FANCA Polymorphism Is Associated with the Rate of Proliferation in Uterine Leiomyoma in Korea
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Subjects
2.2. Isolation of Uterine Leiomyoma Cells and Culture
2.3. Doubling Time Measurement
2.4. Preparation of Genomic DNA and Identification of SNP
2.5. Statistical Analysis
3. Results
3.1. Anthropometrics, Comorbidities, and Blood Chemistry
3.2. Association of rs2239359 SNP in FANCA with the Proliferation Rate of Leiomyoma
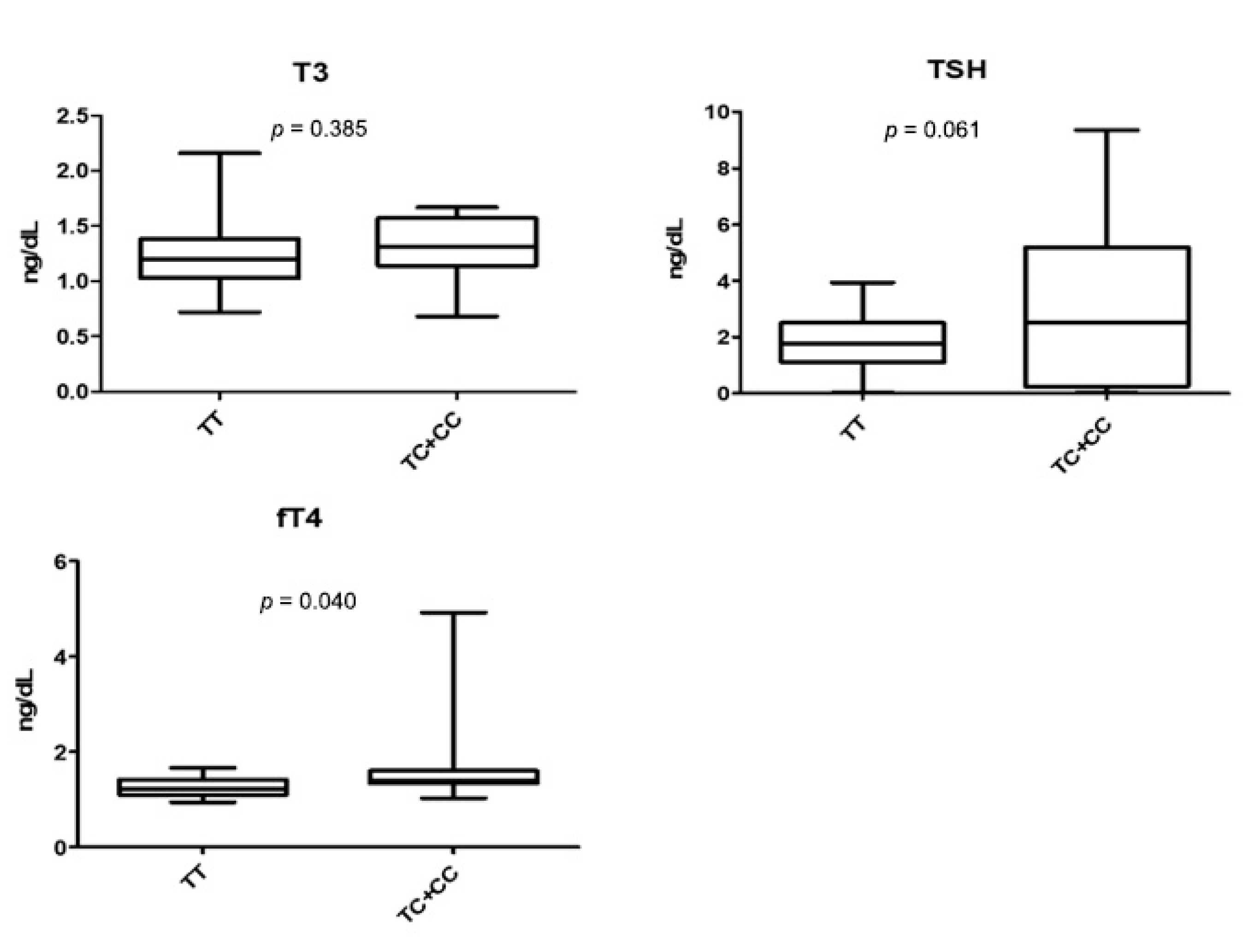
3.3. rs2239359 in FANCA and Thyroid Function
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Buttram, V.C., Jr.; Reiter, R.C. Uterine leiomyomata: Etiology, symptomatology, and management. Fertil. Steril. 1981, 36, 433–445. [Google Scholar] [PubMed]
- Kolankaya, A.; Arici, A. Myomas and assisted reproductive technologies: When and how to act? Obstet. Gynecol. Clin. N. Am. 2006, 33, 145–152. [Google Scholar] [CrossRef]
- Age of Mothers at Childbirth and Age-Specific Fertility. Available online: http://www.oecd.org/els/soc/SF_2_3_Age_mothers_childbirth.pdf (accessed on 6 September 2018).
