Association of UBE3C Variants with Reduced Kidney Function in Patients with Diabetic Kidney Disease
Abstract
1. Introduction
2. Materials and Methods
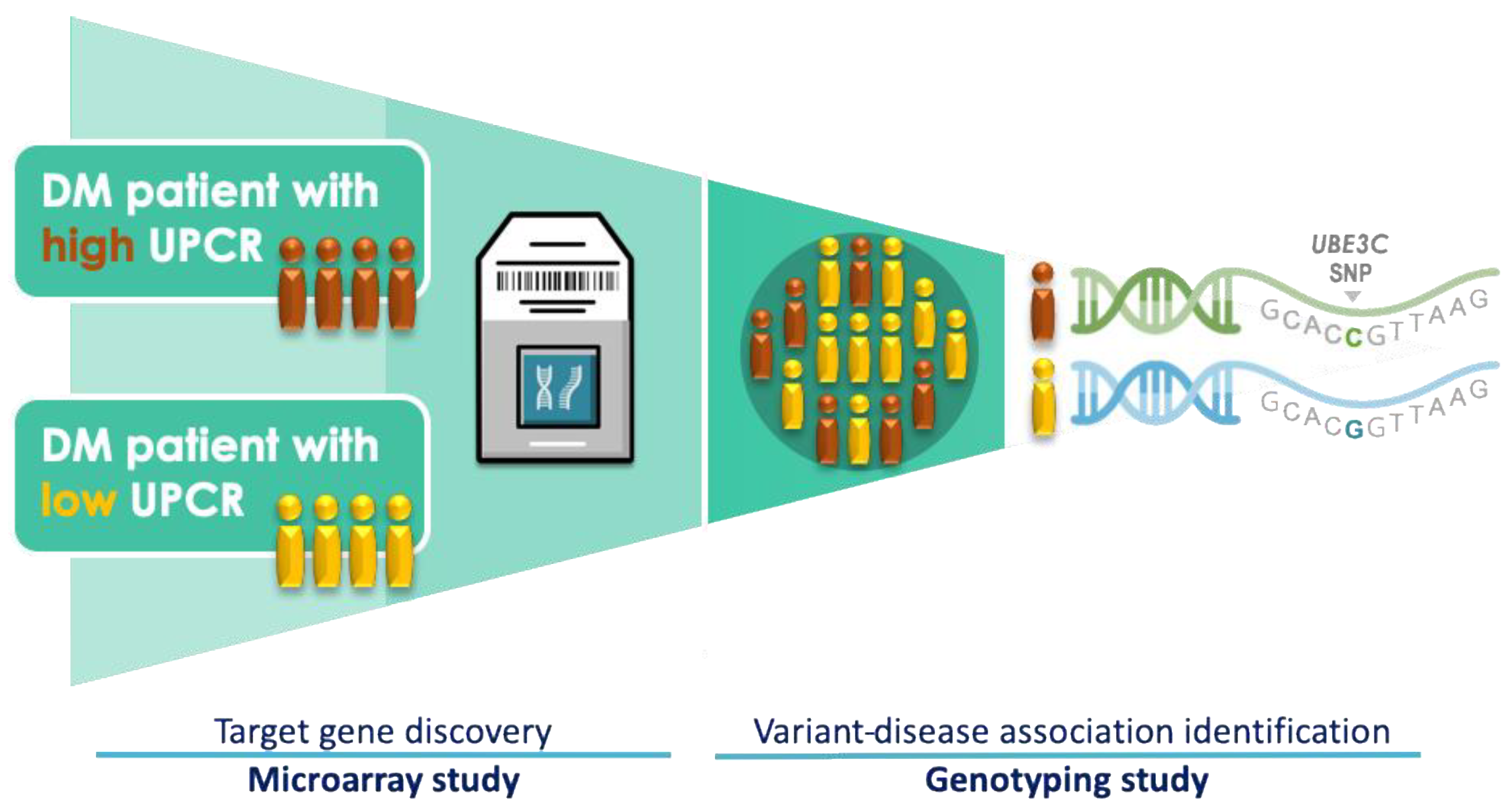
2.1. Study Subjects
2.2. Microarray Experiment
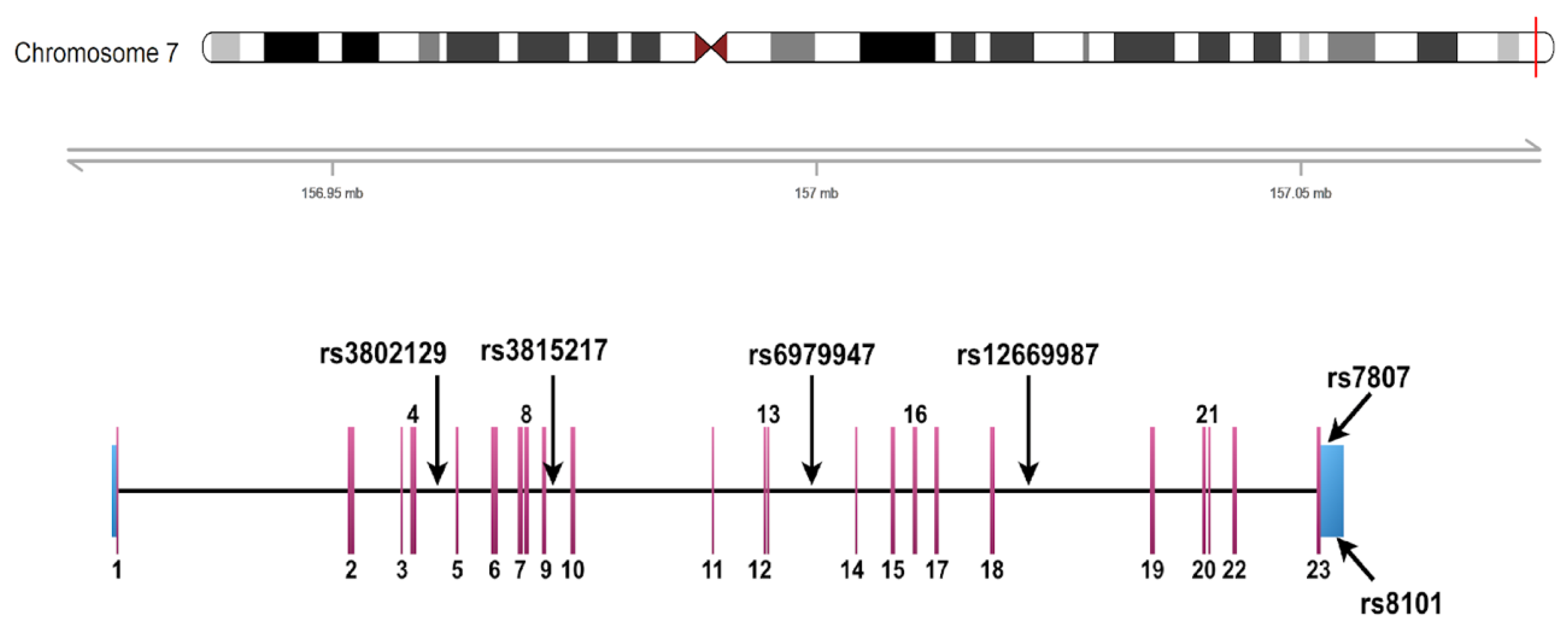
2.3. Criteria for the Selection of UBE3C Variants
2.4. Genotyping and eQTL Analysis
2.5. Statistical Methods and Analytical Tools
3. Results
3.1. Demographic and Clinical Characteristics of the Study Sample
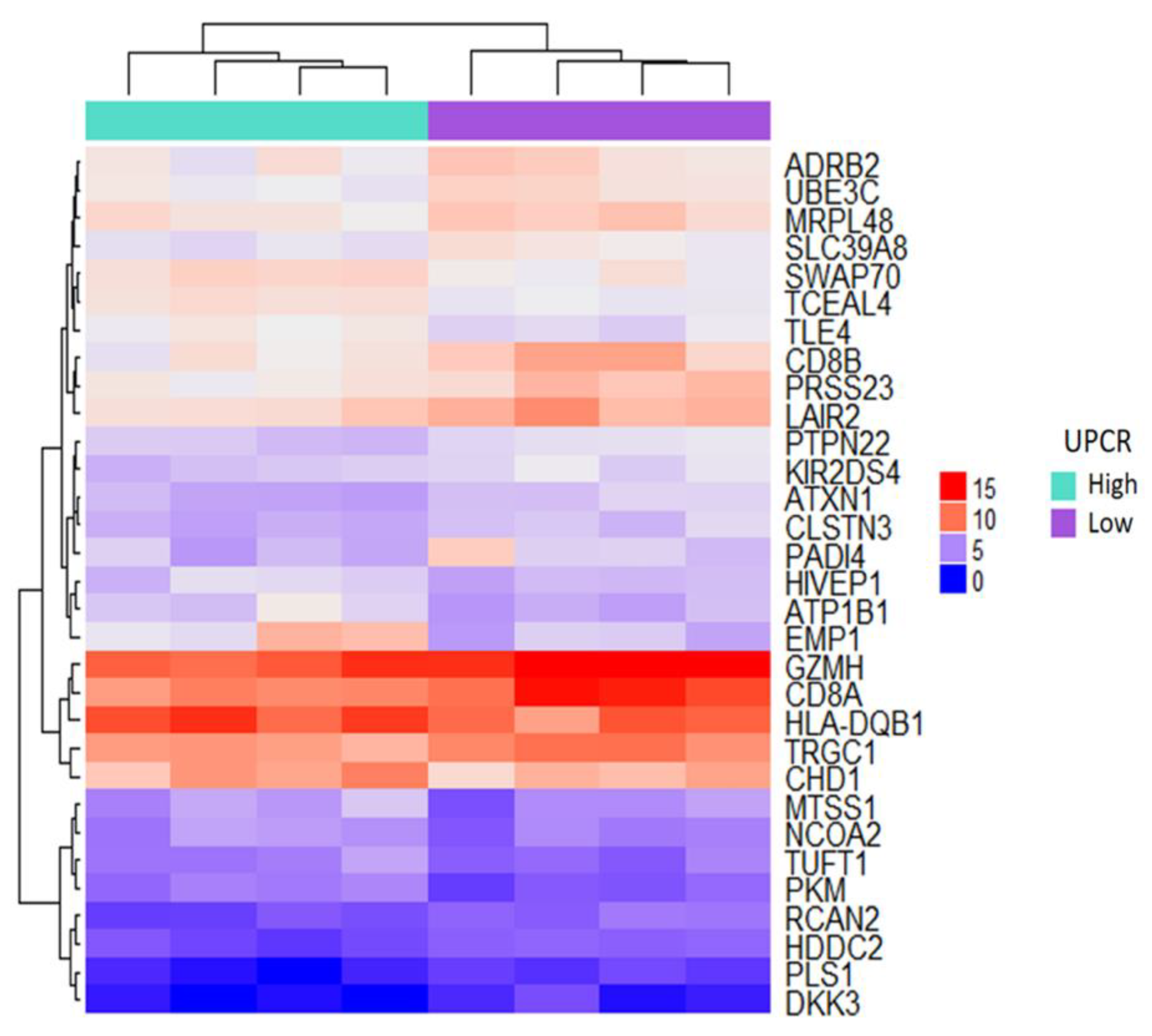
3.2. Microarray-Based Candidate Genes Selection from Eight Patients
3.3. Association of rs3802129 and rs7807 with Reduced Kidney Function
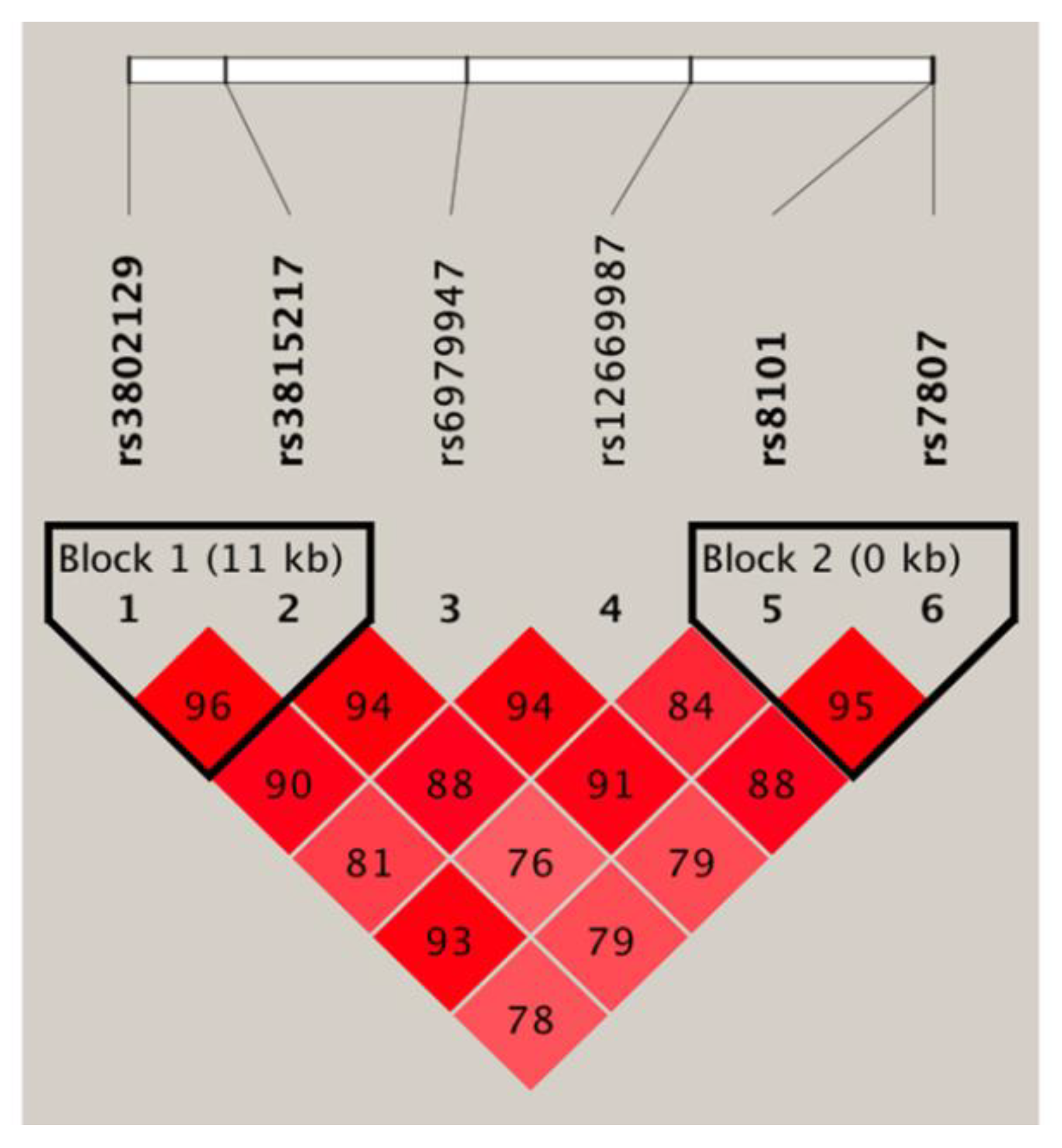
3.4. Haplotype Association Analysis and Expression Quantitative Trait Locus Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zelnick, L.R.; Weiss, N.S.; Kestenbaum, B.R.; Robinson-Cohen, C.; Heagerty, P.J.; Tuttle, K.; Hall, Y.N.; Hirsch, I.B.; de Boer, I.H. Diabetes and ckd in the united states population, 2009–2014. Clin. J. Am. Soc. Nephrol. 2017, 12, 1984–1990. [Google Scholar] [CrossRef] [PubMed]
- Bailey, R.A.; Wang, Y.; Zhu, V.; Rupnow, M.F. Chronic kidney disease in us adults with type 2 diabetes: An updated national estimate of prevalence based on kidney disease: Improving global outcomes (kdigo) staging. BMC Res. Notes 2014, 7, 415. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.C.; Hwang, S.J. Taiwan Society of Nephrology. Incidence, prevalence and mortality trends of dialysis end-stage renal disease in Taiwan from 1990 to 2001: The impact of national health insurance. Nephrol. Dial. Transpl. 2008, 23, 3977–3982. [Google Scholar] [CrossRef] [PubMed]
- Neal, B.; Perkovic, V.; Mahaffey, K.W.; de Zeeuw, D.; Fulcher, G.; Erondu, N.; Shaw, W.; Law, G.; Desai, M.; Matthews, D.R. Canagliflozin and cardiovascular and renal events in type 2 diabetes. N. Engl. J. Med. 2017, 377, 644–657. [Google Scholar] [CrossRef]
