Effects of Growth Hormone Treatment and Rehabilitation in Incomplete Chronic Traumatic Spinal Cord Injury: Insight from Proteome Analysis
Abstract
1. Introduction
2. Materials and Methods
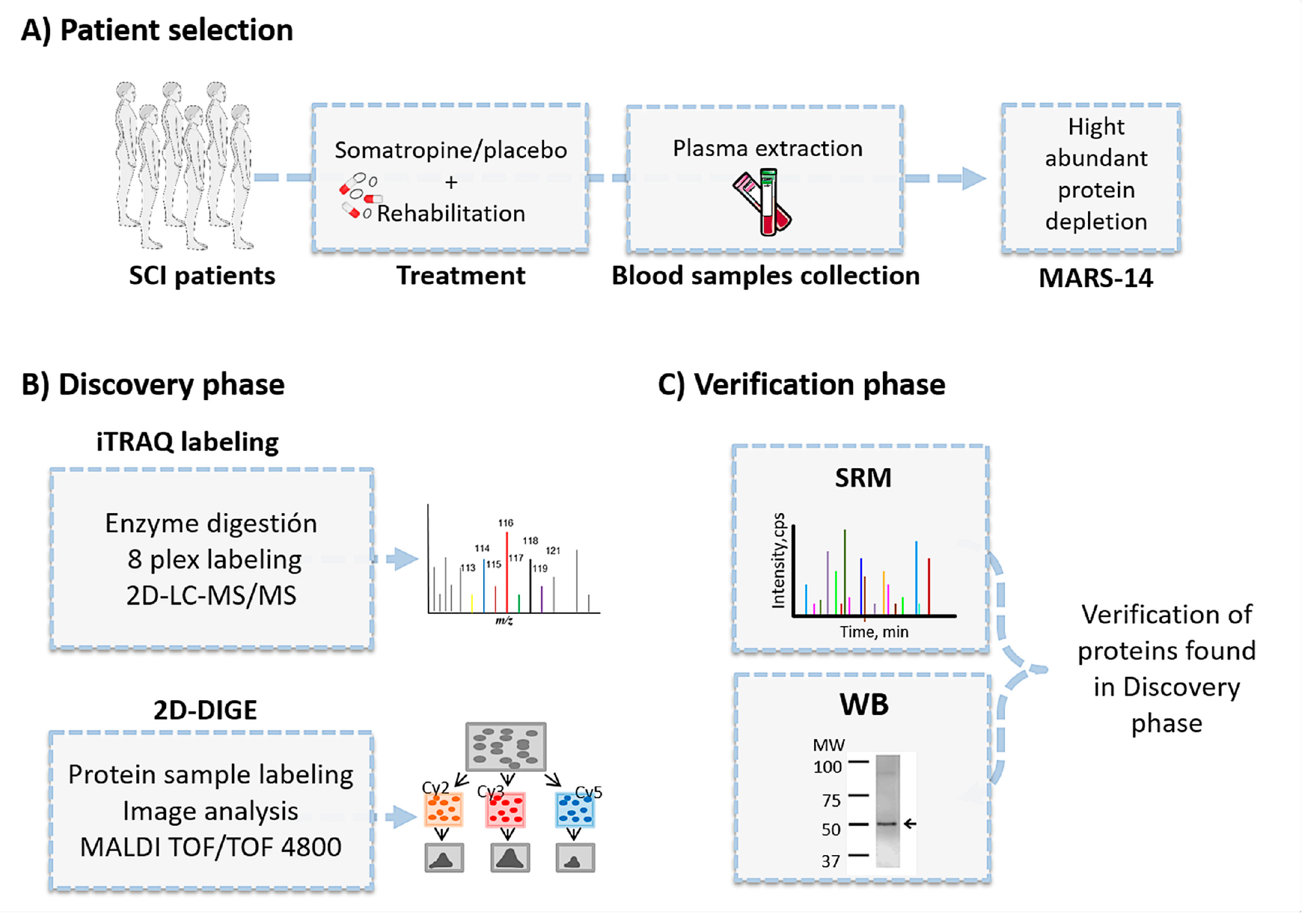
2.1. Patient Recruitment and Study Design
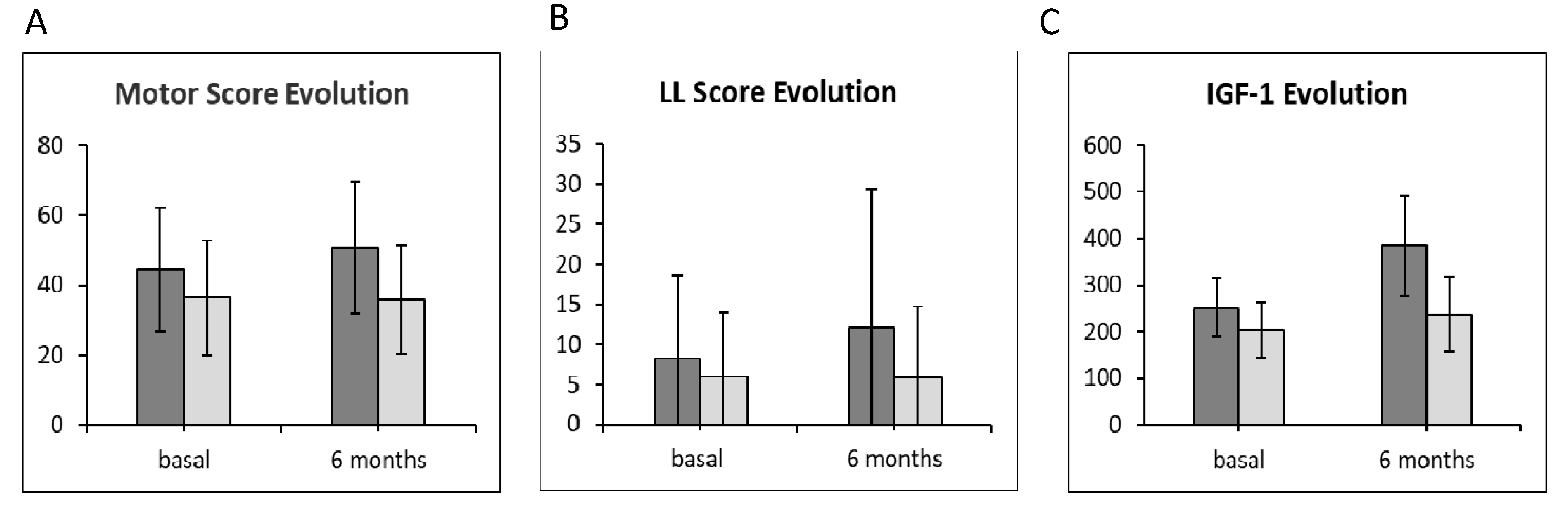
2.2. Motor Score Evolution and Measurement of Insulin-Like Growth Factor-1 Plasma Levels
2.3. Proteomics Overview
2.3.1. Sample Preparation for Proteomic Analyses
2.3.2. iTRAQ
Identification by LC-MS/MS
iTRAQ Data Analysis
2.4. Two-Dimensional Differential Gel Electrophoresis
2.4.1. Sample Labeling
2.4.2. Two-Dimensional Electrophoresis
2.4.3. Image Acquisition and Analysis
2.4.4. Protein Identification by Mass Spectrometry
2.5. Western Blotting
2.6. Selected Reaction Monitoring
2.6. Selected Reaction Monitoring
2.7. Statistical Analysis
3. Results
4. Discussion
4.1. Panel Indicator of Response to Somatropin Treatment
4.2. Panel Indicator of Positive Response to Rehabilitation
Author Contributions
Funding
Conflicts of Interest
References
- Bárbara-Bataller, E.; Méndez-Suárez, J.L.; Alemán-Sánchez, C.; Sánchez-Enríquez, J.; Sosa-Henríquez, M. Change in the profile of traumatic spinal cord injury over 15 years in Spain. Scand. J. Trauma Resusc. Emerg. Med. 2018, 26, 27. [Google Scholar] [CrossRef] [PubMed]
- Schuld, C.; Franz, S.; Brüggemann, K.; Heutehaus, L.; Weidner, N.; Kirshblum, S.C.; Rupp, R. International standards for neurological classification of spinal cord injury: Impact of the revised worksheet (revision 02/13) on classification performance. J. Spinal. Cord Med. 2016, 39, 504–512. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh, A.; Dyck, S.M.; Karimi-Abdolrezaee, S. Traumatic spinal cord injury: An overview of pathophysiology, models and acute injury mechanisms. Front. Neurol. 2019, 10, 282. [Google Scholar] [CrossRef] [PubMed]
- Jutzeler, C.R.; Streijger, F.; Aguilar, J.; Shortt, K.; Manouchehri, N.; Okon, E.; Hupp, M.; Curt, A.; Kwon, B.K.; Kramer, J.L.K. Sensorimotor plasticity after spinal cord injury: A longitudinal and translational study. Ann. Clin. Transl. Neurol. 2019, 6, 68–82. [Google Scholar] [CrossRef]
- Fawcett, J.W.; Curt, A.; Steeves, J.D.; Coleman, W.P.; Tuszynski, M.H.; Lammertse, D.; Bartlett, P.F.; Blight, A.R.; Dietz, V.; Ditunno, J.; et al. Guidelines for the conduct of clinical trials for spinal cord injury as developed by the ICCP panel: Spontaneous recovery after spinal cord injury and statistical power needed for therapeutic clinical trials. Spinal Cord 2007, 45, 190–205. [Google Scholar] [CrossRef]
- Huang, H.; Young, W.; Skaper, S.; Chen, L.; Moviglia, G.; Saberi, H.; Al-Zoubi, Z.; Sharma, H.S.; Muresanu, D.; Sharma, A.; et al. Clinical neurorestorative therapeutic guidelines for spinal cord injury (IANR/CANR version 2019). J. Orthop. Transl. 2020, 20, 14–24. [Google Scholar] [CrossRef]