- Dokal, I. Fanconi’s anaemia and related bone marrow failure syndromes. Br. Med. Bull. 2006, 77, 37–53. [Google Scholar] [CrossRef] [PubMed]
- Reuter, T.Y.; Medhurst, A.L.; Waisfisz, Q.; Zhi, Y.; Herterich, S.; Hoehn, H.; Gross, H.J.; Joenje, H.; Hoatlin, M.E.; Mathew, C.G.; et al. Yeast two-hybrid screens imply involvement of Fanconi anemia proteins in transcription regulation, cell signaling, oxidative metabolism, and cellular transport. Exp. Cell Res. 2003, 289, 211–221. [Google Scholar] [CrossRef]
- Yin, J.; Liu, H.; Liu, Z.; Wang, L.E.; Chen, W.V.; Zhu, D.; Amos, C.I.; Fang, S.; Lee, J.E.; Wei, Q. Genetic variants in fanconi anemia pathway genes BRCA2 and FANCA predict melanoma survival. J. Invest. Dermatol. 2015, 135, 542–550. [Google Scholar] [CrossRef]
- Juko-Pecirep, I.; Ivansson, E.L.; Gyllensten, U.B. Evaluation of Fanconi anaemia genes FANCA, FANCC and FANCL in cervical cancer susceptibility. Gynecol. Oncol. 2011, 122, 377–381. [Google Scholar] [CrossRef]
- Haiman, C.A.; Hsu, C.; De Bakker, P.I.; Frasco, M.; Sheng, X.; Van Den Berg, D.; Casagrande, J.T.; Kolonel, L.N.; Le Marchand, L.; Hankinson, S.E.; et al. Comprehensive association testing of common genetic variation in DNA repair pathway genes in relationship with breast cancer risk in multiple populations. Hum. Mol. Genet. 2008, 17, 825–834. [Google Scholar] [CrossRef] [PubMed]
- Michiels, S.; Laplanche, A.; Boulet, T.; Dessen, P.; Guillonneau, B.; Méjean, A.; Desgrandchamps, F.; Lathrop, M.; Sarasin, A.; Benhamou, S. Genetic polymorphisms in 85 DNA repair genes and bladder cancer risk. Carcinogenesis 2009, 30, 763–768. [Google Scholar] [CrossRef]
- Pyun, J.A.; Kim, S.; Cha, D.H.; Kwack, K. Polymorphisms within the FANCA gene associate with premature ovarian failure in Korean women. Menopause 2014, 21, 530–533. [Google Scholar] [CrossRef]
- ClinVar: NM_000135.2(FANCA):c.1501G>A (p.Gly501Ser). Available online: https://www.ncbi.nlm.nih.gov/clinvar/variation/134244/ (accessed on 7 September 2018).