- Wanner, C.; Inzucchi, S.E.; Lachin, J.M.; Fitchett, D.; von Eynatten, M.; Mattheus, M.; Johansen, O.E.; Woerle, H.J.; Broedl, U.C.; Zinman, B. Empagliflozin and progression of kidney disease in type 2 diabetes. N. Engl. J. Med. 2016, 375, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Parving, H.H.; Lehnert, H.; Brochner-Mortensen, J.; Gomis, R.; Andersen, S.; Arner, P. The effect of irbesartan on the development of diabetic nephropathy in patients with type 2 diabetes. N. Engl. J. Med. 2001, 345, 870–878. [Google Scholar] [CrossRef] [PubMed]
- Lewis, E.J.; Hunsicker, L.G.; Clarke, W.R.; Berl, T.; Pohl, M.A.; Lewis, J.B.; Ritz, E.; Atkins, R.C.; Rohde, R.; Raz, I. Renoprotective effect of the angiotensin-receptor antagonist irbesartan in patients with nephropathy due to type 2 diabetes. N. Engl. J. Med. 2001, 345, 851–860. [Google Scholar] [CrossRef]
- Brenner, B.M.; Cooper, M.E.; de Zeeuw, D.; Keane, W.F.; Mitch, W.E.; Parving, H.H.; Remuzzi, G.; Snapinn, S.M.; Zhang, Z.; Shahinfar, S. Effects of losartan on renal and cardiovascular outcomes in patients with type 2 diabetes and nephropathy. N. Engl. J. Med. 2001, 345, 861–869. [Google Scholar] [CrossRef] [PubMed]
- Care, D. Standards of medical care in diabetes 2019. Diabetes Care 2019, 42, S81. [Google Scholar]
- Gerstein, H.C.; Miller, M.E.; Genuth, S.; Ismail-Beigi, F.; Buse, J.B.; Goff, D.C., Jr.; Probstfield, J.L.; Cushman, W.C.; Ginsberg, H.N.; Bigger, J.T.; et al. Long-term effects of intensive glucose lowering on cardiovascular outcomes. N. Engl. J. Med. 2011, 364, 818–828. [Google Scholar] [PubMed]
- Krolewski, A.S.; Skupien, J.; Rossing, P.; Warram, J.H. Fast renal decline to end-stage renal disease: An unrecognized feature of nephropathy in diabetes. Kidney Int. 2017, 91, 1300–1311. [Google Scholar] [CrossRef]
- Sandholm, N.; Van Zuydam, N.; Ahlqvist, E.; Juliusdottir, T.; Deshmukh, H.A.; Rayner, N.W.; Di Camillo, B.; Forsblom, C.; Fadista, J.; Ziemek, D.; et al. The genetic landscape of renal complications in type 1 diabetes. J. Am. Soc. Nephrol. 2017, 28, 557–574. [Google Scholar] [CrossRef]
