Why Does COVID-19 Affect Patients with Spinal Cord Injury Milder? A Case-Control Study: Results from Two Observational Cohorts
Abstract
1. Introduction
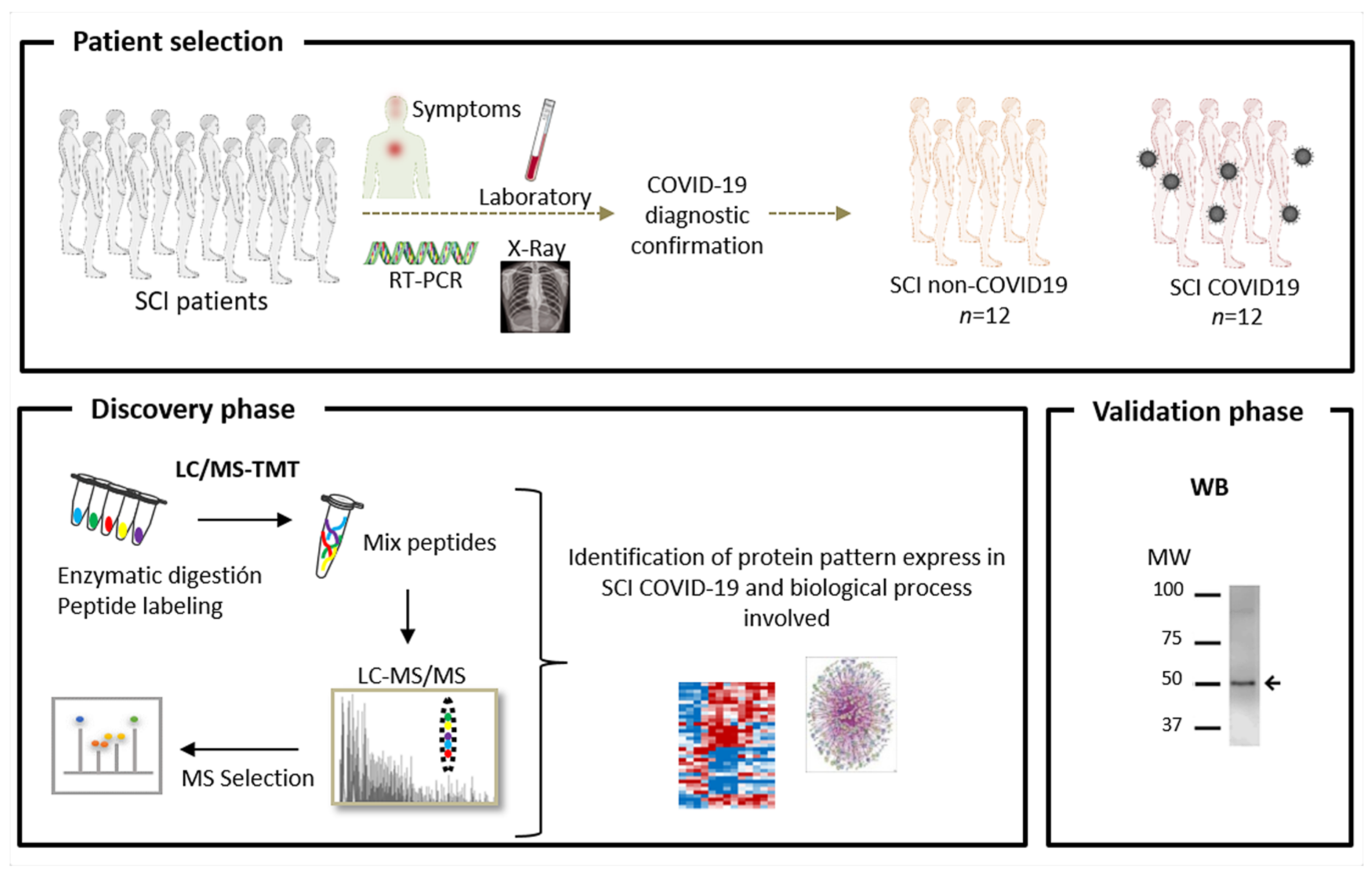
2. Materials and Methods
2.1. Study Population and Sample Preparation
2.2. Quantitative Proteomic Analysis
2.2.1. Protein Digestion and Isobaric Labelling
2.2.2. Protein Identification and Quantitation
2.3. Functional Group Analysis
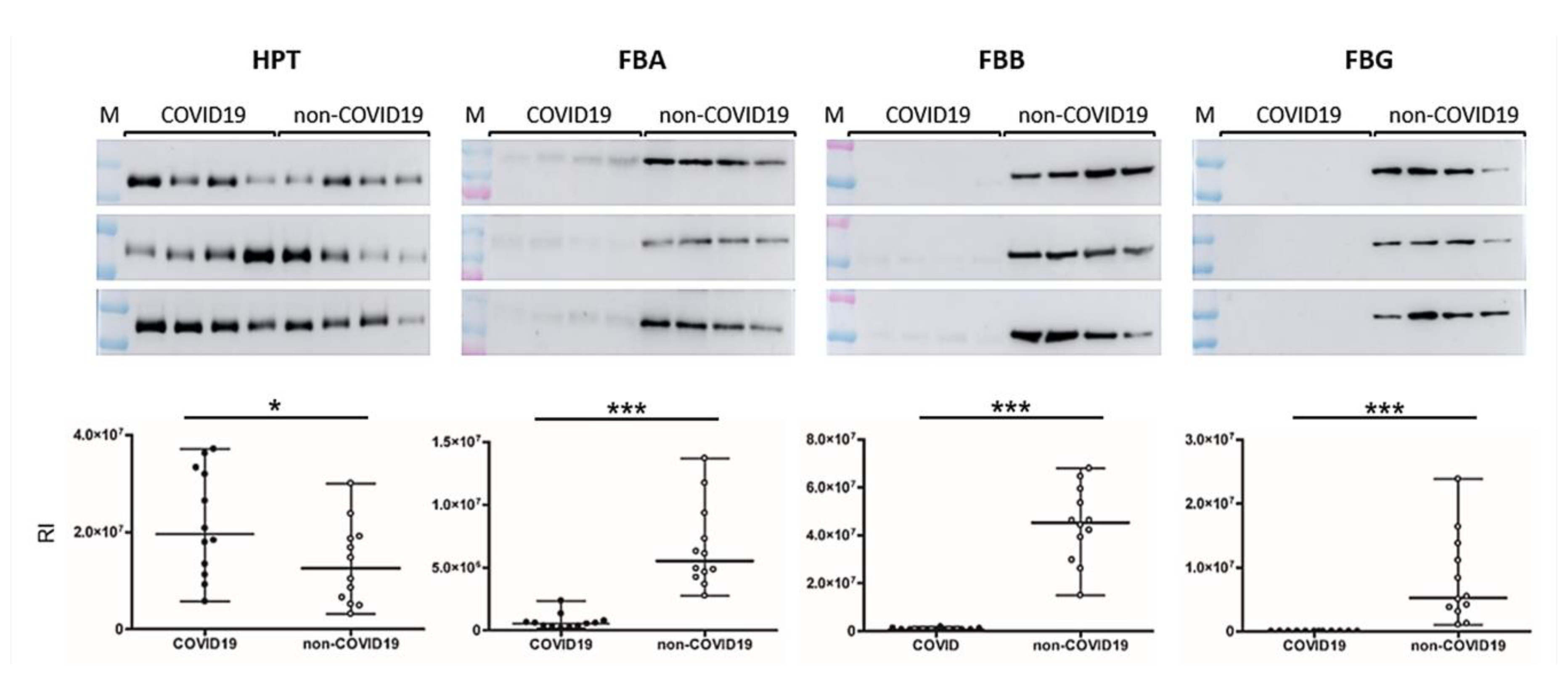
2.4. Western Blotting
2.5. Statistics
2.6. Study Approval
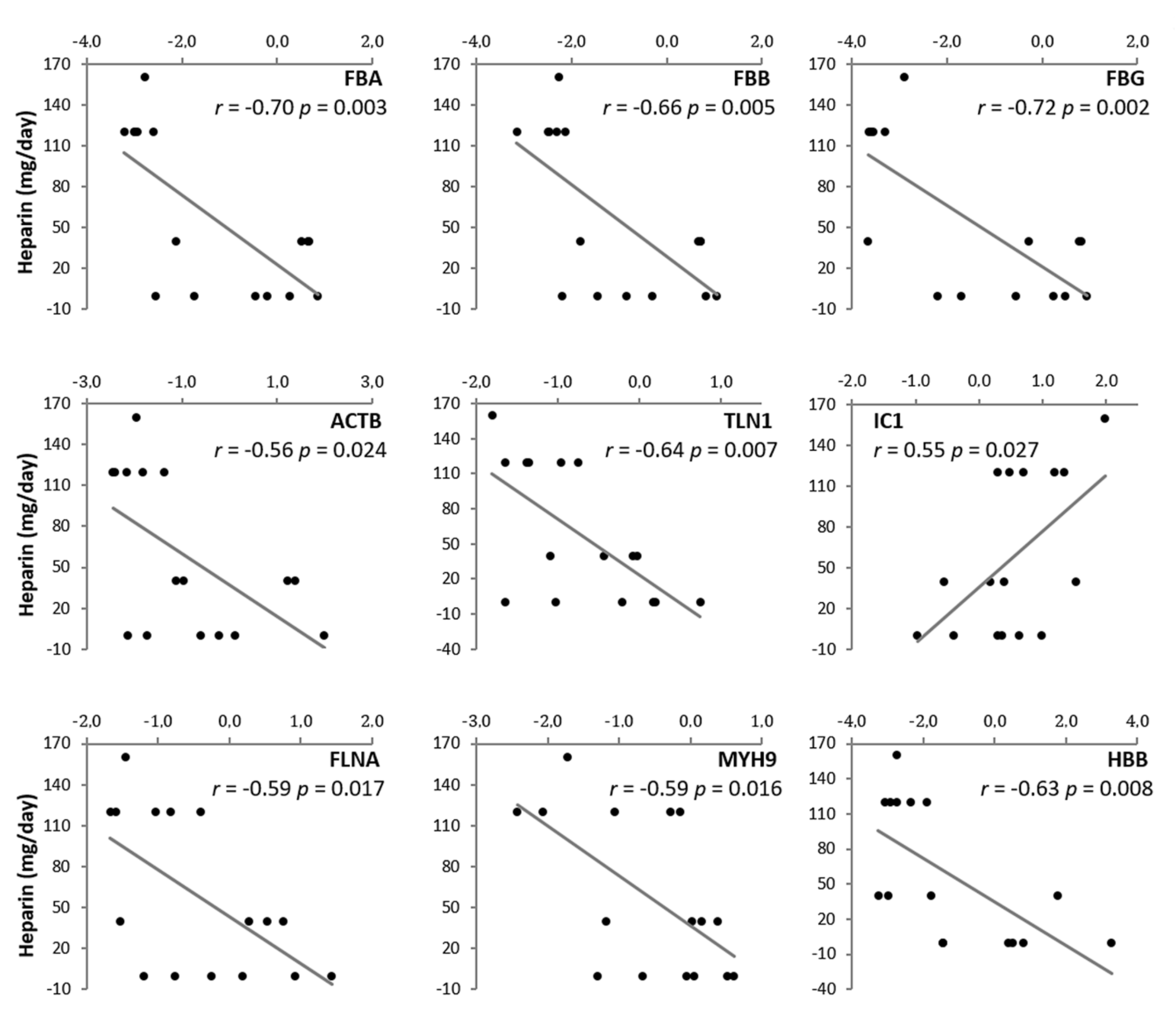
3. Results
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Guan, W.-J.; Ni, Z.-Y.; Hu, Y.; Liang, W.-H.; Ou, C.-Q.; He, J.-X.; Liu, L.; Shan, H.; Lei, C.-L.; Hui, D.S.C.; et al. Clinical characteristics ofcoronavirus disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef] [PubMed]
- Paules, C.I.; Marston, H.D.; Fauci, A.S. Coronavirus infections—More than just the common cold. J. Am. Med. Assoc. 2020, 323, 707–708. [Google Scholar] [CrossRef] [PubMed]
- Hui, D.S.I.; Azhar, E.; Madani, T.A.; Ntoumi, F.; Kock, R.; Dar, O.; Ippolito, G.; McHugh, T.D.; Memish, Z.A.; Drosten, C.; et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—The latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020, 91, 264–266. [Google Scholar] [CrossRef] [PubMed]
- Fauci, A.S.; Lane, H.C.; Redfield, R.R. Covid-19—Navigating the Uncharted. N. Engl. J. Med. 2020, 382, 1268–1269. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Jones, Z.B.; Chen, X.M.; Zhou, L.; So, K.F.; Ren, Y. Multiple organ dysfunction and systemic inflammation after spinal cord injury: A complex relationship. J. Neuroinflamm. 2016, 13, 260. [Google Scholar] [CrossRef] [PubMed]
- Center, J.H.C.R. Coronavirus COVID-19 Global Cases by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU). Available online: https://www.arcgis.com/apps/opsdashboard/index.html (accessed on 15 July 2020).