- van de Vrugt, H.J.; Grmpe, M. Fanconi anemia. In Inborn Errors of Development, 2nd ed.; Epstein, C.J., Erickson, R.P., Erickson, A., Eds.; Oxford University Press: New York, NY, USA, 2008; pp. 1230–1236. [Google Scholar]
- Wong, J.C.; Alon, N.; McKerlie, C.; Huang, J.R.; Meyn, M.S.; Buchwald, M. Targeted disruption of exons 1 to 6 of the Fanconi Anemia group A gene leads to growth retardation, strain-specific microphthalmia, meiotic defects and primordial germ cell hypoplasia. Hum. Mol. Genet. 2003, 12, 2063–2076. [Google Scholar] [CrossRef]
- Giri, N.; Batista, D.L.; Alter, B.P.; Stratakis, C.A. Endocrine abnormalities in patients with Fanconi anemia. J. Clin. Endocrinol. Metab. 2007, 92, 2624–2631. [Google Scholar] [CrossRef] [PubMed]
- Alter, B.P.; Frissora, C.L.; Halpéirin, D.S.; Freedman, M.H.; Chitkara, U.; Alvarez, E.; Lynch, L.; Adler-Brecher, B.; Auerbach, A.D. Fanconi’s anaemia and pregnancy. Br. J. Haematol. 1991, 77, 410–418. [Google Scholar] [CrossRef] [PubMed]
- Cheng, N.C.; van de Vrugt, H.J.; van der Valk, M.A.; Oostra, A.B.; Krimpenfort, P.; de Vries, Y.; Joenje, H.; Berns, A.; Arwert, F. Mice with a targeted disruption of the Fanconi anemia homolog Fanca. Hum. Mol. Genet. 2000, 9, 1805–1811. [Google Scholar] [CrossRef]
- Noll, M.; Battaile, K.P.; Bateman, R.; Lax, T.P.; Rathbun, K.; Reifsteck, C.; Bagby, G.; Finegold, M.; Olson, S.; Grompe, M. Fanconi anemia group A and C double-mutant mice: Functional evidence for a multi-protein Fanconi anemia complex. Exp. Hematol. 2002, 30, 679–688. [Google Scholar] [CrossRef]
- Cirilo, P.D.R.; Marchi, F.A.; de Camargo Barros Filho, M.; Rocha, R.M.; Domingues, M.A.C.; Jurisica, I.; Pontes, A.; Rogatto, S.R. An integrative genomic and transcriptomic analysis reveals potential targets associated with cell proliferation in uterine leiomyomas. PLoS ONE 2013, 8, e57901. [Google Scholar] [CrossRef] [PubMed]
- Välimäki, N.; Kuisma, H.; Pasanen, A.; Heikinheimo, O.; Sjöberg, J.; Bützow, R.; Sarvilinna, N.; Heinonen, H.R.; Tolvanen, J.; Bramante, S.; et al. Genetic predisposition to uterine leiomyoma is determined by loci for genitourinary development and genome stability. Elife 2018, 7. [Google Scholar] [CrossRef]
- Guleria, A.; Chandna, S. ATM kinase: Much more than a DNA damage responsive protein. DNA Repair 2016, 39, 1–20. [Google Scholar] [CrossRef]
- Lee, P.J.; Yoo, N.S.; Hagemann, I.S.; Pfeifer, J.D.; Cottrell, C.E.; Abel, H.J.; Duncavage, E.J. Spectrum of mutations in leiomyosarcomas identified by clinical targeted next-generation sequencing. Exp. Mol. Pathol. 2017, 102, 156–161. [Google Scholar] [CrossRef]