- Bleyer, A.J.; Sedor, J.R.; Freedman, B.I.; O’Brien, A.; Russell, G.B.; Graley, J.; Schelling, J.R. Risk factors for development and progression of diabetic kidney disease and treatment patterns among diabetic siblings of patients with diabetic kidney disease. Am. J. Kidney Dis. 2008, 51, 29–37. [Google Scholar] [CrossRef]
- Woroniecka, K.I.; Park, A.S.; Mohtat, D.; Thomas, D.B.; Pullman, J.M.; Susztak, K. Transcriptome analysis of human diabetic kidney disease. Diabetes 2011, 60, 2354–2369. [Google Scholar] [CrossRef]
- Baelde, H.J.; Eikmans, M.; Doran, P.P.; Lappin, D.W.; de Heer, E.; Bruijn, J.A. Gene expression profiling in glomeruli from human kidneys with diabetic nephropathy. Am. J. Kidney Dis. 2004, 43, 636–650. [Google Scholar] [CrossRef]
- Nam, J.S.; Cho, M.H.; Lee, G.T.; Park, J.S.; Ahn, C.W.; Cha, B.S.; Lim, S.K.; Kim, K.R.; Ha, H.J.; Lee, H.C. The activation of NF-kappaB and AP-1 in peripheral blood mononuclear cells isolated from patients with diabetic nephropathy. Diabetes Res. Clin. Pract. 2008, 81, 25–32. [Google Scholar] [CrossRef]
- Tanaka, N.; Babazono, T.; Saito, S.; Sekine, A.; Tsunoda, T.; Haneda, M.; Tanaka, Y.; Fujioka, T.; Kaku, K.; Kawamori, R.; et al. Association of solute carrier family 12 (sodium/chloride) member 3 with diabetic nephropathy, identified by genome-wide analyses of single nucleotide polymorphisms. Diabetes 2003, 52, 2848–2853. [Google Scholar] [CrossRef]
- Shimazaki, A.; Kawamura, Y.; Kanazawa, A.; Sekine, A.; Saito, S.; Tsunoda, T.; Koya, D.; Babazono, T.; Tanaka, Y.; Matsuda, M.; et al. Genetic variations in the gene encoding elmo1 are associated with susceptibility to diabetic nephropathy. Diabetes 2005, 54, 1171–1178. [Google Scholar] [CrossRef] [PubMed]
- Iyengar, S.K.; Sedor, J.R.; Freedman, B.I.; Kao, W.H.; Kretzler, M.; Keller, B.J.; Abboud, H.E.; Adler, S.G.; Best, L.G.; Bowden, D.W.; et al. Genome-wide association and trans-ethnic meta-analysis for advanced diabetic kidney disease: Family investigation of nephropathy and diabetes (find). PLoS Genet. 2015, 11, e1005352. [Google Scholar] [CrossRef] [PubMed]