- van Middendorp, J.J.; Goss, B.; Urquhart, S.; Atresh, S.; Williams, R.P.; Schuetz, M. Diagnosis and prognosis of traumatic spinal cord injury. Glob. Spine J. 2011, 1, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Baldan-Martin, M.; Martin-Rojas, T.; Corbacho-Alonso, N.; Lopez, J.A.; Sastre-Oliva, T.; Gil-Dones, F.; Vazquez, J.; Arevalo, J.M.; Mourino-Alvarez, L.; Barderas, M.G. Comprehensive proteomic profiling of pressure ulcers in patients with spinal cord injury identifies a specific protein pattern of pathology. Adv. Wound Care (New Rochelle) 2020, 9, 277–294. [Google Scholar] [CrossRef]
- Galeiras Vázquez, R.; RascadoSedes, P.; MoureloFariña, M.; MontotoMarqués, A.; Ferreiro Velasco, M.E. Respiratory management in the patient with spinal cord injury. Biomed. Res. Int. 2013, 2013, 168757. [Google Scholar] [CrossRef]
- Kasinathan, N.; Vanathi, M.B.; Subrahmanyam, V.M.; Rao, J.V. A review on response of immune system in spinal cord injury and therapeutic agents useful in treatment. Curr. Pharm. Biotechnol. 2015, 16, 26–34. [Google Scholar] [CrossRef]
- Riegger, T.; Conrad, S.; Liu, K.; Schluesener, H.J.; Adibzahdeh, M.; Schwab, J.M. Spinal cord injury-induced immune depression syndrome (SCI-IDS). Eur. J. Neurosci. 2007, 25, 1743–1747. [Google Scholar] [CrossRef]
- Arevalo-Martin, A.; Grassner, L.; Garcia-Ovejero, D.; Paniagua-Torija, B.; Barroso-Garcia, G.; Arandilla, A.G.; Mach, O.; Turrero, A.; Vargas, E.; Alcobendas, M.; et al. Elevated autoantibodies in subacute human spinal cord injury are naturally occurring antibodies. Front. Immunol. 2018, 9, 2365. [Google Scholar] [CrossRef]
- Kopp, M.A.; Watzlawick, R.; Martus, P.; Failli, V.; Finkenstaedt, F.W.; Chen, Y.; DeVivo, M.J.; Dirnagl, U.; Schwab, J.M. Long-term functional outcome in patients with acquired infections after acute spinal cord injury. Neurology 2017, 88, 892–900. [Google Scholar] [CrossRef]
- Betz, R.; Biering-Sørensen, F.; Burns, S.P.; Donovan, W.; Graves, D.E.; Guest, J.; Jones, L.; Kirshblum, S.; Krassioukov, A.; Mulcahey, M.J.; et al. The 2019 revision of the International Standards for Neurological Classification of Spinal Cord Injury (ISNCSCI)—What’s new? Spinal Cord. 2019, 57, 815–817. [Google Scholar] [CrossRef]