- Larder, R.; Karali, D.; Nelson, N.; Brown, P. Fanconi anemia A is a nucleocytoplasmic shuttling molecule required for gonadotropin-releasing hormone (GnRH) transduction of the GnRH receptor. Endocrinology 2006, 147, 5676–5689. [Google Scholar] [CrossRef]
- Borahay, M.A.; Al-Hendy, A.; Kilic, G.S.; Boehning, D. Signaling Pathways in Leiomyoma: Understanding Pathobiology and Implications for Therapy. Mol. Med. 2015, 21, 242–256. [Google Scholar] [CrossRef]
- Santin, A.P.; Furlanetto, T.W. Role of estrogen in thyroid function and growth regulation. J. Thyroid Res. 2011, 2011. [Google Scholar] [CrossRef] [PubMed]
- Kawabata, W.M.; Suzuki, T.; Moriya, T.; Fujimori, K.; Naganuma, H.; Inoue, S.; Kinouchi, Y.; Kameyama, K.; Takami, H.; Shimosegawa, T. Estrogen receptors (alpha and beta) and 17beta-hydroxysteroid dehydrogenase type 1 and 2 in thyroid disorders: Possible in situ estrogen synthesis and actions. Mod. Pathol. 2003, 16, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Han, E.J.; Song, H.D.; Yang, J.H.; Park, S.Y.; Kim, S.H.; Yoon, H.K.; Yim, C.H. Thyroid dysfunction associated with administration of the long-acting gonadotropin-releasing hormone agonist. Endocrinol. Metab. 2013, 28, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Kasayama, S.; Miyake, S.; Samejima, Y. Transient thyrotoxicosis and hypothyroidism following administration of the GnRH agonist leuprolide acetate. Endocr. J. 2000, 47, 783–785. [Google Scholar] [CrossRef][Green Version]

| Slow | Fast | p 1 | |
|---|---|---|---|
| N | 26 | 16 | |
| Anthropometric characteristics | |||
| Age (years) | 39.58 ± 7.22 | 42.75 ± 8.82 | 0.211 |
| Weight (kg) | 59.83 ± 7.74 | 58.36 ± 10.49 | 0.820 |
| Height (cm) | 158.28 ± 6.47 | 157.82 ± 6.13 | 0.606 |
| BMI (kg/m2) | 23.92 ± 3.20 | 23.45 ± 4.22 | 0.683 |
| Smoking (None/Yes) | 26/0 | 15/1 | 0.381 |
| Drinking (None/Yes) | 20/6 | 14/2 | 0.688 |
| Myoma number | 3.23 ± 3.02 | 2.88 ± 4.40 | 0.757 |
| Myoma size (mm) | 81.69 ± 24.26 | 92.06 ± 31.16 | 0.235 |
| Disease history | |||
| Recurrence of leiomyoma | 2 (7.7%) | 1 (6.3%) | 1.000 |
| Hypertension | 1 (3.8%) | 1 (6.3%) | 1.000 |
| Diabetes mellitus | 0 (0.0%) | 1 (6.3%) | 0.381 |
| Thyroid disease | 2 (7.7%) | 3 (18.8%) | 0.352 |
| hyperlipidemia | 0 (0.0%) | 1 (6.3%) | 0.381 |
| Previous operation | 2 (7.7%) | 1 (6.3%) | 1.000 |
| Slow | Fast | p | |
|---|---|---|---|
| WBC (103/μL) | 5.32 ± 1.33 | 5.89 ± 1.70 | 0.139 |
| RBC (106/μL) | 4.36 ± 0.33 | 4.27 ± 0.65 | 0.506 |
| Hb (g/dL) | 12.47 ± 1.30 | 12.06 ± 2.04 | 0.385 |
| HCT (%) | 38.57 ± 3.03 | 34.92 ± 6.34 | 0.085 |