- van Zuydam, N.R.; Ahlqvist, E.; Sandholm, N.; Deshmukh, H.; Rayner, N.W.; Abdalla, M.; Ladenvall, C.; Ziemek, D.; Fauman, E.; Robertson, N.R.; et al. A genome-wide association study of diabetic kidney disease in subjects with type 2 diabetes. Diabetes 2018, 67, 1414–1427. [Google Scholar] [CrossRef]
- Guan, M.; Keaton, J.M.; Dimitrov, L.; Hicks, P.J.; Xu, J.; Palmer, N.D.; Ma, L.; Das, S.K.; Chen, Y.I.; Coresh, J.; et al. Genome-wide association study identifies novel loci for type 2 diabetes-attributed end-stage kidney disease in african americans. Hum. Genom. 2019, 13, 21. [Google Scholar] [CrossRef]
- Pattaro, C.; Teumer, A.; Gorski, M.; Chu, A.Y.; Li, M.; Mijatovic, V.; Garnaas, M.; Tin, A.; Sorice, R.; Li, Y.; et al. Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nat. Commun. 2016, 7, 10023. [Google Scholar] [CrossRef]
- Gonzalez, J.R.; Armengol, L.; Sole, X.; Guino, E.; Mercader, J.M.; Estivill, X.; Moreno, V. Snpassoc: An r package to perform whole genome association studies. Bioinformatics 2007, 23, 644–645. [Google Scholar] [CrossRef]
- Hochberg, Y.; Benjamini, Y. More powerful procedures for multiple significance testing. Stat. Med. 1990, 9, 811–818. [Google Scholar] [CrossRef] [PubMed]
- Zeller, T.; Wild, P.; Szymczak, S.; Rotival, M.; Schillert, A.; Castagne, R.; Maouche, S.; Germain, M.; Lackner, K.; Rossmann, H.; et al. Genetics and beyond--the transcriptome of human monocytes and disease susceptibility. PLoS ONE 2010, 5, e10693. [Google Scholar] [CrossRef]
- Dixon, A.L.; Liang, L.; Moffatt, M.F.; Chen, W.; Heath, S.; Wong, K.C.; Taylor, J.; Burnett, E.; Gut, I.; Farrall, M.; et al. A genome-wide association study of global gene expression. Nat. Genet. 2007, 39, 1202–1207. [Google Scholar] [CrossRef]
