From Medical Imaging to Radiomics: Role of Data Science for Advancing Precision Health
Abstract
1. Introduction
2. Imaging Biomarkers in Personalized Medicine
3. Imaging Biobanking
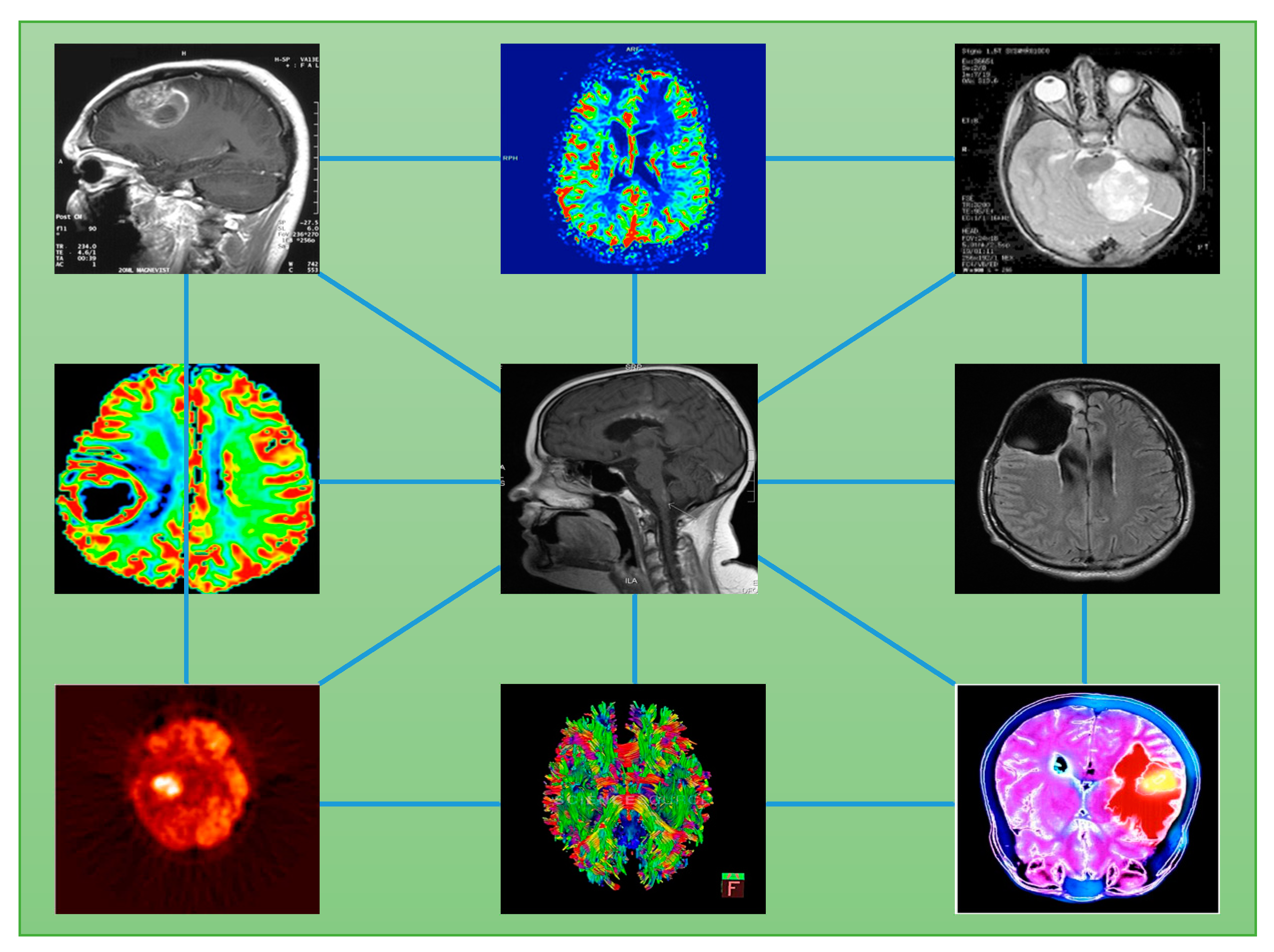
4. Image-to-Data Science Driven Research
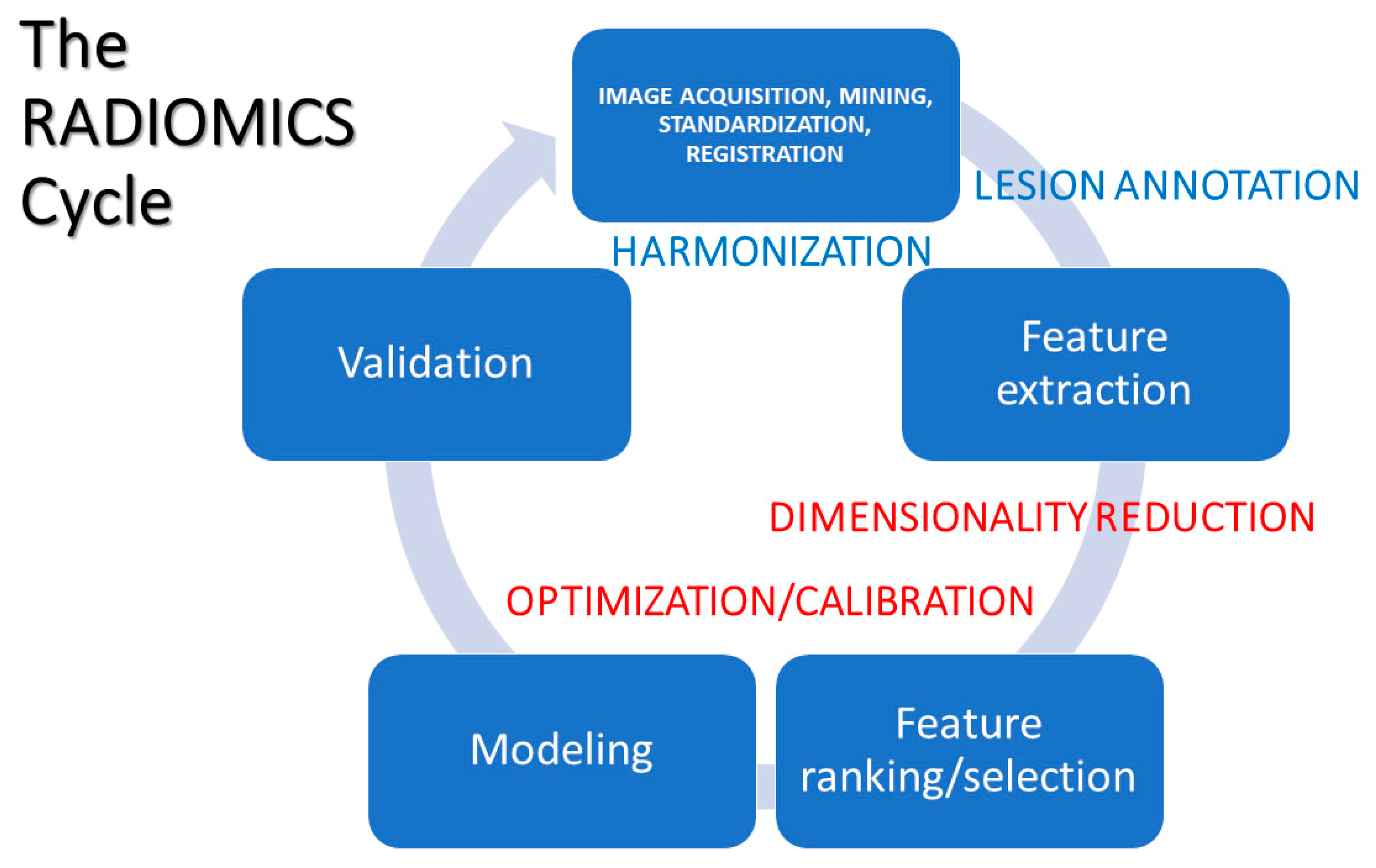
5. Radiomic Profiles: Construction and Interpretation
5.1. Data Acquisition, Mining, and Assimilation
5.2. Data Pre-Processing
5.3. Feature Extraction
5.4. Feature Ranking and/or Selection
5.5. Modeling
5.6. Validation
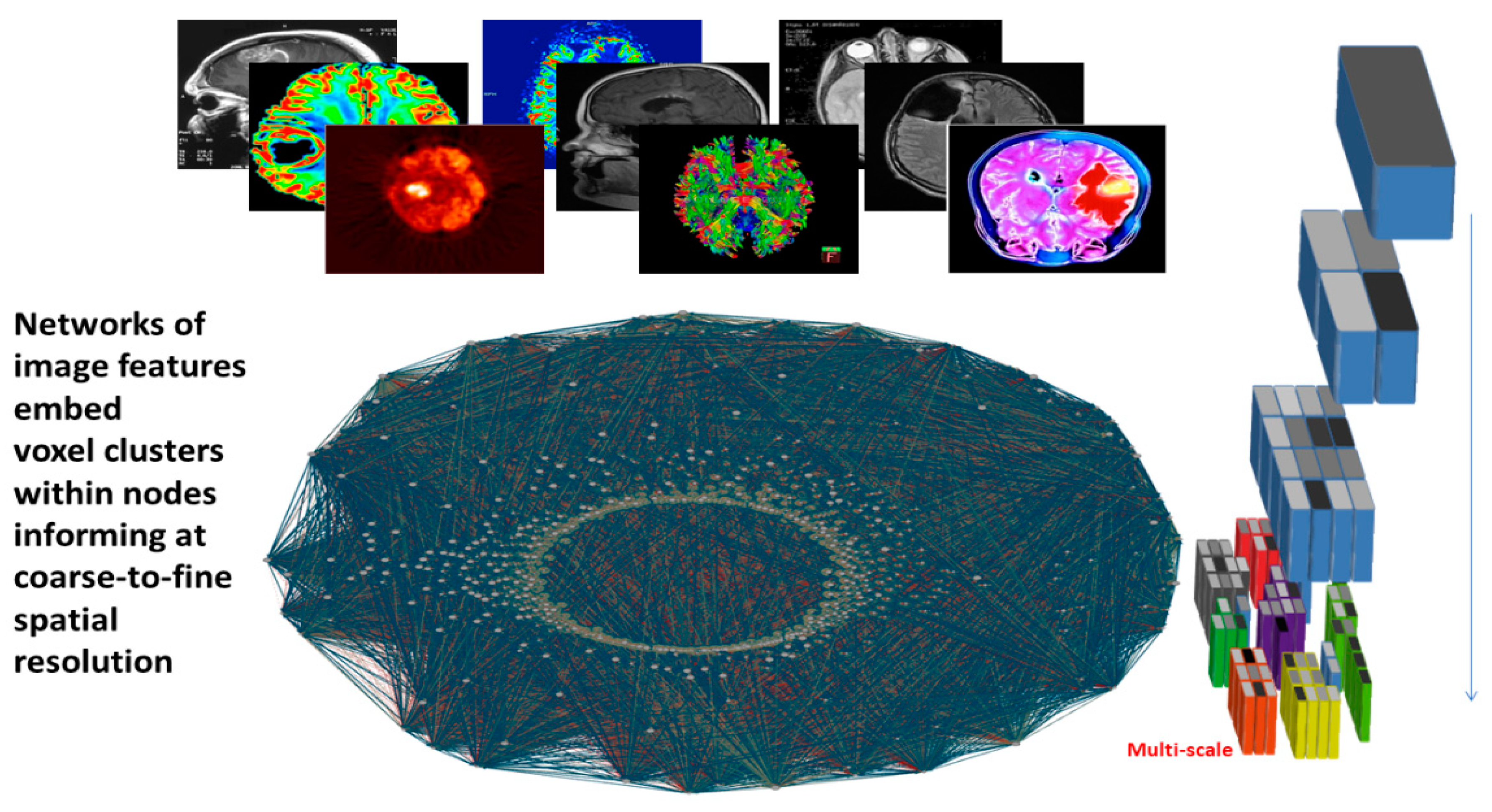
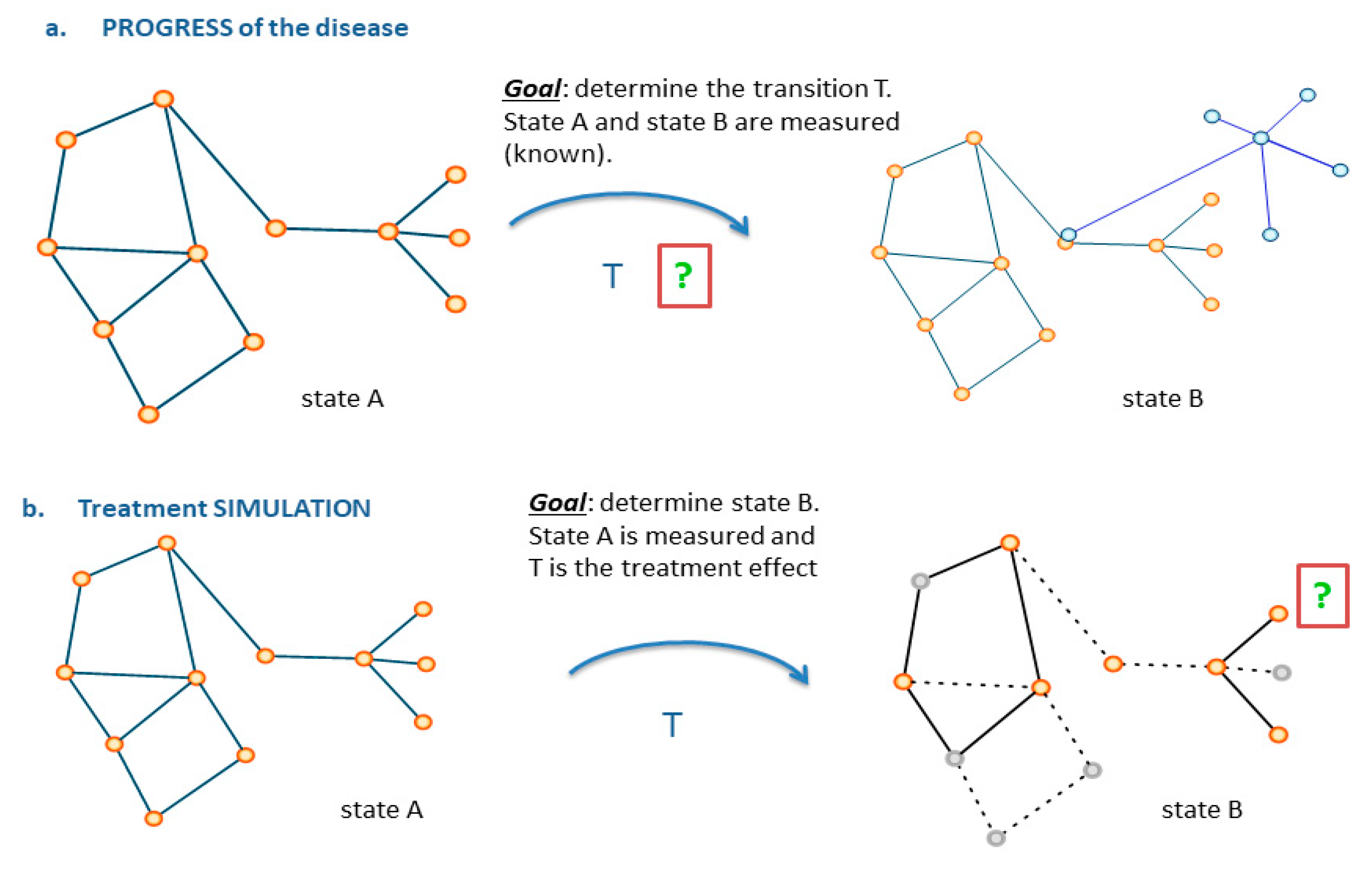
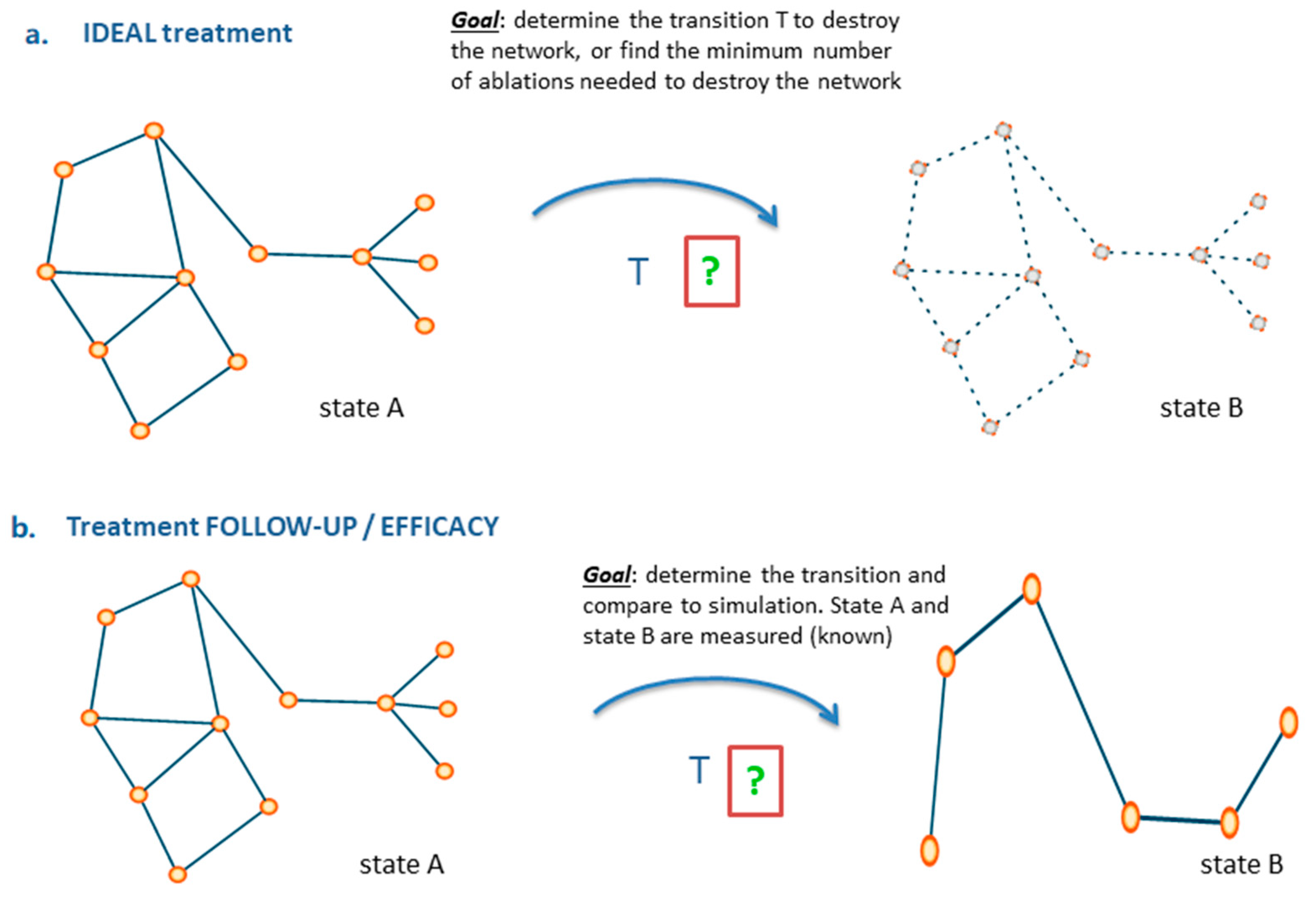
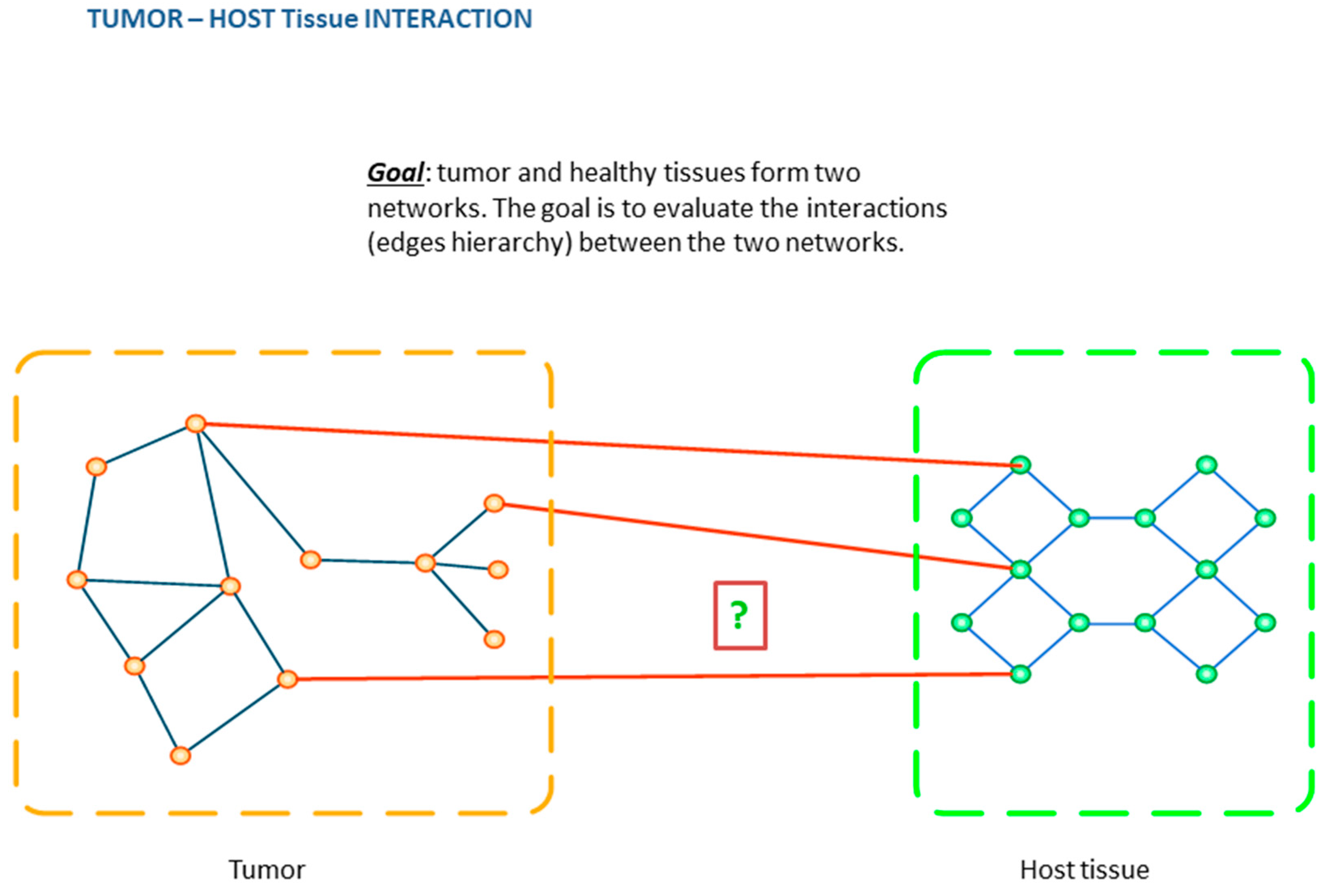
6. From Images to Networks
7. Impact Domains
8. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- O’Connor, J.P. Rethinking the role of clinical imaging. eLife 2017, 6, e30563. [Google Scholar] [CrossRef] [PubMed]
- Ranjbar, S.; Velgos, S.N.; Dueck, A.C.; Geda, Y.E.; Mitchell, J.R.; Alzheimer’s Disease Neuroimaging Initiative. Brain MR Radiomics to Differentiate Cognitive Disorders. J. Neuropsychiatry Clin. Neurosci. 2019, 31, 210–219. [Google Scholar] [CrossRef]
- Li, Y.; Jiang, J.; Lu, J.; Jiang, J.; Zhang, H.; Zuo, C. Radiomics: A novel feature extraction method for brain neuron degeneration disease using 18F-FDG PET imaging and its implementation for Alzheimer’s disease and mild cognitive impairment. Ther. Adv. Neurol. Disord. 2019, 12. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Song, Q.; Wang, M.; Pang, P.; Liao, Z.; Jiang, H.; Shen, D.; Ding, Z. Hippocampus Radiomic Biomarkers for the Diagnosis of Amnestic Mild Cognitive Impairment: A Machine Learning Method. Front. Aging Neurosci. 2019, 11, 323. [Google Scholar] [CrossRef] [PubMed]