- Baldan-Martin, M.; Lopez, J.A.; Corbacho-Alonso, N.; Martinez, P.J.; Rodriguez-Sanchez, E.; Mourino-Alvarez, L.; Sastre-Oliva, T.; Martin-Rojas, T.; Rincon, R.; Calvo, E.; et al. Potential role of new molecular plasma signatures on cardiovascular risk stratification in asymptomatic individuals. Sci. Rep. 2018, 8, 4802. [Google Scholar] [CrossRef]
- Martinez-Bartolome, S.; Navarro, P.; Martin-Maroto, F.; Lopez-Ferrer, D.; Ramos-Fernandez, A.; Villar, M.; Garcia-Ruiz, J.P.; Vazquez, J. Properties of average score distributions of SEQUEST. Mol. Cell. Proteom. 2008, 7, 1135–1145. [Google Scholar] [CrossRef]
- Navarro, P.; Vazquez, J. A refined method to calculate false discovery rates for peptide identification using decoy databases. J. Proteome. Res. 2009, 8, 1792–1796. [Google Scholar] [CrossRef] [PubMed]
- García-Marqués, F.; Trevisan-Herraz, M.; Martínez-Martínez, S.; Camafeita, E.; Jorge, I.; Lopez, J.A.; Méndez-Barbero, N.; Méndez-Ferrer, S.; Del Pozo, M.A.; Ibáñez, B.; et al. A novel systems-biology algorithm for the analysis of coordinated protein responses using quantitative proteomics. Mol. Cell. Proteom. 2016, 15, 1740–1760. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Korupolu, R.; Stampas, A.; Gibbons, C.; Hernandez Jimenez, I.; Skelton, F.; Verduzco-Gutierrez, M. COVID-19: Screening and triage challenges in people with disability due to Spinal Cord Injury. Spinal Cord Ser. Cases 2020, 6, 35. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Cola, M.; Jiménez-Velasco, I.; Gutiérrez-Henares, F.; López-Dolado, E.; Gambarrutta-Malfatti, C.; Vargas-Baquero, E.; Gil-Agudo, Á. Clinical features of coronavirus disease 2019 (COVID-19) in a cohort of patients with disability due to spinal cord injury. Spinal Cord Ser. Cases 2020, 6, 39. [Google Scholar] [CrossRef]