- Westra, H.J.; Peters, M.J.; Esko, T.; Yaghootkar, H.; Schurmann, C.; Kettunen, J.; Christiansen, M.W.; Fairfax, B.P.; Schramm, K.; Powell, J.E.; et al. Systematic identification of trans eqtls as putative drivers of known disease associations. Nat. Genet. 2013, 45, 1238–1243. [Google Scholar] [CrossRef]
- Metzger, M.B.; Pruneda, J.N.; Klevit, R.E.; Weissman, A.M. RING-type E3 ligases: Master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys. Acta 2014, 1843, 47–60. [Google Scholar] [CrossRef]
- Kaneko, M.; Iwase, I.; Yamasaki, Y.; Takai, T.; Wu, Y.; Kanemoto, S.; Matsuhisa, K.; Asada, R.; Okuma, Y.; Watanabe, T.; et al. Genome-wide identification and gene expression profiling of ubiquitin ligases for endoplasmic reticulum protein degradation. Sci. Rep. 2016, 6, 30955. [Google Scholar] [CrossRef]
- Cybulsky, A.V. The intersecting roles of endoplasmic reticulum stress, ubiquitin- proteasome system, and autophagy in the pathogenesis of proteinuric kidney disease. Kidney Int. 2013, 84, 25–33. [Google Scholar] [CrossRef]
- Meyer-Schwesinger, C. The ubiquitin-proteasome system in kidney physiology and disease. Nat. Rev. Nephrol. 2019, 15, 393–411. [Google Scholar] [CrossRef] [PubMed]
- Beeken, M.; Lindenmeyer, M.T.; Blattner, S.M.; Radón, V.; Oh, J.; Meyer, T.N.; Hildebrand, D.; Schlüter, H.; Reinicke, A.T.; Knop, J.H.; et al. Alterations in the ubiquitin proteasome system in persistent but not reversible proteinuric diseases. J. Am. Soc. Nephrol. 2014, 25, 2511–2525. [Google Scholar] [CrossRef]
- Aghdam, S.Y.; Gurel, Z.; Ghaffarieh, A.; Sorenson, C.M.; Sheibani, N. High glucose and diabetes modulate cellular proteasome function: Implications in the pathogenesis of diabetes complications. Biochem. Biophys. Res. Commun. 2013, 432, 339–344. [Google Scholar] [CrossRef][Green Version]
- Otoda, T.; Yuasa, T.; Aihara, K. Evaluating the ubiquitin–proteasome system as a therapeutic target in diabetic kidney disease. Adv. Clin. Transl. Res. 2018, 2, 100010. [Google Scholar]
- Wen, J.L.; Wen, X.F.; Li, R.B.; Jin, Y.C.; Wang, X.L.; Zhou, L.; Chen, H.X. Ube3c promotes growth and metastasis of renal cell carcinoma via activating wnt/beta-catenin pathway. PLoS ONE 2015, 10, e0115622. [Google Scholar] [CrossRef]