- Mo, J.; Liu, Z.; Sun, K.; Ma, Y.; Hu, W.; Zhang, C.; Wang, Y.; Wang, X.; Liu, C.; Zhao, B.; et al. Automated detection of hippocampal sclerosis using clinically empirical and radiomics features. Epilepsia 2019, 60, 2519–2529. [Google Scholar] [CrossRef]
- Fan, Y.; Feng, M.; Wang, R. Application of Radiomics in Central Nervous System Diseases: A Systematic literature review. Clin. Neurol. Neurosurg. 2019, 187, 105565. [Google Scholar] [CrossRef]
- Currie, G.; Iqbal, B.; Kiat, H. Intelligent Imaging: Radiomics and Artificial Neural Networks in Heart Failure. J. Med. Imaging Radiat. Sci. 2019, 50, 571–574. [Google Scholar] [CrossRef]
- Lo, C.-M.; Hung, P.-H.; Hsieh, K.L.-C. Computer-Aided Detection of Hyperacute Stroke Based on Relative Radiomic Patterns in Computed Tomography. Appl. Sci. (2076–3417) 2019, 9, 1668. [Google Scholar] [CrossRef]
- Wu, Q.; Yao, K.; Liu, Z.; Li, L.; Zhao, X.; Wang, S.; Shang, H.; Lin, Y.; Wen, Z.; Tian, J.; et al. Radiomics analysis of placenta on T2WI facilitates prediction of postpartum haemorrhage: A multicentre study. EBioMedicine 2019, 50, 355–365. [Google Scholar] [CrossRef]
- Rossi, F.; Bignotti, B.; Bianchi, L.; Picasso, R.; Martinoli, C.; Tagliafico, A.S. Radiomics of peripheral nerves MRI in mild carpal and cubital tunnel syndrome. Radiol. Med. 2019. [Google Scholar] [CrossRef]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images are more than Pictures, they are Data. Radiology 2015, 278, 2. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, S.; Botta, F.; Raimondi, S.; Origgi, D.; Fanciullo, C.; Morganti, A.G.; Belllomi, M. Radiomics: The facts and the challenges of image analysis. Eur. Radiol. Exp. 2018, 2, 36. [Google Scholar] [CrossRef] [PubMed]