| MCV (fL) | 89.02 ± 6.31 | 86.13 ± 7.28 | 0.209 |
| MCH (pg) | 28.65 ± 2.87 | 28.43 ± 3.65 | 0.825 |
| MCHC (g/dL) | 43.99 ± 58.62 | 32.89 ± 2.20 | 0.411 |
| Platelet (103/μL) | 291.76 ± 68.19 | 324.60 ± 103.04 | 0.341 |
| MPV (fL) | 10.35 ± 13.71 | 7.64 ± 1.01 | 0.489 |
| Neutrophil (%) | 56.90 ± 9.40 | 58.53 ± 12.74 | 0.578 |
| Lymphocyte (%) | 32.69 ± 8.78 | 36.87 ± 17.60 | 0.333 |
| Monocyte (%) | 5.49 ± 1.22 | 5.67 ± 1.67 | 0.596 |
| Eosinophil (%) | 2.52 ± 1.61 | 2.75 ± 2.68 | 0.728 |
| Basophil (%) | 0.64 ± 0.30 | 0.54 ± 0.31 | 0.311 |
| LUC (%) | 1.76 ± 0.53 | 1.66 ± 0.58 | 0.614 |
| aPTT (sec) | 27.72 ± 1.68 | 27.61 ± 2.35 | 0.914 |
| PT (INR) | 1.02 ± 0.05 | 1.00 ± 0.05 | 0.574 |
| Sodium (mmol/L) | 138.48 ± 1.76 | 138.20 ± 2.21 | 0.988 |
| Potassium (mmol/L) | 4.08 ± 0.37 | 4.13 ± 0.27 | 0.794 |
| Chloride (mmol/L) | 104.76 ± 1.51 | 98.51 ± 24.33 | 0.258 |
| Total Cholesterol (mg/dL) | 197.76 ± 33.17 | 193.67 ± 23.54 | 0.606 |
| Total calcium (mg/dL) | 13.00 ± 18.54 | 9.43 ± 0.35 | 0.403 |
| Phosphorus (mg/dL) | 3.65 ± 0.46 | 3.75 ± 0.50 | 0.885 |
| Glucose (mg/dL) | 97.52 ± 14.12 | 102.67 ± 28.00 | 0.756 |
| BUN (mg/dL) | 12.96 ± 3.13 | 12.80 ± 5.20 | 0.641 |
| Creatinine (mg/dL) | 0.62 ± 0.08 | 0.69 ± 0.21 | 0.268 |
| Total Protein (g/dL) | 7.36 ± 0.39 | 7.58 ± 0.46 | 0.201 |
| Albumin (g/dL) | 4.42 ± 0.21 | 4.43 ± 0.18 | 0.911 |
| Total Bilirubin (mg/dL) | 0.57 ± 0.15 | 0.60 ± 0.17 | 0.494 |
| ALP (U/L) | 54.44 ± 15.66 | 60.00 ± 12.21 | 0.613 |
| AST (U/L) | 19.36 ± 5.31 | 20.20 ± 5.07 | 0.554 |
| ALT (U/L) | 13.40 ± 7.16 | 13.47 ± 5.01 | 0.756 |
| eGFR (mL/min/1.73 m2) | 108.66 ± 17.46 | 101.21 ± 24.21 | 0.459 |
| T3 (ng/mL) | 1.25 ± 0.30 | 1.29 ± 0.27 | 0.554 |
| TSH (uIU/mL) | 1.96 ± 1.09 | 3.05 ± 2.84 | 0.219 |
| free T4 (ng/dL) | 1.43 ± 0.76 | 1.36 ± 0.17 | 0.670 |
| Slow Group | Fast Group | OR (95% CI) | p | ||
|---|---|---|---|---|---|
| allele | |||||
| T | 48 (88.89%) | 22(64.71%) | 4.04 (1.18–13.86) | 0.0266 | |
| C | 6 (11.11%) | 12(35.29%) | |||
| genotype | |||||
| TT | 21 (77.8%) | 6 (35.3%) | 6.44 (1.90–31.96) | 0.0231 | |
| TC+CC | 6 (22.2%) | 11(64.7%) |
| TT | TC + CC | p | |
|---|---|---|---|
| WBC (103/μL) | 5.57 ± 1.49 | 5.47 ± 1.53 | 0.897 |
| RBC (106/μL) | 4.31 ± 0.51 | 4.35 ± 0.40 | 0.347 |
| Hb (g/dL) | 12.45 ± 1.66 | 12.09 ± 1.54 | 0.884 |
| HCT (%) | 37.50 ± 5.20 | 36.70 ± 4.26 | 0.737 |
| MCV (fL) | 88.74 ± 5.88 | 86.60 ± 8.03 | 0.368 |
| MCH (pg) | 28.90 ± 2.62 | 28.02 ± 3.89 | 0.334 |
| MCHC (g/dL) | 44.25 ± 58.56 | 32.47 ± 2.48 | 0.244 |
| Platelet (103/μL) | 297.00 ± 83.72 | 315.87 ± 84.01 | 0.507 |
