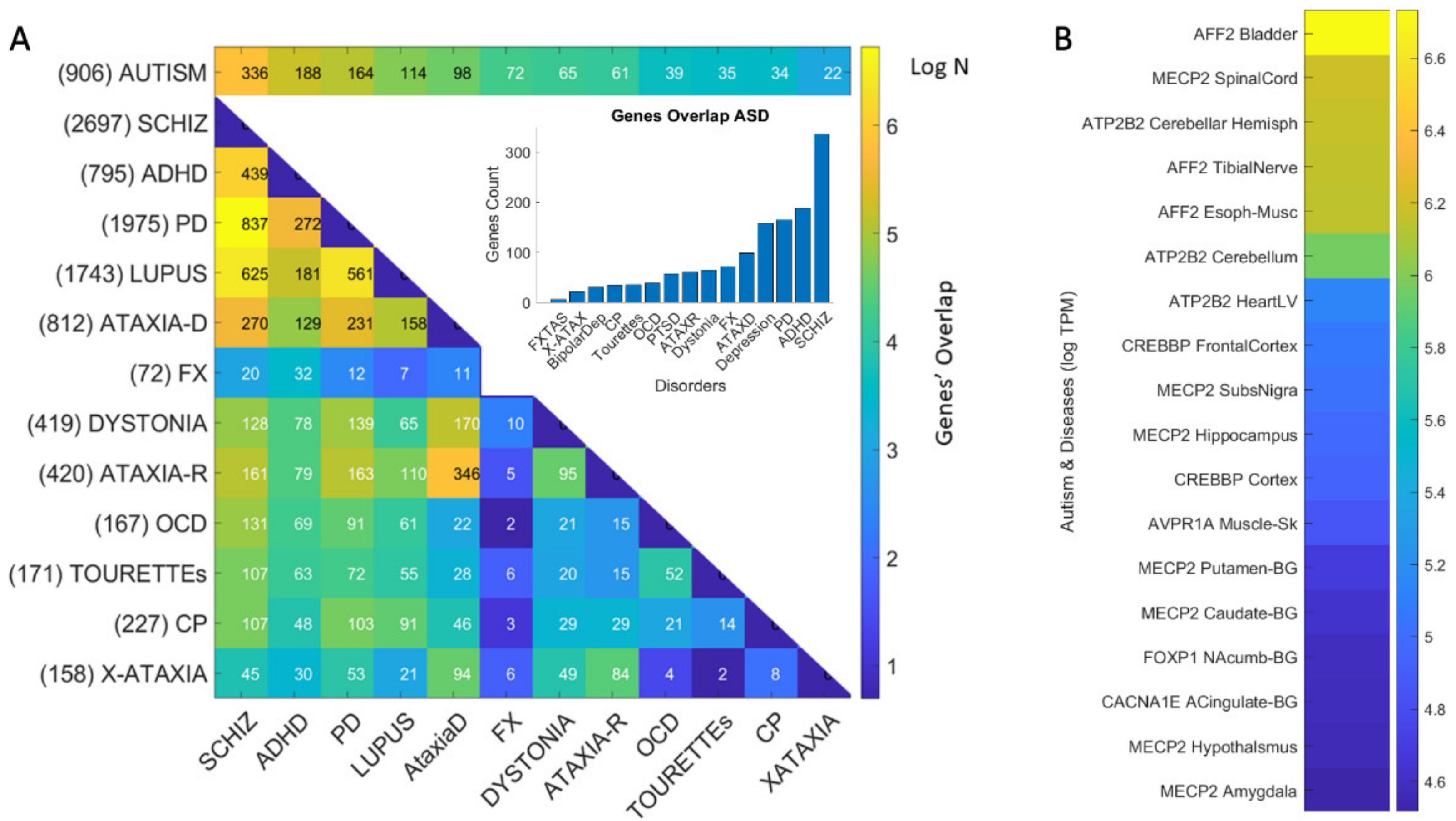
3.4. Genomic Stratification of Neurological and Neuropsychiatric Diseases Making Up Autism Today
4. Discussion
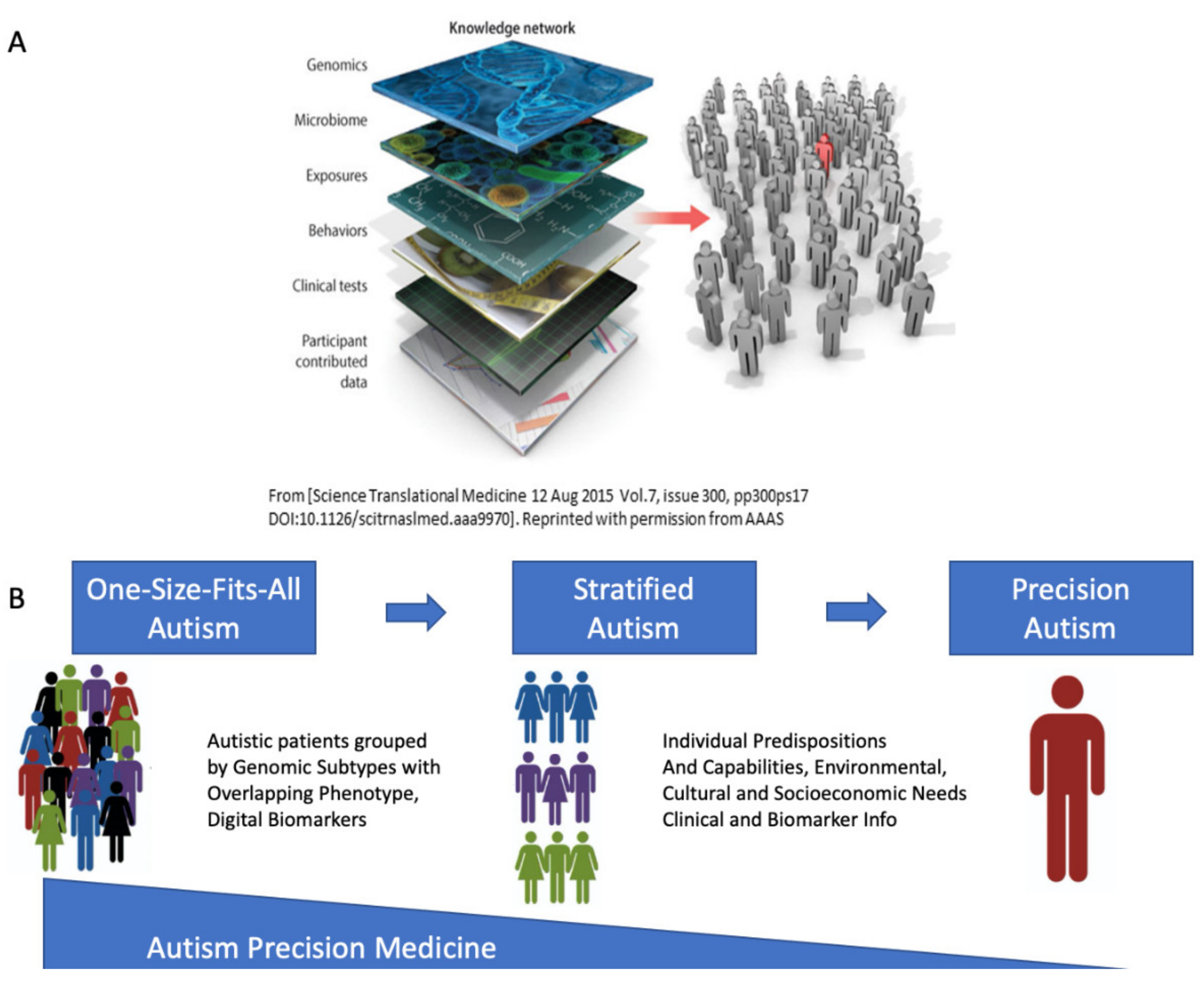
4.1. The Genetically Informed Autism Subtypes
4.2. The Genes Common to Autism and Each Subtype
4.3. The Genes Common to Autism and All Subtypes
4.4. Implications of This Genomic Categorization for Treatment Selection in Autism
4.5. The Importance of Reframing Autism under the Precision Medicine Paradigm
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maenner, M.J.; Shaw, K.A.; Baio, J.; Washington, A.; Patrick, M.; DiRienzo, M.; Christensen, D.L.; Wiggins, L.D.; Pettygrove, S.; Andrews, J.G.; et al. Prevalence of Autism Spectrum Disorder Among Children Aged 8 Years—Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2016. MMWR Surveill. Summ. 2020, 69, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Mandy, W.; Chilvers, R.; Chowdhury, U.; Salter, G.; Seigal, A.; Skuse, D. Sex differences in autism spectrum disorder: Evidence from a large sample of children and adolescents. J. Autism Dev. Disord. 2012, 42, 1304–1313. [Google Scholar] [CrossRef] [PubMed]
- Russell, G.; Stapley, S.; Newlove-Delgado, T.; Salmon, A.; White, R.; Warren, F.; Pearson, A.; Ford, T. Time trends in autism diagnosis over 20 years: A UK population-based cohort study. J. Child Psychol. Psychiatry 2021, 69, 1–12. [Google Scholar] [CrossRef]
- Torres, E.B.; Isenhower, R.W.; Yanovich, P.; Rehrig, G.; Stigler, K.; Nurnberger, J.; Jose, J.V. Strategies to develop putative biomarkers to characterize the female phenotype with autism spectrum disorders. J. Neurophysiol. 2013, 110, 1646–1662. [Google Scholar] [CrossRef][Green Version]
- Torres, E.B.; Denisova, K. Motor noise is rich signal in autism research and pharmacological treatments. Sci. Rep. 2016, 6, 37422. [Google Scholar] [CrossRef]
- Torres, E.B.; Nguyen, J.; Mistry, S.; Whyatt, C.; Kalampratsidou, V.; Kolevzon, A. Characterization of the Statistical Signatures of Micro-Movements Underlying Natural Gait Patterns in Children with Phelan McDermid Syndrome: Towards Precision-Phenotyping of Behavior in ASD. Front. Integr. Neurosci. 2016, 10, 22. [Google Scholar] [CrossRef]
- Torres, E.B.; Mistry, S.; Caballero, C.; Whyatt, C.P. Stochastic Signatures of Involuntary Head Micro-movements Can Be Used to Classify Females of ABIDE into Different Subtypes of Neurodevelopmental Disorders. Front. Integr. Neurosci. 2017, 11, 10. [Google Scholar] [CrossRef] [PubMed]
- Bokadia, H.; Rai, R.; Torres, E.B. Digitized Autism Observation Diagnostic Schedule: Social Interactions beyond the Limits of the Naked Eye. J. Pers. Med. 2020, 10, 159. [Google Scholar] [CrossRef] [PubMed]
- Torres, E.B.; Rai, R.; Mistry, S.; Gupta, B. Hidden Aspects of the Research ADOS Are Bound to Affect Autism Science. Neural Comput. 2020, 32, 515–561. [Google Scholar] [CrossRef] [PubMed]
- Lord, C.; Risi, S.; Lambrecht, L.; Cook, E.H., Jr.; Leventhal, B.L.; DiLavore, P.C.; Pickles, A.; Rutter, M. The autism diagnostic observation schedule-generic: A standard measure of social and communication deficits associated with the spectrum of autism. J. Autism Dev. Disord. 2000, 30, 205–223. [Google Scholar] [CrossRef]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 5th ed.; American Psychiatric Association: Arlington, VA, USA, 2013. [Google Scholar] [CrossRef]