- Martinez-Moreno, C.G.; Calderon-Vallejo, D.; Harvey, S.; Aramburo, C.; Quintanar, J.L. Growth Hormone (GH) and Gonadotropin-Releasing Hormone (GnRH) in the central nervous system: A potential neurological combinatory therapy? Int. J. Mol. Sci. 2018, 19, 375. [Google Scholar] [CrossRef]
- Lopez, J.; Quan, A.; Budihardjo, J.; Xiang, S.; Wang, H.; Kiron, K.; Cashman, C.; Lee, W.P.A.; Hoke, A.; Tuffaha, S.; et al. Growth Hormone improves nerve regeneration, muscle re-innervation, and functional outcomes after chronic denervation injury. Sci. Rep. 2019, 9, 3117. [Google Scholar] [CrossRef]
- Hua, K.; Forbes, M.E.; Lichtenwalner, R.J.; Sonntag, W.E.; Riddle, D.R. Adult-onset deficiency in growth hormone and insulin-like growth factor-I alters oligodendrocyte turnover in the corpus callosum. Glia 2009, 57, 1062–1071. [Google Scholar] [CrossRef]
- Gibney, J.; Healy, M.L.; Sönksen, P.H. The growth hormone/insulin-like growth factor-I axis in exercise and sport. Endocr. Rev. 2007, 28, 603–624. [Google Scholar] [CrossRef]
- Walser, M.; Samà, M.T.; Wickelgren, R.; Aberg, M.; Bohlooly, Y.M.; Olsson, B.; Törnell, J.; Isgaard, J.; Aberg, N.D. Local overexpression of GH and GH/IGF1 effects in the adult mouse hippocampus. J. Endocrinol. 2012, 215, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Rezaei, M.; Zarkesh-Esfahani, S.H. Optimization of production of recombinant human growth hormone in Escherichia coli. J. Res. Med Sci. Off. J. Isfahan Univ. Med Sci. 2012, 17, 681–685. [Google Scholar]
- Liang, G.; Cline, G.W.; Macica, C.M. IGF-1 stimulates de novo fatty acid biosynthesis by Schwann cells during myelination. Glia 2007, 55, 632–641. [Google Scholar] [CrossRef] [PubMed]
- Cuatrecasas, G.; Kumru, H.; Coves, M.J.; Vidal, J. GH deficiency in patients with spinal cord injury: Efficacy/safety of GH replacement, a pilot study. Endocr. Connect. 2018, 7, 1031–1039. [Google Scholar] [CrossRef] [PubMed]
- Barakat-Walter, I.; Kraftsik, R. Stimulating effect of thyroid hormones in peripheral nerve regeneration: Research history and future direction toward clinical therapy. Neural Regen. Res. 2018, 13, 599–608. [Google Scholar] [CrossRef]
- Tator, C.H.; van der Jagt, R.H. The effect of exogenous thyroid hormones on functional recovery of the rat after acute spinal cord compression injury. J. Neurosurg 1980, 53, 381–384. [Google Scholar] [CrossRef]
- Hwang, D.H.; Park, H.H.; Shin, H.Y.; Cui, Y.; Kim, B.G. Insulin-like Growth Factor-1 Receptor dictates beneficial effects of treadmill training by regulating survival and migration of neural stem cell grafts in the injured spinal cord. Exp. Neurobiol. 2018, 27, 489–507. [Google Scholar] [CrossRef]