| MPV (fL) | 10.36 ± 13.71 | 7.62 ± 1.09 | 0.542 |
| Neutrophil (%) | 56.06 ± 7.44 | 59.92 ± 14.52 | 0.226 |
| Lymphocyte (%) | 33.64 ± 7.06 | 35.28 ± 19.14 | 0.754 |
| Monocyte (%) | 5.50 ± 1.21 | 5.65 ± 1.67 | 0.799 |
| Eosinophil (%) | 2.42 ± 1.53 | 2.90 ± 2.74 | 0.920 |
| Basophil (%) | 0.63 ± 0.31 | 0.55 ± 0.30 | 0.494 |
| LUC (%) | 1.74 ± 0.48 | 1.69 ± 0.67 | 0.519 |
| aPTT (sec) | 27.87 ± 1.76 | 27.37 ± 2.21 | 0.922 |
| PT (INR) | 1.02 ± 0.05 | 1.00 ± 0.03 | 0.485 |
| Sodium (mmol/L) | 138.52 ± 1.73 | 138.13 ± 2.23 | 0.937 |
| Potassium (mmol/L) | 4.04 ± 0.26 | 4.21 ± 0.42 | 0.151 |
| Chloride (mmol/L) | 104.84 ± 1.31 | 98.38 ± 24.31 | 0.335 |
| Total Cholesterol (mg/dL) | 198.68 ± 31.88 | 192.13 ± 26.08 | 0.481 |
| Total calcium (mg/dL) | 13.02 ± 18.54 | 9.39 ± 0.36 | 0.261 |
| Phosphorus (mg/dL) | 3.61 ± 0.44 | 3.82 ± 0.52 | 0.428 |
| Glucose (mg/dL) | 97.16 ± 15.09 | 103.27 ± 26.98 | 0.724 |
| BUN (mg/dL) | 13.12 ± 3.19 | 12.53 ± 5.11 | 0.728 |
| Creatinine (mg/dL) | 0.64 ± 0.09 | 0.66 ± 0.21 | 0.707 |
| Total Protein (g/dL) | 7.35 ± 0.39 | 7.61 ± 0.45 | 0.076 |
| Albumin (g/dL) | 4.40 ± 0.19 | 4.47 ± 0.21 | 0.204 |
| Total Bilirubin (mg/dL) | 0.57 ± 0.16 | 0.60 ± 0.16 | 0.417 |
| ALP (U/L) | 55.60 ± 16.68 | 58.07 ± 10.44 | 0.666 |
| AST (U/L) | 19.12 ± 4.59 | 20.60 ± 6.09 | 0.869 |
| ALT (U/L) | 13.36 ± 7.42 | 13.53 ± 4.31 | 0.509 |
| eGFR (mL/min/1.73 m2) | 106.60 ± 17.28 | 104.65 ± 25.13 | 0.858 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ha, E.; Lee, S.; Lee, S.M.; Jung, J.; Chung, H.; Choi, E.; Kwon, S.Y.; Cha, M.H.; Shin, S.-J. FANCA Polymorphism Is Associated with the Rate of Proliferation in Uterine Leiomyoma in Korea. J. Pers. Med. 2020, 10, 228. https://doi.org/10.3390/jpm10040228
Ha E, Lee S, Lee SM, Jung J, Chung H, Choi E, Kwon SY, Cha MH, Shin S-J. FANCA Polymorphism Is Associated with the Rate of Proliferation in Uterine Leiomyoma in Korea. Journal of Personalized Medicine. 2020; 10(4):228. https://doi.org/10.3390/jpm10040228
Chicago/Turabian StyleHa, Eunyoung, Seungmee Lee, So Min Lee, Jeeyeon Jung, Hyewon Chung, Eunsom Choi, Sun Young Kwon, Min Ho Cha, and So-Jin Shin. 2020. "FANCA Polymorphism Is Associated with the Rate of Proliferation in Uterine Leiomyoma in Korea" Journal of Personalized Medicine 10, no. 4: 228. https://doi.org/10.3390/jpm10040228
APA StyleHa, E., Lee, S., Lee, S. M., Jung, J., Chung, H., Choi, E., Kwon, S. Y., Cha, M. H., & Shin, S.-J. (2020). FANCA Polymorphism Is Associated with the Rate of Proliferation in Uterine Leiomyoma in Korea. Journal of Personalized Medicine, 10(4), 228. https://doi.org/10.3390/jpm10040228