- Nie, K.; Al-Hallaq, H.; Li, X.A.; Benedict, S.H.; Sohn, J.W.; Moran, J.M.; Fan, Y.; Huang, M.; Knopp, M.V.; Michalski, J.M.; et al. NCTN Assessment on Current Applications of Radiomics in Oncology. Int. J. Radiat. Oncol. Biol. Phys. 2019, 104, 302–315. [Google Scholar] [CrossRef] [PubMed]
- Limkin, E.J.; Sun, R.; Dercle, L.; Zacharaki, E.I.; Robert, C.; Reuzé, S.; Schernberg, A.; Paragios, N.; Deutsch, E.; Ferté, C. Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann. Oncol. 2017, 28, 1191–1206. [Google Scholar] [CrossRef]
- Chen, C.-M.; Chou, Y.-H.; Tagawa, N.; Do, Y. Computer Aided Detection and Diagnosis in Medical Imaging. Comput. Math. Meth. Med. 2013, 790608. [Google Scholar] [CrossRef]
- Van Ginneken, B.; Schaefer-Prokop, C.M.; Prokop, M. Computer-Aided Diagnosis: How to move from the Laboratory to the Clinic. Radiology 2011, 261, 3. [Google Scholar] [CrossRef]
- Hu, Z.; Tang, J.; Wang, Z.; Zhang, K.; Zhang, L.; Sun, Q. Deep learning for image-based cancer detection and diagnosis - A survey. Patt. Recogn. 2018, 83, 134–149. [Google Scholar] [CrossRef]
- Grossmann, P.; Stringfield, O.; El-Hachem, N.; Bui, M.M.; Velazquez, E.M.; Parmar, C.; Leijenaar, R.T.H.; Haibe-Kains, B.; Lambin, P.; Gillies, R.J.; et al. Defining the biological basis of radiomic phenotypes in lung cancer. eLife 2017, 6, e23421. [Google Scholar] [CrossRef]
- Wu, J.; Tha, K.K.; Xing, L.; Li, R. Radiomics and radiogenomics for precision radiotherapy. J. Radiat. Res. 2018, 59, i25–i31. [Google Scholar] [CrossRef]
- Colin, T. Integration of radiomic, genomic and clinical data to support decision making for lung cancer. J. Clin. Oncol. 2019, 37, e14607. [Google Scholar] [CrossRef]