- Mottron, L. A radical change in our autism research strategy is needed: Back to prototypes. Autism Res. 2021, 14, 2213–2220. [Google Scholar] [CrossRef]
- Council, N.R. Educating Children with Autism; National Academy of Science: Washington, DC, USA, 2001. [Google Scholar]
- Constantino, J.N. Response to “A Radical Change in Our Autism Research Strategy is Needed: Back to Prototypes” by Mottron et al. Autism Res. 2021, 14, 2221–2223. [Google Scholar] [CrossRef]
- Gotham, K.; Pickles, A.; Lord, C. Standardizing ADOS scores for a measure of severity in autism spectrum disorders. J. Autism Dev. Disord. 2009, 39, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Esler, A.N.; Bal, V.H.; Guthrie, W.; Wetherby, A.; Ellis Weismer, S.; Lord, C. The Autism Diagnostic Observation Schedule, Toddler Module: Standardized Severity Scores. J. Autism Dev. Disord. 2015, 45, 2704–2720. [Google Scholar] [CrossRef] [PubMed]
- Bermperidis, T.; Rai, R.; Ryu, J.; Torres, E.B. Optimal Time Lags from Causal Prediction Model Help Stratify and Forecast Nervous System Pathology. Sci. Rep. 2021, 11, 20904. [Google Scholar] [CrossRef]
- Sanger, T.D.; Kukke, S.N. Abnormalities of tactile sensory function in children with dystonic and diplegic cerebral palsy. J. Child. Neurol. 2007, 22, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Pham, R.; Mol, B.W.; Gecz, J.; MacLennan, A.H.; MacLennan, S.C.; Corbett, M.A.; van Eyk, C.L.; Webber, D.L.; Palmer, L.J.; Berry, J.G. Definition and diagnosis of cerebral palsy in genetic studies: A systematic review. Dev. Med. Child. Neurol. 2020, 62, 1024–1030. [Google Scholar] [CrossRef]
- Lin, J.P.; Nardocci, N. Recognizing the Common Origins of Dystonia and the Development of Human Movement: A Manifesto of Unmet Needs in Isolated Childhood Dystonias. Front. Neurol. 2016, 7, 226. [Google Scholar] [CrossRef] [PubMed]
- Xiao, R.; Zhong, H.; Li, X.; Ma, Y.; Zhang, R.; Wang, L.; Zang, Z.; Fan, X. Abnormal Cerebellar Development Is Involved in Dystonia-Like Behaviors and Motor Dysfunction of Autistic BTBR Mice. Front. Cell Dev. Biol. 2020, 8, 231. [Google Scholar] [CrossRef] [PubMed]
- Geurts, H.M.; Grasman, R.P.; Verte, S.; Oosterlaan, J.; Roeyers, H.; van Kammen, S.M.; Sergeant, J.A. Intra-individual variability in ADHD, autism spectrum disorders and Tourette’s syndrome. Neuropsychologia 2008, 46, 3030–3041. [Google Scholar] [CrossRef] [PubMed]
- Zablotsky, B.; Bramlett, M.D.; Blumberg, S.J. The Co-Occurrence of Autism Spectrum Disorder in Children With ADHD. J. Atten. Disord. 2020, 24, 94–103. [Google Scholar] [CrossRef]
- Bell, L.; Wittkowski, A.; Hare, D.J. Movement Disorders and Syndromic Autism: A Systematic Review. J. Autism Dev. Disord. 2019, 49, 54–67. [Google Scholar] [CrossRef] [PubMed]
- Torres, E.B. Reframing Psychiatry for Precision Medicine. J. Pers. Med. 2020, 10, 144. [Google Scholar] [CrossRef] [PubMed]
- Hagerman, R.; Hoem, G.; Hagerman, P. Fragile X and autism: Intertwined at the molecular level leading to targeted treatments. Mol. Autism 2010, 1, 12. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.W.; Berry-Kravis, E.; Hagerman, R.J. Fragile X: Leading the way for targeted treatments in autism. Neurotherapeutics 2010, 7, 264–274. [Google Scholar] [CrossRef]