- Dicks, M.A.; Clements, N.D.; Gibbons, C.R.; Verduzco-Gutierrez, M.; Trbovich, M. Atypical presentation of Covid-19 in persons with spinal cord injury. Spinal Cord Ser. Cases 2020, 6, 38. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in different types of clinical specimens. J. Am. Med. Assoc. 2020, 323, 1843–1844. [Google Scholar] [CrossRef] [PubMed]
- Budnik, I.; Shenkman, B.; Savion, N. Synergistic effect of signaling from receptors of soluble platelet agonists and outside-in signaling in formation of a stable fibrinogen-integrin αIIbβ3-actin cytoskeleton complex. Thromb. Res. 2015, 135, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Haling, J.R.; Monkley, S.J.; Critchley, D.R.; Petrich, B.G. Talin—Dependent integrin activation is required for fibrin clot retraction by platelets. Blood 2011, 117, 1719–1722. [Google Scholar] [CrossRef] [PubMed]
- Cubedo, J.; Blasco, A.; Padro, T.; Ramaiola, I.; Juan-Babot, O.; Goicolea, J.; Fernández-Díaz, J.A.; Oteo, J.F.; Badimon, L. Molecular signature of coronary stent thrombosis: Oxidative stress and innate immunity cells. Thromb. Haemost. 2017, 117, 1816–1827. [Google Scholar] [CrossRef] [PubMed]
- Falet, H. New insights into the versatile roles of platelet FlnA. Platelets 2013, 24, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Rosa, J.P.; Raslova, H.; Bryckaert, M. FilaminA: Key actor in platelet biology. Blood 2019, 134, 1279–1288. [Google Scholar] [CrossRef]
- Chaudhary, H.; Jindal, A.; Guleria, S.; Sharma, S.; Sachdeva, M.U.S.; Ahluwalia, J. Familial macro thrombocytopenia: Role of genetics where morphology fails. Blood Coagul. Fibrinolysis 2020, 31, 333–334. [Google Scholar] [CrossRef]
- Zetterberg, E.; CarlssonAlle, M.S.; Najm, J.; Greinacher, A. Thrombin generation in two families with MYH9-related platelet disorder. Platelets 2016, 27, 264–267. [Google Scholar] [CrossRef][Green Version]
- Van Nostrand, W.E.; McKay, L.D.; Baker, J.B.; Cunningham, D.D. Functional and structural similarities between protease nexin I and C1 inhibitor. J. Biol. Chem. 1988, 263, 3979–3983. [Google Scholar]