- Sanduleanu, S.; Woodruff, H.C.; de Jong, E.E.C.; van Timmeren, J.E.; Jochems, A.; Dubois, L.; Lambin, P. Tracking tumor biology with radiomics: A systematic review utilizing a radiomics quality score. Radioth. Oncol. 2018, 127, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Dregely, I.; Prezzi, D.; Kelly-Morland, C.; Roccia, E.; Neji, R.; Goh, V. Imaging biomarkers in oncology: Basics and application to MRI. J. Magn. Reson. Imaging. 2018, 48, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Sala, E.; Mema, E.; Himoto, Y.; Veeraraghavan, H.; Brenton, J.D.; Snyder, A.; Weigelt, B.; Vargas, H.A. Unravelling tumour heterogeneity using next-generation imaging: Radiomics, radiogenomics, and habitat imaging. Clin. Radiol. 2017, 72, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Alic, L.; Niessen, W.J.; Veenland, J.F. Quantification of Heterogeneity as a Biomarker in Tumor Imaging: A Systematic Review. PLoS ONE 2014, 9, e110300. [Google Scholar] [CrossRef]
- Daye, D.; Kamran, S.C.; Tabari, A.; Michalski, M.; Clark, J.W.; Ryan, D.P.; Allen, J.N.; Murphy, J.E.; Hong, T.S.; Gee, M.S. Quantitative MR imaging biomarkers of tumor heterogeneity predict prognosis in metastatic colorectal lesions. J. Clin. Oncol. 2017, 35, e15121. [Google Scholar] [CrossRef]
- O’Connor, J.P.B.; Rose, C.J.; Waterton, J.C.; Carano, R.A.D.; Parker, G.J.M.; Jackson, A. Imaging Intratumor Heterogeneity: Role in Therapy Response, Resistance, and Clinical Outcome. Clin. Cancer Res. 2015, 21, 249–257. [Google Scholar] [CrossRef]
- Lu, L.; Ehmke, R.C.; Schwartz, L.H.; Zhao, B. Assessing Agreement between Radiomic Features Computed for Multiple CT Imaging Settings. PLoS ONE 2016, 11, e0166550. [Google Scholar] [CrossRef]
- Orlhac, F.; Boughdad, S.; Philippe, C.; Stalla-Bourdillon, H.; Nioche, C.; Champion, L.; Soussan, M.; Frouin, F.; Frouin, V.; Buvat, I. A Postreconstruction Harmonization Method for Multicenter Radiomic Studies in PET. J. Nucl. Med. 2018, 59, 1321–1328. [Google Scholar] [CrossRef]