- Torres, E.B.; Brincker, M.; Isenhower, R.W.; Yanovich, P.; Stigler, K.A.; Nurnberger, J.I.; Metaxas, D.N.; Jose, J.V. Autism: The micro-movement perspective. Front. Integr. Neurosci. 2013, 7, 32. [Google Scholar] [CrossRef] [PubMed]
- Torres, E.B. Atypical signatures of motor variability found in an individual with ASD. Neurocase 2011, 19, 150–165. [Google Scholar] [CrossRef] [PubMed]
- Torres, E.B.; Caballero, C.; Mistry, S. Aging with Autism Departs Greatly from Typical Aging. Sensors 2020, 20, 572. [Google Scholar] [CrossRef]
- Torres, E.B.; Whyatt, C. Autism: The Movement Sensing Perspective; CRC Press/Taylor & Francis Group: Boca Raton, FL, USA, 2018; p. xviii. 386p. [Google Scholar]
- Whyatt, C.P.; Torres, E.B. Autism Research: An Objective Quantitative Review of Progress and Focus Between 1994 and 2015. Front. Psychol. 2018, 9, 1526. [Google Scholar] [CrossRef] [PubMed]
- Hawgood, S.; Hook-Barnard, I.G.; O’Brien, T.C.; Yamamoto, K.R. Precision medicine: Beyond the inflection point. Sci. Transl. Med. 2015, 7, 300ps317. [Google Scholar] [CrossRef] [PubMed]
- Scott, W.K.; Nance, M.A.; Watts, R.L.; Hubble, J.P.; Koller, W.C.; Lyons, K.; Pahwa, R.; Stern, M.B.; Colcher, A.; Hiner, B.C.; et al. Complete genomic screen in Parkinson disease: Evidence for multiple genes. JAMA 2001, 286, 2239–2244. [Google Scholar] [CrossRef] [PubMed]
- Klein, C.; Westenberger, A. Genetics of Parkinson’s disease. Cold Spring Harb. Perspect. Med. 2012, 2, a008888. [Google Scholar] [CrossRef] [PubMed]
- Starkstein, S.; Gellar, S.; Parlier, M.; Payne, L.; Piven, J. High rates of parkinsonism in adults with autism. J. Neurodev. Disord. 2015, 7, 29. [Google Scholar] [CrossRef] [PubMed]
- Pahlman, M.; Gillberg, C.; Wentz, E.; Himmelmann, K. Autism spectrum disorder and attention-deficit/hyperactivity disorder in children with cerebral palsy: Results from screening in a population-based group. Eur. Child. Adolesc. Psychiatry 2020, 29, 1569–1579. [Google Scholar] [CrossRef] [PubMed]
- Riccio, M.P.; Siracusano, R.; d’Alessandro, I.; Marino, M.; Bravaccio, C. Dystonic Movement Disorder as Symptom of Catatonia in Autism Spectrum Disorder. Case Rep. Psychiatry 2020, 2020, 8832075. [Google Scholar] [CrossRef]
- Dell’Osso, L.; Carpita, B.; Cremone, I.M.; Gesi, C.; D’Ermo, A.; De Iorio, G.; Massimetti, G.; Aguglia, E.; Bucci, P.; Carpiniello, B.; et al. Autism spectrum in patients with schizophrenia: Correlations with real-life functioning, resilience, and coping styles. CNS Spectr. 2021, 1–11. [Google Scholar] [CrossRef]
- Gecz, J.; Berry, J.G. Cerebral palsy with autism and ADHD: Time to pay attention. Dev. Med. Child. Neurol. 2021, 63, 247–248. [Google Scholar] [CrossRef]
- Ghaziuddin, M.; Ghaziuddin, N. Bipolar Disorder and Psychosis in Autism. Psychiatr. Clin. 2021, 44, 1–9. [Google Scholar] [CrossRef]
- Pan, P.Y.; Bolte, S.; Kaur, P.; Jamil, S.; Jonsson, U. Neurological disorders in autism: A systematic review and meta-analysis. Autism 2021, 25, 812–830. [Google Scholar] [CrossRef]