- Mosesson, M.W. Fibrinogen functions and fibrin assembly. Fibrinolysis Proteolysis 2000, 14, 182–186. [Google Scholar] [CrossRef]
- Messner, C.B.; Demichev, V.; Wendisch, D.; Michalick, L.; White, M.; Freiwald, A.; Textoris-Taube, K.; Vernardis, S.I.; Egger, A.S.; Kreidl, M.; et al. Ultra-high-throughput clinical proteomics reveals classifiers of COVID-19 infection. Cell Syst. 2020. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.J.; Tham, P.Y.; Chan, D.Z.; Chou, C.F.; Shen, S.; Fielding, B.C.; Tan, T.H.; Lim, S.G.; Hong, W. The severe acute respiratory syndrome coronavirus 3a protein up-regulates expression of fibrinogen in lung epithelial cells. J. Virol. 2005, 79, 10083–10087. [Google Scholar] [CrossRef] [PubMed]
- Becker, R.C. COVID-19 update: Covid-19-associated coagulopathy. J. Thromb. Thrombolysis 2020, 50, 54–67. [Google Scholar] [CrossRef]
- Cure, E.; Cure, M.C. COVID-19 may predispose to thrombosis by affecting both vascular endothelium and platelets. Clin. Appl. Thromb. Hemost. 2020, 26, 1076029620933945. [Google Scholar] [CrossRef]
- Zvi, B.; Levy, A.P. Haptoglobin phenotypes, which one is better and when? Clin. Lab. 2006, 52, 29–35. [Google Scholar]
- Wan, J.; Sun, W.; Li, X.; Ying, W.; Dai, J.; Kuai, X.; Wei, H.; Gao, X.; Zhu, Y.; Jiang, Y.; et al. Inflammation inhibitors were remarkably up-regulated in plasma of severe acute respiratory syndrome patients at progressive phase. Proteomics 2006, 6, 2886–2894. [Google Scholar] [CrossRef]
- Sengupta, M.B.; Basu, M.; Iswarari, S.; Mukhopadhyay, K.K.; Sardar, K.P.; Acharyya, B.; Mohanty, P.K.; Mukhopadhyay, D. CSF proteomics of secondary phase spinal cord injury in human subjects: Perturbed molecular pathways post injury. PLoS ONE 2014, 9, e110885. [Google Scholar] [CrossRef]
- Lim, S.K.; Ferraro, B.; Moore, K.; Halliwell, B. Role of haptoglobin in free hemoglobin metabolism. Redox Rep. Commun. Free Radic. Res. 2001, 6, 219–227. [Google Scholar] [CrossRef]