- Neri, E.; Regge, D. Imaging biobanks in oncology: European perspective. Future Oncol. 2017, 13, 433–441. [Google Scholar] [CrossRef]
- Coppola, L.; Cianflone, A.; Grimaldi, A.M.; Incoronato, M.; Bevilacqua, P.; Messina, F.; Baselice, S.; Soricelli, A.; Mirabelli, P.; Salvatore, M. Biobanking in health care: Evolution and future directions. J. Transl. Med. 2019, 17, 172. [Google Scholar] [CrossRef]
- Gatidis, S.; Heber, S.D.; Storz, C.; Bamberg, F. Population-based imaging biobanks as source of big data. Radiol. Med. 2017, 122, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Lieu, C.H.; Corcoran, R.B.; Overman, M.J. Integrating Biomarkers and Targeted Therapy Into Colorectal Cancer Management. Am. Soc. Clin. Oncol. Educ. Book 2019, 39, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Yue, T.; Jia, X.; Petrosino, J.; Sun, L.; Fan, Z.; Fine, J.; Davis, R.; Galster, S.; Kuret, J.; Scharre, D.W.; et al. Computational integration of nanoscale physical biomarkers and cognitive assessments for Alzheimer’s disease diagnosis and prognosis. Sci. Adv. 2017, 3, E1700669. [Google Scholar] [CrossRef] [PubMed]
- Murphy, K.; Murphy, B.T.; Boyce, S.; Flynn, L.; Gilgunn, S.; O’Rourke, C.J.; Rooney, C.; Stöckmann, H.; Walsh, A.L.; Finn, S.; et al. Integrating biomarkers across omic platforms: An approach to improve stratification of patients with indolent and aggressive prostate cancer. Mol. Oncol. 2018, 12, 9. [Google Scholar] [CrossRef] [PubMed]
- Zanfardino, M.; Franzese, M.; Pane, K.; Cavaliere, C.; Monti, S.; Esposito, G.; Salvatore, M.; Aiello, M. Bringing radiomics into a multi-omics framework for a comprehensive genotype–phenotype characterization of oncological diseases. J. Transl. Med. 2019, 17. [Google Scholar] [CrossRef] [PubMed]