- Lovejoy, J.C.; Smith, S.R.; Zachwieja, J.J.; Bray, G.A.; Windhauser, M.M.; Wickersham, P.J.; Veldhuis, J.D.; Tulley, R.; de la Bretonne, J.A. Low-dose T(3) improves the bed rest model of simulated weightlessness in men and women. Am. J. Physiol. 1999, 277, E370–E379. [Google Scholar] [CrossRef]
- Sun, Y.; Braga-Neto, U.; Dougherty, E.R. A systematic model of the LC-MS proteomics pipeline. BMC Genom. 2012, 13, S2. [Google Scholar] [CrossRef]
- Iwamoto, N.; Shimada, T. Recent advances in mass spectrometry-based approaches for proteomics and biologics: Great contribution for developing therapeutic antibodies. Pharmacol. Ther. 2018, 185, 147–154. [Google Scholar] [CrossRef]
- Toby, T.K.; Fornelli, L.; Srzentić, K.; DeHart, C.J.; Levitsky, J.; Friedewald, J.; Kelleher, N.L. A comprehensive pipeline for translational top-down proteomics from a single blood draw. Nat. Protoc. 2019, 14, 119–152. [Google Scholar] [CrossRef]
- Doucette, A.A.; Nickerson, J.L. Developing front-end devices for improved sample preparation in MS-based proteome analysis. J. Mass Spectrom. 2020, 55, e4494. [Google Scholar] [CrossRef] [PubMed]
- Dardé, V.; Barderas, M.G.; Vivanco, F. Depletion of high-abundance proteins in plasma by immunoaffinity subtraction for two-dimensional difference gel electrophoresis analysis. Methods Mol. Biol. 2007, 357, 351–364. [Google Scholar] [PubMed]
- Alonso-Orgaz, S.; Martin-Rojas, T.; Calvo, E.; López, J.A.; Vivanco, F.; Barderas, M.G. Differential protein expression analysis of degenerative aortic stenosis by iTRAQ labeling. Methods Mol. Biol. 2013, 1005, 109–117. [Google Scholar] [CrossRef]
- Dardé, V.M.; Barderas, M.G.; Vivanco, F. Multiple reaction monitoring (MRM) of plasma proteins in cardiovascular proteomics. Methods Mol. Biol. 2013, 1000, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Bonzon-Kulichenko, E.; Perez-Hernández, D.; Nuñez, E.; Martinez-Acedo, P.; Navarro, P.; Trevisan-Herraz, M.; Ramos Mdel, C.; Sierra, S.; Martinez-Martinez, S.; Ruiz-Meana, M.; et al. A robust method for quantitative high-throughput analysis of proteomes by 18O labeling. Mol. Cell Proteom. 2011, 10, M110.003335. [Google Scholar] [CrossRef]
- Martinez-Bartolome, S.; Navarro, P.; Martin-Maroto, F.; Lopez-Ferrer, D.; Ramos-Fernandez, A.; Villar, M.; Garcia-Ruiz, J.P.; Vazquez, J. Properties of average score distributions of SEQUEST. Mol. Cell Proteom. 2008, 7, 1135–1145. [Google Scholar] [CrossRef]
- Navarro, P.; Vazquez, J. A refined method to calculate false discovery rates for peptide identification using decoy databases. J. Proteome Res. 2009, 8, 1792–1796. [Google Scholar] [CrossRef]
- Navarro, P.; Trevisan-Herraz, M.; Bonzon-Kulichenko, E.; Nuñez, E.; Martinez-Acedo, P.; Perez-Hernandez, D.; Jorge, I.; Mesa, R.; Calvo, E.; Carrascal, M.; et al. General statistical framework for quantitative proteomics by stable isotope labeling. J. Proteome Res. 2014, 13, 1234–1247. [Google Scholar] [CrossRef]