- Tseng, C.F.; Lin, C.C.; Huang, H.Y.; Liu, H.C.; Mao, S.J. Antioxidant role of human haptoglobin. Proteomics 2004, 4, 2221–2228. [Google Scholar] [CrossRef]
- Yuan, X.; Huang, W.; Ye, B.; Chen, C.; Huang, R.; Wu, F.; Wei, Q.; Zhang, W.; Hu, J. Changes of hematological and immunological parameters in COVID-19 patients. Int. J. Hematol. 2020, 1–7. [Google Scholar] [CrossRef]
- Lippi, G.; Mattiuzzi, C. Hemoglobin value may be decreased in patients with severe coronavirus disease 2019. Hematol. Transfus. Cell Ther. 2020, 42, 116–117. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, J.; Arnold, K.; Pawlinski, R.; Key, N.S. Using heparin molecules to manage COVID-2019. Res. Pract. Thromb. Haemost. 2020, 4, 518–523. [Google Scholar] [CrossRef] [PubMed]
- Thachil, J. The versatile heparin in COVID-19. J. Thromb. Haemost. 2020, 18, 1020–1022. [Google Scholar] [CrossRef]
- Kim, S.Y.; Jin, W.; Sood, A.; Montgomery, D.W.; Grant, O.C.; Fuster, M.M.; Fu, L.; Dordick, J.S.; Woods, R.J.; Zhang, F.; et al. Characterization of heparin and severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) spike glycoprotein binding interactions. Antiviral. Res. 2020, 181, 104873. [Google Scholar] [CrossRef]
- Kwon, P.S.; Oh, H.; Kwon, S.-J.; Jin, W.; Zhang, F.; Fraser, K.; Hong, J.J.; Linhardt, R.J.; Dordick, J.S. Sulfated polysaccharides effectively inhibit SARS-CoV-2 in vitro. Cell Discov. 2020, 6, 50. [Google Scholar] [CrossRef]
- Gozzo, L.; Viale, P.; Longo, L.; Vitale, D.C.; Drago, F. The potential role of heparin in patients with COVID-19: Beyond the anticoagulant effect. A review. Front. Pharmacol. 2020, 11. [Google Scholar] [CrossRef]

| Non COVID | COVID | p-Value | ||
|---|---|---|---|---|
| Age (years) | 52.7 ± 17.1 | 61.1 ± 14.4 | 0.22 | |
| Gender (%M/F) | 72/28 | 73/27 | 0.86 | |
| ASIA Impairment scale (%) | A | 54 | 45 | NA |
| B | 8 | 9 | NA | |
| C | 23 | 9 | NA | |
| D | 15 | 18 | NA | |
| Heparin (%) | 46 | 82 | 0.15 | |
| Heparin dose (mg/day) | 40 | 98 | 0.03 | |
| Term | t-test | FC | Proteins | |