- Seyakula, R.K.; Singh, V.; Verma, N.K.; Kumar, C. Transfer learning for molecular cancer classification using deep neural networks. Trans. Comp. Biol. Bioinf. 2018. [Google Scholar] [CrossRef]
- Kensen, A.; Harrison, P.J.; Spjuth, O. Transfer learning with deep convolutional neural networks for classifying cellular morphological changes. SLAS Discov. 2019, 24, 466–475. [Google Scholar] [CrossRef]
- Zhen, X.; Chen, J.; Zhong, Z.; Hrycushko, B.; Zhou, L.; Jiang, S.; Albuquerque, K.; Gu, X. Deep convolutional neural network with transfer learning for rectum toxicity prediction in cervical cancer radiotherapy: A feasibility study. Phys. Med. Biol. 2017, 62, 8246–8263. [Google Scholar] [CrossRef]
- Dhruba, S.R.; Rahman, R.; Matlock, K.; Ghosh, S.; Pal, R. Application of transfer learning for cancer drug sensitivity prediction. BMC Bioinf. 2018, 19, 497. [Google Scholar] [CrossRef]
- Xu, Y.; Hosny, A.; Zaleznik, R.; Parmar, C.; Coreller, T.; Franco, I.; Mak, R.H.; Aerts, H.J.W.L. Deep learning predicts lung cancer treatment response from serial medical imaging. Clin. Can. Res. 2019, 25, 11. [Google Scholar] [CrossRef]
- De Matos, J.; Britto, A.; De, S.; Oliveira, L.E.S.; Koerich, A.L. Double transfer learning for breast cancer histopathological image classification. arXiv 2019, arXiv:1904.07834. [Google Scholar]
- Yang, Y.; Yan, L.-F.; Zhang, X.; Han, Y.; Nan, H.-Y.; Hu, Y.-C.; Hu, B.; Yah, S.-L.; Zhang, J.; Cheng, D.-L.; et al. Glioma grading on conventional MR images: A deep learning study with transfer learning. Front. Neurosc. 2018, 12, 804. [Google Scholar] [CrossRef]
- Hosny, A.; Parmar, C.; Coreller, T.P.; Grossmann, P.; Zeleznik, R.; Kumar, A.; Bussink, J.; Gillies, R.J.; Mak, R.H.; Aerts, H.J.W.L. Deep learning for lung cancer prognostication: A retrospective multi-cohort radiomics study. PLoS Med. 2018, 15, e1002711. [Google Scholar] [CrossRef]