- Bradford, M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Martín-Rojas, T.; Gil-Dones, F.; Lopez-Almodovar, L.F.; Padial, L.R.; Vivanco, F.; Barderas, M.G. Proteomic profile of human aortic stenosis: Insights into the degenerative process. J. Proteome Res. 2012, 11, 1537–1550. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Mann, M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996, 68, 850–858. [Google Scholar] [CrossRef] [PubMed]
- Perkins, D.N.; Pappin, D.J.C.; Creasy, D.M.; Cottrell, J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999, 20, 3551–3567. [Google Scholar] [CrossRef]
- Mourino-Alvarez, L.; Baldan-Martin, M.; Sastre-Oliva, T.; Martin-Lorenzo, M.; Maroto, A.S.; Corbacho-Alonso, N.; Rincon, R.; Martin-Rojas, T.; Lopez-Almodovar, L.F.; Alvarez-Llamas, G.; et al. A comprehensive study of calcific aortic stenosis: From rabbit to human samples. Dis. Models Mech. 2018, 11, dmm033423. [Google Scholar] [CrossRef] [PubMed]
- Martin-Rojas, T.; Mourino-Alvarez, L.; Gil-Dones, F.; de la Cuesta, F.; Rosello-Lleti, E.; Laborde, C.M.; Rivera, M.; Lopez-Almodovar, L.F.; Lopez, J.A.; Akerstrom, F.; et al. A clinical perspective on the utility of alpha 1 antichymotrypsin for the early diagnosis of calcific aortic stenosis. Clin. Proteom. 2017, 14, 12. [Google Scholar] [CrossRef] [PubMed]
- Baldan-Martin, M.; Mourino-Alvarez, L.; Gonzalez-Calero, L.; Moreno-Luna, R.; Sastre-Oliva, T.; Ruiz-Hurtado, G.; Segura, J.; Lopez, J.A.; Vazquez, J.; Vivanco, F.; et al. Plasma molecular signatures in hypertensive patients with renin-angiotensin system suppression: New predictors of renal damage and de novo albuminuria indicators. Hypertension 2016, 68, 157–166. [Google Scholar] [CrossRef][Green Version]
- Lindsey, M.L.; Mayr, M.; Gomes, A.V.; Delles, C.; Arrell, D.K.; Murphy, A.M.; Lange, R.A.; Costello, C.E.; Jin, Y.F.; Laskowitz, D.T.; et al. Transformative impact of proteomics on cardiovascular health and disease: A scientific statement from the American Heart Association. Circulation 2015, 132, 852–872. [Google Scholar] [CrossRef]
- Li, S.Y.; Li, Z.X.; He, Z.G.; Wang, Q.; Li, Y.J.; Yang, Q.; Wu, D.Z.; Zeng, H.L.; Xiang, H.B. Quantitative proteomics reveal the alterations in the spinal cord after myocardial ischemia-reperfusion injury in rats. Int. J. Mol. Med. 2019, 44, 1877–1887. [Google Scholar] [CrossRef]
- Sarkis, G.A.; Mangaonkar, M.D.; Moghieb, A.; Lelling, B.; Guertin, M.; Yadikar, H.; Yang, Z.; Kobeissy, F.; Wang, K.K. The Application of proteomics to traumatic brain and spinal cord injuries. Curr. Neurol. Neurosci. Rep. 2017, 17, 23. [Google Scholar] [CrossRef]