- Torres, E.B.; Yanovich, P.; Metaxas, D.N. Give spontaneity and self-discovery a chance in ASD: Spontaneous peripheral limb variability as a proxy to evoke centrally driven intentional acts. Front. Integr. Neurosci. 2013, 7, 46. [Google Scholar] [CrossRef]
- Wu, D.; Jose, J.V.; Nurnberger, J.I.; Torres, E.B. A Biomarker Characterizing Neurodevelopment with applications in Autism. Sci. Rep. 2018, 8, 614. [Google Scholar] [CrossRef] [PubMed]
- Caballero, C.; Mistry, S.; Vero, J.; Torres, E.B. Characterization of Noise Signatures of Involuntary Head Motion in the Autism Brain Imaging Data Exchange Repository. Front. Integr. Neurosci. 2018, 12, 7. [Google Scholar] [CrossRef] [PubMed]
- Caballero, C.; Mistry, S.; Torres, E.B. Age-Dependent Statistical Changes of Involuntary Head Motion Signatures Across Autism and Controls of the ABIDE Repository. Front. Integr. Neurosci. 2020, 14, 23. [Google Scholar] [CrossRef]
- Consortium, G.T. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 2020, 369, 1318–1330. [Google Scholar] [CrossRef] [PubMed]
- Torres, E.B.; Isenhower, R.W.; Nguyen, J.; Whyatt, C.; Nurnberger, J.I.; Jose, J.V.; Silverstein, S.M.; Papathomas, T.V.; Sage, J.; Cole, J. Toward Precision Psychiatry: Statistical Platform for the Personalized Characterization of Natural Behaviors. Front. Neurol. 2016, 7, 8. [Google Scholar] [CrossRef]
- Haruvi-Lamdan, N.; Horesh, D.; Golan, O. PTSD and autism spectrum disorder: Co-morbidity, gaps in research, and potential shared mechanisms. Psychol. Trauma 2018, 10, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Pettersson, E.; Lichtenstein, P.; Larsson, H.; Song, J.; Attention Deficit/Hyperactivity Disorder Working Group of the iPSYCH-Broad-PGC Consortium; Autism Spectrum Disorder Working Group of the iPSYCH-Broad-PGC Consortium; Agrawal, A.; Borglum, A.D.; Bulik, C.M.; Daly, M.J.; et al. Genetic influences on eight psychiatric disorders based on family data of 4 408 646 full and half-siblings, and genetic data of 333 748 cases and controls. Psychol. Med. 2019, 49, 1166–1173. [Google Scholar] [CrossRef] [PubMed]
- Monge, G. Memoire sur la Theorie des Deblais et des Remblais. In Histoire de l’ Academie Royale des Science; Avec les Memoires de Mathematique et de Physique; De L’imprimerie Royale: Paris, France, 1781. [Google Scholar]
- Rubner, Y.; Tomasi, C.; Guibas, L.J. Metric for Distributions with Applications to Image Databases. In Proceedings of the ICCV, Bombay, India, 4–7 January 1998. [Google Scholar]
- Stolfi, J.; Guibas, L.J. The earth mover’s distance as a metric for image retrieval. Int. J. Comput. Vis. 2000, 40, 99–121. [Google Scholar]
- Hoover, D.W.; Kaufman, J. Adverse childhood experiences in children with autism spectrum disorder. Curr. Opin. Psychiatry 2018, 31, 128–132. [Google Scholar] [CrossRef]
- Stack, A.; Lucyshyn, J. Autism Spectrum Disorder and the Experience of Traumatic Events: Review of the Current Literature to Inform Modifications to a Treatment Model for Children with Autism. J. Autism Dev. Disord. 2019, 49, 1613–1625. [Google Scholar] [CrossRef]
- Yousef Yengej, F.A.; van Royen-Kerkhof, A.; Derksen, R.; Fritsch-Stork, R.D.E. The development of offspring from mothers with systemic lupus erythematosus. A systematic review. Autoimmun. Rev. 2017, 16, 701–711. [Google Scholar] [CrossRef]