- Merkwirth, C.; Wichard, J.; Ogorzałek, M.J. Ensemble Modeling for Bio-medical Applications. In Modelling Dynamics in Processes and Systems; Mitkowski, W., Kacprzyk, J., Eds.; Studies in Computational Intelligence; Springer: Berlin/Heidelberg, Germany, 2009; Volume 180. [Google Scholar] [CrossRef]
- Lu, H.; Arshad, M.; Thornton, A.; Avesani, G.; Cunnea, P.; Curry, E.; Kanavati, F.; Liang, J.; Nixon, K.; Williams, S.; et al. A mathematical-descriptor of tumor-mesoscopic-structure from computed-tomography images annotates prognostic- and molecular-phenotypes of epithelial ovarian cancer. Nat. Comm. 2019, 10, 764. [Google Scholar] [CrossRef] [PubMed]
- Schindlbeck, K.A.; Eidelberg, D. Network imaging biomarkers: Insights and clinical applications in Parkinson’s disease. Lancet Neurol. 2018, 17, 629–640. [Google Scholar] [CrossRef]
- Yan, K.; Wang, X.; Lu, L.; Summers, R.M. DeepLesion: Automated mining of large-scale lesion annotations and universal lesion detection with deep learning. J. Med. Imag. 2018, 5, 036501. [Google Scholar] [CrossRef]
- Aiello, M.; Cavaliere, C.; D’Albore, A.; Salvatore, M. The Challenges of Diagnostic Imaging in the Era of Big Data. J. Clin. Med. 2019, 8, 316. [Google Scholar] [CrossRef]
- Jaffray, D.A.; Das, S.; Jacobs, P.M.; Jeray, R.; Lambin, P. How advances in Imaging will affect Precision Radiation Oncology. Int. J. Radiat. Oncol. Biol. Phys. 2018, 101, 292–298. [Google Scholar] [CrossRef]
- Schafer, N.; Gielen, G.H.; Rauschenbach, L.; Kebir, S.; Till, A.; Reinartz, R.; Simon, M.; Niehusmann, P.; Kleinschnitz, C.; Herrlinger, U.; et al. Longitudinal heterogeneity in glioblastoma: Moving targets in recurrent versus primary tumors. J. Trans. Med. 2019, 17, 96. [Google Scholar] [CrossRef]