- Kasperska-Zajac, A.; Grzanka, A.; Machura, E.; Misiolek, M.; Mazur, B.; Jochem, J. Increased serum complement C3 and C4 concentrations and their relation to severity of chronic spontaneous urticaria and CRP concentration. J. Inflamm. 2013, 10, 22. [Google Scholar] [CrossRef]
- Carpanini, S.M.; Torvell, M.; Morgan, B.P. Therapeutic inhibition of the complement system in diseases of the central nervous system. Front. Immunol. 2019, 10, 362. [Google Scholar] [CrossRef] [PubMed]
- Barthel, D.; Schindler, S.; Zipfel, P.F. Plasminogen is a complement inhibitor. J. Biol. Chem. 2012, 287, 18831–18842. [Google Scholar] [CrossRef] [PubMed]
- Amara, U.; Flierl, M.A.; Rittirsch, D.; Klos, A.; Chen, H.; Acker, B.; Brückner, U.B.; Nilsson, B.; Gebhard, F.; Lambris, J.D.; et al. Molecular intercommunication between the complement and coagulation systems. J. Immunol. 2010, 185, 5628–5636. [Google Scholar] [CrossRef] [PubMed]
- Madathil, S.K.; Evans, H.N.; Saatman, K.E. Temporal and regional changes in IGF-1/IGF-1R signaling in the mouse brain after traumatic brain injury. J. Neurotrauma 2010, 27, 95–107. [Google Scholar] [CrossRef]
- Kiernan, J.A. Hypotheses concerned with axonal regeneration in the mammalian nervous system. Biol. Rev. Camb. Philos. Soc. 1979, 54, 155–197. [Google Scholar] [CrossRef]
- Seeds, N.W.; Akison, L.; Minor, K. Role of plasminogen activator in spinal cord remodeling after spinal cord injury. Respir. Physiol. Neurobiol. 2009, 169, 141–149. [Google Scholar] [CrossRef][Green Version]
- Rubin, M.R.; Bilezikian, J.P. Hypoparathyroidism: Clinical features, skeletal microstructure and parathyroid hormone replacement. Arq. Bras. Endocrinol. Metabol. 2010, 54, 220–226. [Google Scholar] [CrossRef]
- Jiang, S.D.; Jiang, L.S.; Dai, L.Y. Mechanisms of osteoporosis in spinal cord injury. Clin. Endocrinol. 2006, 65, 555–565. [Google Scholar] [CrossRef]
- Bajwa, N.M.; Kesavan, C.; Mohan, S. Long-term consequences of traumatic brain Injury in bone metabolism. Front. Neurol. 2018, 9, 115. [Google Scholar] [CrossRef]
- Andreassen, T.T.; Oxlund, H. Local anabolic effects of growth hormone on intact bone and healing fractures in rats. Calcif. Tissue Int. 2003, 73, 258–264. [Google Scholar] [CrossRef]
- Bost, F.; Diarra-Mehrpour, M.; Martin, J.P. Inter-alpha-trypsin inhibitor proteoglycan family-a group of proteins binding and stabilizing the extracellular matrix. Eur. J. Biochem. 1998, 252, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y. Lecticans: Organizers of the brain extracellular matrix. Cell Mol. Life Sci. 2000, 57, 276–289. [Google Scholar] [CrossRef]
- Sherman, L.S.; Back, S.A. A ‘GAG’ reflex prevents repair of the damaged CNS. Trends Neurosci. 2008, 31, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, M.B.; Mukhopadhyay, D. Possible role of apolipoprotein A1 in healing and cell death after neuronal injury. Front. Biosci. 2016, 8, 460–477. [Google Scholar] [CrossRef]