- Jiang, J.H.; Liu, Y.F.; Ke, A.W.; Gu, F.M.; Yu, Y.; Dai, Z.; Gao, Q.; Shi, G.M.; Liao, B.Y.; Xie, Y.H.; et al. Clinical significance of the ubiquitin ligase ube3c in hepatocellular carcinoma revealed by exome sequencing. Hepatology 2014, 59, 2216–2227. [Google Scholar] [CrossRef]
- Pasaje, C.F.; Kim, J.H.; Park, B.L.; Park, J.S.; Uh, S.T.; Kim, M.K.; Park, C.S.; Shin, H.D. Ube3c genetic variations as potent markers of nasal polyps in korean asthma patients. J. Hum. Genet. 2011, 56, 797–800. [Google Scholar] [CrossRef][Green Version]
- Lee, J.S.; Kim, J.H.; Bae, J.S.; Kim, J.Y.; Park, T.J.; Pasaje, C.F.; Park, B.L.; Cheong, H.S.; Park, J.S.; Uh, S.T.; et al. Association analysis of ube3c polymorphisms in korean aspirin-intolerant asthmatic patients. Ann. Allergy Asthma Immunol. 2010, 105, 307–312. [Google Scholar] [CrossRef]
- Scott, R.A.; Scott, L.J.; Magi, R.; Marullo, L.; Gaulton, K.J.; Kaakinen, M.; Pervjakova, N.; Pers, T.H.; Johnson, A.D.; Eicher, J.D.; et al. An expanded genome-wide association study of type 2 diabetes in Europeans. Diabetes 2017, 66, 2888–2902. [Google Scholar] [CrossRef]
- Popovic, D.; Vucic, D.; Dikic, I. Ubiquitination in disease pathogenesis and treatment. Nat. Med. 2014, 20, 1242. [Google Scholar] [CrossRef]
- Goru, S.K.; Pandey, A.; Gaikwad, A.B. E3 ubiquitin ligases as novel targets for inflammatory diseases. Pharmacol. Res. 2016, 106, 1–9. [Google Scholar] [CrossRef]
| Characteristics | Case | Control |
|---|---|---|
| Total number | 172 | 91 |
| Male, n (%) | 104 (60.47) | 47 (51.65) |
| Age (years) 1 | 70.47 ± 12.75 | 65.18 ± 11.89 |
| BMI (kg/m2) 1 | 26.94 ± 4.44 | 26.69 ± 4.69 |
| Smoking, n (%) | 36 (20.93) | 21 (23.08) |
| HbA1c (%) 1 | 7 ± 1.33 | 7.17 ± 1.68 |
| eGFR (ml/min/1.73 m2) 1 | 32.81 ± 16.11 | 89.58 ± 24.31 |
| UPCR (mg/g) 1 | 2174.52 ± 3090.89 | 784.51 ± 1461.02 |