- Zhu, Z.; Tang, S.; Deng, X.; Wang, Y. Maternal Systemic Lupus Erythematosus, Rheumatoid Arthritis, and Risk for Autism Spectrum Disorders in Offspring: A Meta-analysis. J. Autism Dev. Disord. 2020, 50, 2852–2859. [Google Scholar] [CrossRef]
- Blaxill, M.; Rogers, T.; Nevison, C. Autism Tsunami: The Impact of Rising Prevalence on the Societal Cost of Autism in the United States. J. Autism Dev. Disord. 2021. [Google Scholar] [CrossRef]
- Bradbury, K.R.; Anderberg, E.I.; Huang-Storms, L.; Vasile, I.; Greene, R.K.; Duvall, S.W. Co-occurring Down Syndrome and Autism Spectrum Disorder: Cognitive, Adaptive, and Behavioral Characteristics. J. Autism Dev. Disord. 2021. [Google Scholar] [CrossRef]
- Salcedo-Arellano, M.J.; Cabal-Herrera, A.M.; Punatar, R.H.; Clark, C.J.; Romney, C.A.; Hagerman, R.J. Overlapping Molecular Pathways Leading to Autism Spectrum Disorders, Fragile X Syndrome, and Targeted Treatments. Neurotherapeutics 2021, 18, 265–283. [Google Scholar] [CrossRef]
- Tylee, D.S.; Sun, J.; Hess, J.L.; Tahir, M.A.; Sharma, E.; Malik, R.; Worrall, B.B.; Levine, A.J.; Martinson, J.J.; Nejentsev, S.; et al. Genetic correlations among psychiatric and immune-related phenotypes based on genome-wide association data. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2018, 177, 641–657. [Google Scholar] [CrossRef] [PubMed]
- Swanberg, S.E.; Nagarajan, R.P.; Peddada, S.; Yasui, D.H.; LaSalle, J.M. Reciprocal co-regulation of EGR2 and MECP2 is disrupted in Rett syndrome and autism. Hum. Mol. Genet. 2009, 18, 525–534. [Google Scholar] [CrossRef]
- Platt, R.J.; Zhou, Y.; Slaymaker, I.M.; Shetty, A.S.; Weisbach, N.R.; Kim, J.A.; Sharma, J.; Desai, M.; Sood, S.; Kempton, H.R.; et al. Chd8 Mutation Leads to Autistic-like Behaviors and Impaired Striatal Circuits. Cell Rep. 2017, 19, 335–350. [Google Scholar] [CrossRef] [PubMed]
- Talkowski, M.E.; Rosenfeld, J.A.; Blumenthal, I.; Pillalamarri, V.; Chiang, C.; Heilbut, A.; Ernst, C.; Hanscom, C.; Rossin, E.; Lindgren, A.M.; et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell 2012, 149, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Mondal, K.; Ramachandran, D.; Patel, V.C.; Hagen, K.R.; Bose, P.; Cutler, D.J.; Zwick, M.E. Excess variants in AFF2 detected by massively parallel sequencing of males with autism spectrum disorder. Hum. Mol. Genet. 2012, 21, 4356–4364. [Google Scholar] [CrossRef]
- Hiatt, S.M.; Thompson, M.L.; Prokop, J.W.; Lawlor, J.M.J.; Gray, D.E.; Bebin, E.M.; Rinne, T.; Kempers, M.; Pfundt, R.; van Bon, B.W.; et al. Deleterious Variation in BRSK2 Associates with a Neurodevelopmental Disorder. Am. J. Hum. Genet. 2019, 104, 701–708. [Google Scholar] [CrossRef]
- Ciammola, A.; Carrera, P.; Di Fonzo, A.; Sassone, J.; Villa, R.; Poletti, B.; Ferrari, M.; Girotti, F.; Monfrini, E.; Buongarzone, G.; et al. X-linked Parkinsonism with Intellectual Disability caused by novel mutations and somatic mosaicism in RAB39B gene. Parkinsonism Relat. Disord. 2017, 44, 142–146. [Google Scholar] [CrossRef] [PubMed]