- Benvenga, S.; Robbins, J. Lipoprotein-thyroid hormone interactions. Trends Endocrinol. Metab. 1993, 4, 194–198. [Google Scholar] [CrossRef]


| Inclusion Criteria | Exclusion Criteria |
|---|---|
| Age between 18 and 75 years Incomplete SCI (AIS B or C) Etiology: traumatic Neurological level between C4 and T12 Time since injury > 18 months | Age < 18 or >75 years Etiology: Non-traumatic spinal cord injury Complete SCI (AIS A) Incomplete SCI (AIS D or E) Neurological level above C4 or below T12 Time since injury <18 months Intensive Care Unit stay for a period of ≥2 months More than 3 urinary tract infections in the last year Pneumonia in the 6 months prior to the study or severe respiratory failure History of head trauma Severe psychiatric disorder History of heart disease, diabetes or hypertension Concomitant neurological diseases Regular use of substances of abuse Severe kidney and/or liver failure. Impossibility to be included in an intensive rehabilitation program Pregnancy or breast-feeding History of malignancy Impossibility to obtain informed consent |
| Characteristics | Mean | GH Group | C I | Placebo Group | C I | p |
|---|---|---|---|---|---|---|
| No of subjects | 23 | 23 | ||||
| Sex (Male) | z | 0786 | ||||
| Male | 82.88 | |||||
| Female | 17.12 | |||||
| Age (y) | 35.00 | 0946 | ||||
| AIS grade | Only baseline | |||||
| A | Not include | NA | ||||
| B | 55.48 | NA | ||||
| C | 44.42 | NA | ||||
| D | Not include | NA | ||||
| IGF-1 | ||||||
| Baseline | 195.83 | 28.50 | 199.66 | 27.48 | 05567 | |
| Motor score | ||||||
| Baseline | 44.44 | 6.99 | 36.38 | 6.95 | 0119 |
| AccN | Protein Name | Abb | Functions | N | Zq Placebo Group | Zq GH Group | Power |
|---|---|---|---|---|---|---|---|
| P08603 | Complement factor H | CFAH | Complement activation/Regulation of complement cascade | 66 | 2.30 | 0.45 | 0.999 |
| P00747 | Plasminogen | PLMN | Blood coagulation/Catalytic activity/Extracellular matrix disassembly | 39 | 2.19 | 0.33 | 0.999 |
| P02760 | Protein AMBP | AMBP | Metabolic process/Catalytic activity/Negative regulation of immune response | 13 | 2.33 | −0.01 | 1.000 |
| P11226 | Mannose-binding protein C | MBL2 | Acute-phase response/Complement activation/Innate immunity | 9 | −0.56 | −3.30 | 0.999 |
| P00739 | Haptoglobin-related protein | HPTR | Acute inflammatory response/ Positive regulation of cell death/Metabolic process | 9 | −2.58 | 6.27 | 0.999 |
| Q01459 | Di-N-acetylchitobiase | DIAC | Glycosidase/Hydrolase | 3 | −0.57 | −1.92 | 0.999 |
| P80188 | Neutrophil gelatinase-associated lipocalin | NGAL | Innate immune response/Transport/Protease binding | 2 | 1.67 | −1.34 | 1.000 |
| Q9Y3D7 | Mitochondrial import inner membrane translocase subunit TIM16 | TIM16 | Ossification/Negative regulation of ATPase activity/Transport | 2 | −1.92 | −0.31 | 0.995 |