|---|---|---|---|---|
| Cluster 1 Enrichment Score: 1.69 | platelet aggregation | 0.000 | 7.3 | fibrinogen alpha chain (FGA); fibrinogen beta chain (FGB); fibrinogen gamma chain (FGG); plasma protease C1 inhibitor (IC1); filamin A (FLNA); myosin heavy chain 9 (MYH9); actin beta (ACTB); hemoglobin subunit beta (HBB); talin 1 (TLN1) |
| positive regulation of substrate adhesion-dependent cell spreading | 0.004 | 10 | ||
| cellular protein complex assembly | 0.016 | 13 | ||
| positive regulation of vasoconstriction | 0.016 | 13 | ||
| positive regulation of peptide hormone secretion | 0.016 | 13 | ||
| negative regulation of endothelial cell apoptotic process | 0.016 | 13 | ||
| negative regulation of extrinsic apoptotic signaling pathway via death domain receptors | 0.016 | 13 | ||
| Platelet activation | 0.018 | 4.1 | ||
| protein binding, bridging | 0.025 | 10 | ||
| positive regulation of heterotypic cell-cell adhesion | 0.030 | 9.6 | ||
| positive regulation of exocytosis | 0.030 | 9.6 | ||
| positive regulation of protein secretion | 0.030 | 9.6 | ||
| response to calcium ion | 0.030 | 9.6 | ||
| protein polymerization | 0.030 | 9.6 | ||
| blood coagulation, fibrin clot formation | 0.030 | 9.6 | ||
| plasminogen activation | 0.048 | 7.7 | ||
| cell adhesion molecule binding | 0.058 | 7 | ||
| fibrinolysis | 0.080 | 3.7 | ||
| fibrinogen complex | 0.087 | 5.7 | ||
| platelet alpha granule | 0.087 | 5.7 | ||
| positive regulation of ERK1 and ERK2 cascade | 0.091 | 5.5 | ||
| Cluster 2 Enrichment Score: 1.55 | haptoglobin-hemoglobin complex | 0.015 | 13 | Haptoglobin (HP); hemoglobin subunit beta (HBB); hemoglobin subunit alpha 1 (HBA1) |
| positive regulation of cell death | 0.016 | 13 | ||
| response to hydrogen peroxide | 0.030 | 9.6 | ||
| cellular oxidant detoxification | 0.091 | 5.5 | ||
| Cluster 3 Enrichment Score: 1.38 | oxygen transporter activity | 0.025 | 10 | Hemoglobin subunit delta (HBD); hemoglobin subunit beta (HBB); hemoglobin subunit alpha 1 (HBA1) |