- Giannandrea, M.; Bianchi, V.; Mignogna, M.L.; Sirri, A.; Carrabino, S.; D’Elia, E.; Vecellio, M.; Russo, S.; Cogliati, F.; Larizza, L.; et al. Mutations in the small GTPase gene RAB39B are responsible for X-linked mental retardation associated with autism, epilepsy, and macrocephaly. Am. J. Hum. Genet. 2010, 86, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Lesage, S.; Bras, J.; Cormier-Dequaire, F.; Condroyer, C.; Nicolas, A.; Darwent, L.; Guerreiro, R.; Majounie, E.; Federoff, M.; Heutink, P.; et al. Loss-of-function mutations in RAB39B are associated with typical early-onset Parkinson disease. Neurol. Genet. 2015, 1, e9. [Google Scholar] [CrossRef] [PubMed]
- Wilson, G.R.; Sim, J.C.; McLean, C.; Giannandrea, M.; Galea, C.A.; Riseley, J.R.; Stephenson, S.E.; Fitzpatrick, E.; Haas, S.A.; Pope, K.; et al. Mutations in RAB39B cause X-linked intellectual disability and early-onset Parkinson disease with alpha-synuclein pathology. Am. J. Hum. Genet. 2014, 95, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Hansen, F.H.; Skjorringe, T.; Yasmeen, S.; Arends, N.V.; Sahai, M.A.; Erreger, K.; Andreassen, T.F.; Holy, M.; Hamilton, P.J.; Neergheen, V.; et al. Missense dopamine transporter mutations associate with adult parkinsonism and ADHD. J. Clin. Investig. 2014, 124, 3107–3120. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, P.J.; Campbell, N.G.; Sharma, S.; Erreger, K.; Herborg Hansen, F.; Saunders, C.; Belovich, A.N.; Consortium, N.A.A.S.; Sahai, M.A.; Cook, E.H.; et al. De novo mutation in the dopamine transporter gene associates dopamine dysfunction with autism spectrum disorder. Mol. Psychiatry 2013, 18, 1315–1323. [Google Scholar] [CrossRef] [PubMed]
- Doege, C.A.; Inoue, K.; Yamashita, T.; Rhee, D.B.; Travis, S.; Fujita, R.; Guarnieri, P.; Bhagat, G.; Vanti, W.B.; Shih, A.; et al. Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature 2012, 488, 652–655. [Google Scholar] [CrossRef] [PubMed]
- Guallar, D.; Bi, X.; Pardavila, J.A.; Huang, X.; Saenz, C.; Shi, X.; Zhou, H.; Faiola, F.; Ding, J.; Haruehanroengra, P.; et al. RNA-dependent chromatin targeting of TET2 for endogenous retrovirus control in pluripotent stem cells. Nat. Genet. 2018, 50, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Shovlin, C.L.; Simmonds, H.A.; Fairbanks, L.D.; Deacock, S.J.; Hughes, J.M.; Lechler, R.I.; Webster, A.D.; Sun, X.M.; Webb, J.C.; Soutar, A.K. Adult onset immunodeficiency caused by inherited adenosine deaminase deficiency. J. Immunol. 1994, 153, 2331–2339. [Google Scholar] [PubMed]
- Hla, T.; Neilson, K. Human cyclooxygenase-2 cDNA. Proc. Natl. Acad. Sci. USA 1992, 89, 7384–7388. [Google Scholar] [CrossRef] [PubMed]
- Samad, T.A.; Moore, K.A.; Sapirstein, A.; Billet, S.; Allchorne, A.; Poole, S.; Bonventre, J.V.; Woolf, C.J. Interleukin-1beta-mediated induction of Cox-2 in the CNS contributes to inflammatory pain hypersensitivity. Nature 2001, 410, 471–475. [Google Scholar] [CrossRef] [PubMed]
- Tamura, S.; Morikawa, Y.; Iwanishi, H.; Hisaoka, T.; Senba, E. Foxp1 gene expression in projection neurons of the mouse striatum. Neuroscience 2004, 124, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Hisaoka, T.; Nakamura, Y.; Senba, E.; Morikawa, Y. The forkhead transcription factors, Foxp1 and Foxp2, identify different subpopulations of projection neurons in the mouse cerebral cortex. Neuroscience 2010, 166, 551–563. [Google Scholar] [CrossRef]