| D6RF35 | Vitamin D-binding protein | D6RF35 | Transport/Actin-binding | 2 | −1.58 | 1.19 | 1.000 |
| Q8TD57 | Dynein heavy chain 3, axonemal | DYH3 | Cellular process/ATP binding/Microtubule-based movement | 1 | −1.39 | 3.73 | 1.000 |
| Q9BXX3 | Ankyrin repeat domain-containing protein 30A | AN30A | Regulation of transcription/DNA binding | 1 | −0.72 | 1.89 | 1.000 |
| P12757 | Ski-like protein | SKIL | Cell division and differentiation/Response to cytokine and growth factor/Positive regulation of axonogenesis | 1 | −0.72 | 1.75 | 0.999 |
| Q7Z6K5 | Arpin | ARPIN | Negative regulation of actin nucleation/Negative regulation of cell migration | 1 | −1.65 | 1.31 | 1.000 |
| Q6P0Q8 | Microtubule-associated serine/threonine-protein kinase 2 | MAST2 | Cytoskeleton organization/Intracellular signal transduction/Regulation of interleukin-12 biosynthetic process | 1 | 2.40 | −0.94 | 1.000 |
| P78347 | General transcription factor II-I | GTF2I | Negative regulation of angiogenesis/Transcription by RNA polymerase II/Signal transduction | 1 | −0.63 | −2.52 | 1.000 |
| P82987 | ADAMTS-like protein 3 | ATL3 | Peptidase activity/Cellular process/Proteolysis | 1 | 0.53 | −10.78 | 0.950 |
| Q9NVT9 | Armadillo repeat-containing protein 1 | ARMC1 | Metal ion transport/Biological regulation/Metabolic process | 1 | 1.63 | −4.12 | 0.996 |
| Q8N138 | ORM1-like protein 3 | ORML3 | Cellular sphingolipid homeostasis/Ceramide metabolic process/Neutrophil degranulation | 1 | 0.19 | −2.17 | 1.000 |
| Q96HL8 | SH3 domain-containing YSC84-like protein 1 | SH3YL1 | Phosphatidylinositol biosynthetic process/Regulation of ruffle assembly | 1 | 2.94 | −0.01 | 0.999 |
| P05109 | Protein S100-A8 | S10A8 | Regulation of inflammatory processes and immune response/Positive regulation of cell growth/Astrocyte development/Wound healing | 1 | 2.41 | −0.70 | 0.999 |
| H0YGM0 | Caseinolytic peptidase B protein homolog | H0YGM0 | Regulatory ATPase/Cellular response to heat/Protein binding | 1 | −0.31 | −1.84 | 0.999 |
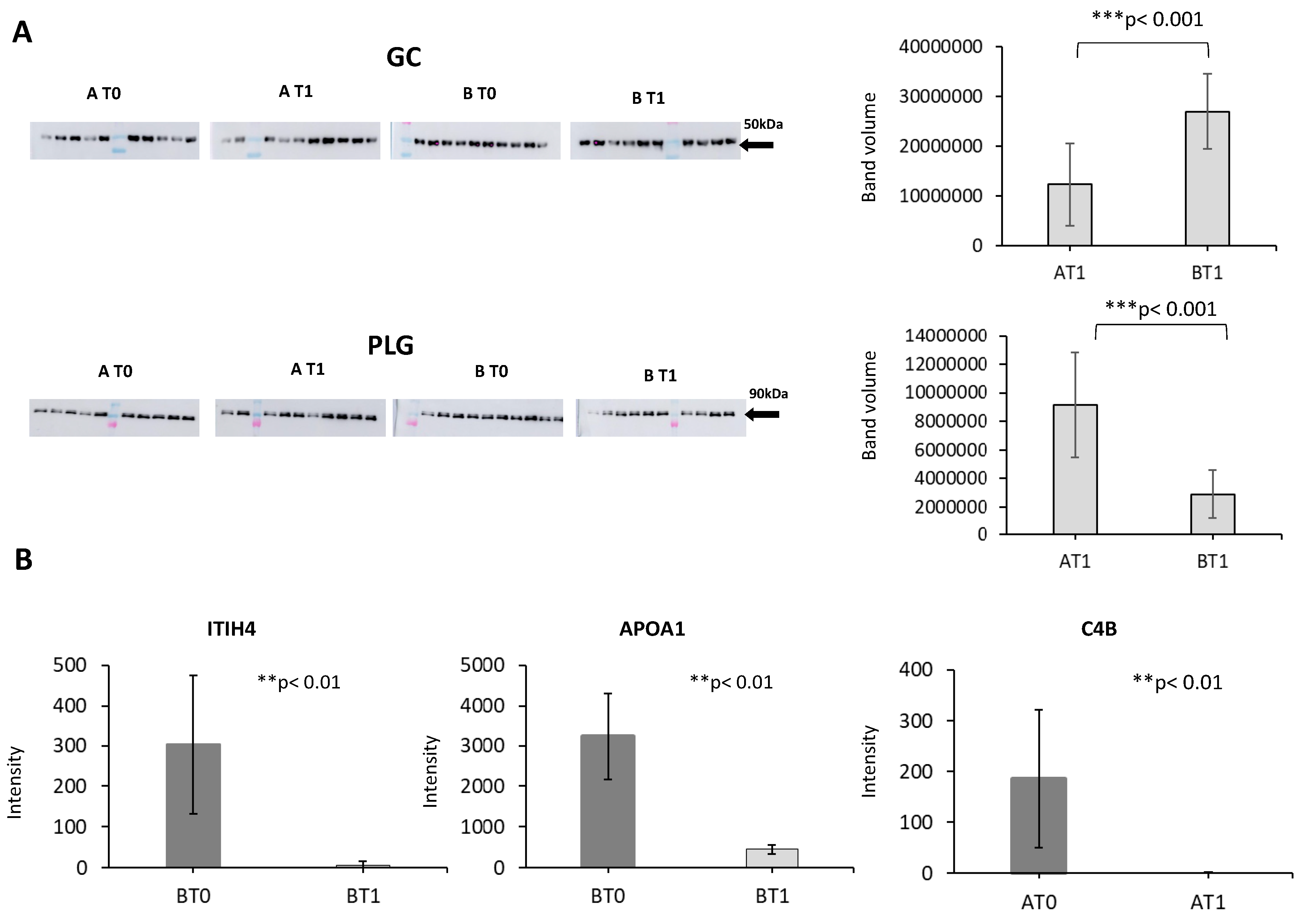
| AccN | Protein Name | Functions | Somatropin t0 vs. t1 | Placebo t0 vs. t1 | |||
|---|---|---|---|---|---|---|---|
| AvR | p-Value | AvR | p-Value | ||||
| 1 | Q14624 | Inter-alpha-trypsin inhibitor heavy chain H4 | Serine-type endopeptidase inhibitor activity | 1.55 | 7.70 × 10−3 | ||
| 2 | Q14624 | Inter-alpha-trypsin inhibitor heavy chain H4 | Serine-type endopeptidase inhibitor activity | −3.82 | 0.0106 | ||
| 3 | P0C0L5 | Complement C4-B | Inflammatory response/Complement activation | −1.24 | 0.0107 | ||
| 4 | P02647 | Apolipoprotein AI | Cholesterol transport | 1.65 | 0.0293 | ||
| 5 | P02647 | Apolipoprotein AI | Cholesterol transport | −1.66 | 0.0474 | ||