| hemoglobin complex | 0.028 | 9.9 | ||
| oxygen transport | 0.030 | 9.6 | ||
| oxygen binding | 0.041 | 8.4 | ||
| iron ion binding | 0.078 | 6 | ||
| heme binding | 0.078 | 6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calvo, E.; Corbacho-Alonso, N.; Sastre-Oliva, T.; Nuñez, E.; Baena-Galan, P.; Hernandez-Fernandez, G.; Rodriguez-Cola, M.; Jimenez-Velasco, I.; Corrales, F.J.; Gambarrutta-Malfati, C.; et al. Why Does COVID-19 Affect Patients with Spinal Cord Injury Milder? A Case-Control Study: Results from Two Observational Cohorts. J. Pers. Med. 2020, 10, 182. https://doi.org/10.3390/jpm10040182
Calvo E, Corbacho-Alonso N, Sastre-Oliva T, Nuñez E, Baena-Galan P, Hernandez-Fernandez G, Rodriguez-Cola M, Jimenez-Velasco I, Corrales FJ, Gambarrutta-Malfati C, et al. Why Does COVID-19 Affect Patients with Spinal Cord Injury Milder? A Case-Control Study: Results from Two Observational Cohorts. Journal of Personalized Medicine. 2020; 10(4):182. https://doi.org/10.3390/jpm10040182
Chicago/Turabian StyleCalvo, Enrique, Nerea Corbacho-Alonso, Tamara Sastre-Oliva, Estefania Nuñez, Patricia Baena-Galan, German Hernandez-Fernandez, Miguel Rodriguez-Cola, Irena Jimenez-Velasco, Fernando J. Corrales, Claudia Gambarrutta-Malfati, and et al. 2020. "Why Does COVID-19 Affect Patients with Spinal Cord Injury Milder? A Case-Control Study: Results from Two Observational Cohorts" Journal of Personalized Medicine 10, no. 4: 182. https://doi.org/10.3390/jpm10040182
APA StyleCalvo, E., Corbacho-Alonso, N., Sastre-Oliva, T., Nuñez, E., Baena-Galan, P., Hernandez-Fernandez, G., Rodriguez-Cola, M., Jimenez-Velasco, I., Corrales, F. J., Gambarrutta-Malfati, C., Gutierrez-Henares, F., Lopez-Dolado, E., Gil-Agudo, A., Vazquez, J., Mourino-Alvarez, L., & Barderas, M. G. (2020). Why Does COVID-19 Affect Patients with Spinal Cord Injury Milder? A Case-Control Study: Results from Two Observational Cohorts. Journal of Personalized Medicine, 10(4), 182. https://doi.org/10.3390/jpm10040182