- Precious, S.V.; Kelly, C.M.; Reddington, A.E.; Vinh, N.N.; Stickland, R.C.; Pekarik, V.; Scherf, C.; Jeyasingham, R.; Glasbey, J.; Holeiter, M.; et al. FoxP1 marks medium spiny neurons from precursors to maturity and is required for their differentiation. Exp. Neurol. 2016, 282, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Zhou, Y.; Wang, K. Multiplex gene and phenotype network to characterize shared genetic pathways of epilepsy and autism. Sci. Rep. 2021, 11, 952. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Li, Y. Genetic associations between voltage-gated calcium channels and autism spectrum disorder: A systematic review. Mol. Brain 2020, 13, 96. [Google Scholar] [CrossRef] [PubMed]
- Yoshino, Y.; Roy, B.; Dwivedi, Y. Altered miRNA landscape of the anterior cingulate cortex is associated with potential loss of key neuronal functions in depressed brain. Eur. Neuropsychopharmacol. 2020, 40, 70–84. [Google Scholar] [CrossRef] [PubMed]
- Bottema-Beutel, K.; Crowley, S.; Sandbank, M.; Woynaroski, T.G. Adverse event reporting in intervention research for young autistic children. Autism 2021, 25, 322–335. [Google Scholar] [CrossRef] [PubMed]
- Thompson-Hodgetts, S.; Labonte, C.; Mazumder, R.; Phelan, S. Helpful or harmful? A scoping review of perceptions and outcomes of autism diagnostic disclosure to others. Res. Autism Spectr. Disord. 2020, 77, 101598. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Chen, Y.L.; Gau, S.S. Suicidality in Children with Elevated Autistic Traits. Autism Res. 2020, 13, 1811–1821. [Google Scholar] [CrossRef] [PubMed]
- Costa, A.P.; Loor, C.; Steffgen, G. Suicidality in Adults with Autism Spectrum Disorder: The Role of Depressive Symptomatology, Alexithymia, and Antidepressants. J. Autism Dev. Disord. 2020, 50, 3585–3597. [Google Scholar] [CrossRef]
- Hand, B.N.; Benevides, T.W.; Carretta, H.J. Suicidal Ideation and Self-inflicted Injury in Medicare Enrolled Autistic Adults with and Without Co-occurring Intellectual Disability. J. Autism Dev. Disord. 2020, 50, 3489–3495. [Google Scholar] [CrossRef]
- Broderick, A.A.; Roscigno, R. Autism, Inc.: The Autism Industrial Complex. J. Disabil. Stud. Educ. 2021, 1–25. [Google Scholar] [CrossRef]
- Bottema-Beutel, K.; Crowley, S.; Sandbank, M.; Woynaroski, T.G. Research Review: Conflicts of Interest (COIs) in autism early intervention research—A meta-analysis of COI influences on intervention effects. J. Child. Psychol. Psychiatry 2021, 62, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Dawson, M.; Fletcher-Watson, S. Commentary: What conflicts of interest tell us about autism intervention research-a commentary on Bottema-Beutel et al. J. Child Psychol. Psychiatry 2021, 62, 16–18. [Google Scholar] [CrossRef] [PubMed]
- Sandbank, M.; Bottema-Beutel, K.; Crowley, S.; Cassidy, M.; Dunham, K.; Feldman, J.I.; Crank, J.; Albarran, S.A.; Raj, S.; Mahbub, P.; et al. Project AIM: Autism intervention meta-analysis for studies of young children. Psychol. Bull. 2020, 146, 1–29. [Google Scholar] [CrossRef]
- Carroll, L.; Braeutigam, S.; Dawes, J.M.; Krsnik, Z.; Kostovic, I.; Coutinho, E.; Dewing, J.M.; Horton, C.A.; Gomez-Nicola, D.; Menassa, D.A. Autism Spectrum Disorders: Multiple Routes to, and Multiple Consequences of, Abnormal Synaptic Function and Connectivity. Neuroscientist 2021, 27, 10–29. [Google Scholar] [CrossRef] [PubMed]