| 6 | P00450 | Ceruloplasmin | Oxidoreductase | −1.15 | 0.0139 | ||
| 7 | P00450 | Ceruloplasmin | Oxidoreductase | −1.5 | 0.0175 | ||
| 8 | P36955 | Pigment epithelium-derived factor | Serine-type endopeptidase inhibitor activity | −1.19 | 0.0472 | ||
| 9 | P02649 | Apolipoprotein E | Cholesterol metabolism | −1.11 | 0.0181 | ||
| 10 | P02790 | Hemopexin | Metalloprotease | −3.13 | 0.0332 | ||
| 11 | P04217 | Alpha-1B-glycoprotein | Neutrophil and platelet degranulation | −1.27 | 0.00545 | ||
| AccN | Function | Abb | Peptide Sequence | Peptide Means | Protein Means | p-Value | |
|---|---|---|---|---|---|---|---|
| Bt0 vs. Bt1 | At0 vs. At1 | ||||||
| Q14624 | Protease inhibitor | ITIH4 | ANTVQEATFQMELPK | 595.12 ± 12.81 | 302.71 ± 6.66 | <0.001 | NA |
| IPKPEASFSPR | 10.30 ± 0.51 | ||||||
| P0C0L5 | Complement activation; Inflammatory response | CO4B | VLSLAQEQVGGSPEK | 31.62 ± 3.03 | 186.82 ± 1.65 | NA | <0.001 |
| AEMADQAAAWLTR | 342.03 ± 0.27 | ||||||
| P02647 | Cholesterol transport | APOA1 | DYVSQFEGSALGK | 3234.20 ± 456.42 | 3234.20 ± 456.42 | <0.001 | NA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martin-Rojas, T.; Sastre-Oliva, T.; Esclarín-Ruz, A.; Gil-Dones, F.; Mourino-Alvarez, L.; Corbacho-Alonso, N.; Moreno-Luna, R.; Hernandez-Fernandez, G.; Lopez, J.A.; Oliviero, A.; et al. Effects of Growth Hormone Treatment and Rehabilitation in Incomplete Chronic Traumatic Spinal Cord Injury: Insight from Proteome Analysis. J. Pers. Med. 2020, 10, 183. https://doi.org/10.3390/jpm10040183
Martin-Rojas T, Sastre-Oliva T, Esclarín-Ruz A, Gil-Dones F, Mourino-Alvarez L, Corbacho-Alonso N, Moreno-Luna R, Hernandez-Fernandez G, Lopez JA, Oliviero A, et al. Effects of Growth Hormone Treatment and Rehabilitation in Incomplete Chronic Traumatic Spinal Cord Injury: Insight from Proteome Analysis. Journal of Personalized Medicine. 2020; 10(4):183. https://doi.org/10.3390/jpm10040183
Chicago/Turabian StyleMartin-Rojas, Tatiana, Tamara Sastre-Oliva, Ana Esclarín-Ruz, Felix Gil-Dones, Laura Mourino-Alvarez, Nerea Corbacho-Alonso, Rafael Moreno-Luna, German Hernandez-Fernandez, Juan Antonio Lopez, Antonio Oliviero, and et al. 2020. "Effects of Growth Hormone Treatment and Rehabilitation in Incomplete Chronic Traumatic Spinal Cord Injury: Insight from Proteome Analysis" Journal of Personalized Medicine 10, no. 4: 183. https://doi.org/10.3390/jpm10040183
APA StyleMartin-Rojas, T., Sastre-Oliva, T., Esclarín-Ruz, A., Gil-Dones, F., Mourino-Alvarez, L., Corbacho-Alonso, N., Moreno-Luna, R., Hernandez-Fernandez, G., Lopez, J. A., Oliviero, A., & Barderas, M. G. (2020). Effects of Growth Hormone Treatment and Rehabilitation in Incomplete Chronic Traumatic Spinal Cord Injury: Insight from Proteome Analysis. Journal of Personalized Medicine, 10(4), 183. https://doi.org/10.3390/jpm10040183