| Frequency | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP | Position a | Ref | Alt | AFR | AMR | ASN | EUR | TWB | Study Cohort b |
| rs3802129 | chr7:156965297 | G | A | 0.18 | 0.44 | 0.48 | 0.33 | 0.44 | 0.42 |
| rs3815217 | chr7:156976981 | A | G | 0.08 | 0.44 | 0.48 | 0.47 | 0.50 | 0.46 |
| rs6979947 | chr7:157005863 | A | G | 0.24 | 0.37 | 0.44 | 0.26 | 0.40 | 0.40 |
| rs12669987 | chr7:157032676 | C | T | 0.00 | 0.05 | 0.14 | 0.00 | 0.15 | 0.15 |
| rs8101 | chr7:157061474 | T | C | 0.86 | 0.57 | 0.52 | 0.66 | 0.54 | 0.52 |
| rs7807 | chr7:157061642 | C | A | 0.71 | 0.72 | 0.6 | 0.79 | 0.63 | 0.63 |
| Number | Recessive Model | |||||
|---|---|---|---|---|---|---|
| SNPs | Genotypes | Case (%) | Control (%) | OR (95% CI) | p Value | q Value |
| rs3802129 | G/G-A/G | 127 (75.6) | 80 (88.9) | 1.00 | 0.008 | 0.024 |
| A/A | 41 (24.4) | 10 (11.1) | 2.66 (1.24–5.68) | |||
| rs3815217 | A/A-A/G | 115 (75.2) | 74 (87.1) | 1.00 | 0.032 | 0.064 |
| G/G | 38 (24.8) | 11 (12.9) | 2.19 (1.04–4.62) | |||
| rs6979947 | A/A-A/G | 128 (77.1) | 74 (87.1) | 1.00 | 0.063 | 0.076 |
| G/G | 38 (22.9) | 11 (12.9) | 1.97 (0.94–4.15) | |||
| rs12669987 | C/C-C/T | 149 (96.1) | 83 (95.4) | 1.00 | 0.641 | 0.641 |
| T/T | 6 (3.9) | 4 (4.6) | 0.73 (0.19–2.74) | |||
| rs8101 | C/C-C/T | 116 (69.9) | 71 (81.6) | 1.00 | 0.045 | 0.068 |
| T/T | 50 (30.1) | 16 (18.4) | 1.92 (1.00–3.68) | |||
| rs7807 | A/A-A/C | 136 (80.5) | 81 (94.2) | 1.00 | 0.003 | 0.018 |
| C/C | 33 (19.5) | 5 (5.8) | 3.76 (1.39–10.13) | |||
| Haplotypes | Frequency | OR (95% CI) | p Value | |
|---|---|---|---|---|
| Case | Control | |||
| Block 1 (rs3802129/rs3815217) | ||||
| A/G | 0.464 | 0.315 | 1.76 (1.19–2.61) | 0.005 |
| G/G | 0.051 | 0.074 | 0.91 (0.39–2.12) | 0.835 |
| A/A | 0.004 | 0.019 | 0.22 (0.02–2.13) | 0.189 |
| G/A | 0.482 | 0.592 | Reference | |
| Block 2 (rs8101/rs7807) | ||||
| T/C | 0.395 | 0.297 | 1.58 (1.06–2.35) | 0.024 |
| C/C | 0.010 | 0.007 | 1.46 (0.23–9.37) | 0.687 |
| T/A | 0.121 | 0.114 | 1.21 (0.68–2.18) | 0.519 |