- Torres, E.B. Objective Biometric Methods for the Diagnosis and Treatment of Nervous System Disorders; Academic Press: London, UK, 2018; 568p. [Google Scholar]

| Neuropsychiatric Condition | Number of Genes in DISGENET | Number of Genes Shared with SFARI ASD-Linked Genes | % (Relative to DisGeNET, Relative to 906 SFARI Genes) |
|---|---|---|---|
| Schizophrenia | 2697 | 336 | (24.58, 37.08) |
| ADHD | 795 | 188 | (23.64, 20.75) |
| Depression | 1407 | 158 | (11.22, 17.43) |
| PTSD | 395 | 55 | (13.92, 6.07) |
| Bipolar Depression | 116 | 33 | (28.34, 3.6) |
| Neurological Disorder | Number of Genes Reported in DISGENET | Number of Genes Shared with SFARI ASD-Linked Genes | % (Relative to DisGeNET, Relative to 906 SFARI Genes) |
|---|---|---|---|
| Parkinson’s | 1975 | 164 | (8.3, 18.10) |
| Ataxia Autosomal Dominant | 812 | 98 | (12.06, 10.81) |
| FX | 72 | 72 | (100.0, 7.94) |
| Dystonia | 419 | 65 | (15.51, 7.17) |
| Ataxia Autosomal Recessive | 420 | 61 | (14.52, 6.73) |
| OCD | 167 | 39 | (23.35, 4.30) |
| Tourette’s | 171 | 35 | (20.46, 3.86) |
| CP | 227 | 34 | (14.97, 3.75) |
| X-Ataxia | 158 | 22 | (13.92, 2.42) |
| FXTAS | 63 | 22 | (34.9, 2.42) |
| Progressive Cerebellar Ataxia | 134 | 13 | (9.70, 1.43) |
| Genes Common to ASD and Neuro-Disorders | Tissue with Max Expression |
|---|---|
| ACTB | Liver |
| AFF2 | Adipose Subcutaneous, Adipose Visceral Omentum, AdrenalGland, Artery Aorta, Artery Coronary, Artery Tibial, Bladder, Breast Mammary Tissue, Cervix Ectocervix, Cervix Endocervix, Colon Sigmoid, Colon Transverse, Esophagus Gastro esophageal Junction, Esophagus Muscularis, Fallopian Tube, Heart AtrialAppendage, Kidney Cortex, Kidney Medulla, Lung, Minor Salivary Gland, Nerve Tibial, Pituitary, Prostate, Small Int ileum, Spleen, Stomach, Thyroid, Uterus, Vagina |
| AKAP9 | Whole Blood |
| ALDH5A1 | Esophagus Mucosa |
| ATP2B2 | Heart Left, Ventricle, Ovary, Cerebellar Hemi, Cerebellum |
| AVPR1A | Skeletal Muscle |
| CACNA1E | Ant Cingulate Cortex |
| CHD7 | Pancreas, Skin not Sun Exposed Suprapubic, Skin Sun Exposed Lower Leg |
| CREBBP | Cortex, Frontal Cortex, Fibroblasts, Cell-Lymphocytes |
| FOXP1 | Nucleus Accumbens of the Basal Ganglia (BG) |
| MECP2 | Amygdala, Caudate-BG, Hippocampus, Hypothalamus, Putamen-BG, Spinal Cord-Cervical, Substantia Nigra |
| SMAD4 | Testis |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Torres, E.B. Precision Autism: Genomic Stratification of Disorders Making Up the Broad Spectrum May Demystify Its “Epidemic Rates”. J. Pers. Med. 2021, 11, 1119. https://doi.org/10.3390/jpm11111119
Torres EB. Precision Autism: Genomic Stratification of Disorders Making Up the Broad Spectrum May Demystify Its “Epidemic Rates”. Journal of Personalized Medicine. 2021; 11(11):1119. https://doi.org/10.3390/jpm11111119
Chicago/Turabian StyleTorres, Elizabeth B. 2021. "Precision Autism: Genomic Stratification of Disorders Making Up the Broad Spectrum May Demystify Its “Epidemic Rates”" Journal of Personalized Medicine 11, no. 11: 1119. https://doi.org/10.3390/jpm11111119
APA StyleTorres, E. B. (2021). Precision Autism: Genomic Stratification of Disorders Making Up the Broad Spectrum May Demystify Its “Epidemic Rates”. Journal of Personalized Medicine, 11(11), 1119. https://doi.org/10.3390/jpm11111119