| C/A | 0.474 | 0.582 | Reference | |
| SNPs | Ref | Alt | Gencode ID (ENSG00000-) | Gene | p Value | Effect Size | Tissue |
|---|---|---|---|---|---|---|---|
| rs3802129 | G | A | 9335.13 | UBE3C | 0.0000033 | 0.11 | Esophagus-Muscularis |
| 9335.13 | UBE3C | 0.00001 | 0.11 | Muscle-Skeletal | |||
| rs3815217 | A | G | 229660.1 | RP5–1142J19.1 | 3.10 × 10−9 | −0.37 | Skin–Suprapubic 1 |
| 229660.1 | RP5–1142J19.1 | 0.0000041 | −0.29 | Skin–Lower leg 2 | |||
| 229660.1 | RP5–1142J19.1 | 0.000016 | −0.33 | Adipose-Visceral | |||
| rs8101 | T | C | 9335.13 | UBE3C | 0.000008 | −0.1 | Esophagus-Muscularis |
| rs7807 | C | A | 9335.13 | UBE3C | 1.40 × 10−8 | −0.15 | Esophagus-Muscularis |
| 9335.13 | UBE3C | 0.000024 | −0.15 | Artery-Tibial |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.-C.; Wu, M.-Y.; Yu, Z.-L.; Chou, W.-H.; Lai, Y.-T.; Kao, C.-C.; Faridah, I.N.; Wu, M.-S.; Chang, W.-C. Association of UBE3C Variants with Reduced Kidney Function in Patients with Diabetic Kidney Disease. J. Pers. Med. 2020, 10, 210. https://doi.org/10.3390/jpm10040210
Chen Y-C, Wu M-Y, Yu Z-L, Chou W-H, Lai Y-T, Kao C-C, Faridah IN, Wu M-S, Chang W-C. Association of UBE3C Variants with Reduced Kidney Function in Patients with Diabetic Kidney Disease. Journal of Personalized Medicine. 2020; 10(4):210. https://doi.org/10.3390/jpm10040210
Chicago/Turabian StyleChen, Ying-Chun, Mei-Yi Wu, Zhi-Lei Yu, Wan-Hsuan Chou, Yi-Ting Lai, Chih-Chin Kao, Imaniar Noor Faridah, Mai-Szu Wu, and Wei-Chiao Chang. 2020. "Association of UBE3C Variants with Reduced Kidney Function in Patients with Diabetic Kidney Disease" Journal of Personalized Medicine 10, no. 4: 210. https://doi.org/10.3390/jpm10040210
APA StyleChen, Y.-C., Wu, M.-Y., Yu, Z.-L., Chou, W.-H., Lai, Y.-T., Kao, C.-C., Faridah, I. N., Wu, M.-S., & Chang, W.-C. (2020). Association of UBE3C Variants with Reduced Kidney Function in Patients with Diabetic Kidney Disease. Journal of Personalized Medicine, 10(4), 210. https://doi.org/10.3390/jpm10040210