- Fisher, R.; Pusztai, L.; Swanton, C. Cancer heterogeneity: Implications for targeted therapeutics. Br. J Cancer 2013, 108, 479–485. [Google Scholar] [CrossRef]
- Lin, G.; Keshari, K.R.; Park, J.M. Cancer Metabolism and Tumor Heterogeneity: Imaging Perspectives Using MR Imaging and Spectroscopy. Contrast Media Mol. Imaging. 2017, 2017, 6053879. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Li, H.; Guo, W.; Drukker, K.; Lan, L.; Giger, M.L.; Ji, Y. Deciphering Genomic Underpinnings of Quantitative MRI-based Radiomic Phenotypes of Invasive Breast Carcinoma. Sci. Rep. 2015, 5, 17787. [Google Scholar] [CrossRef] [PubMed]
- Yankeelov, T.E.; Mankoff, D.A.; Schwartz, L.H.; Lieberman, F.S.; Buatti, J.M.; Mountz, J.M.; Erickson, B.J.; Fennessy, F.M.M.; Huang, W.; Kalpathy-Cramer, J.; et al. Quantitative Imaging in Cancer Clinical Trials. Clin. Can. Res. 2016, 22, 2. [Google Scholar] [CrossRef] [PubMed]
- Soldatos, T.G.; Kaduthanam, S.; Jackson, D.B. Precision Oncology - The Quest for Evidence. J Person. Med. 2019, 9, 43. [Google Scholar] [CrossRef]
- Hormuth, D.A., II; Sorace, A.G.; Virostko, J.; Abramson, R.G.; Bhujwalla, Z.M.; Enriquez-Navas, P.; Gillies, R.; Hazle, J.D.; Mason, R.P.; Quarles, C.C.; et al. Translating preclinical MRI methods to clinical oncology. JMRI 2019. [Google Scholar] [CrossRef]
- Park, J.E.; Park, S.Y.; Kim, H.J.; Kim, H.S. Reproducibility and Generalizability in Radiomics Modeling: Possible strategies in Radiologic and Statistical perspectives. KJR 2019, 20, 1124–1137. [Google Scholar] [CrossRef]
- Traverso, A.; Wee, L.; Dekker, A.; Gillies, R. Repeatability and Reproducibility of Radiomic Features: A Systematic Review. Int. J. Radiat. Oncol. Biol. Phys. 2018, 102, 1143–1158. [Google Scholar] [CrossRef]
- Bi, W.L.; Hosny, A.; Schabath, M.B.; Giger, M.L.; Birkbak, N.J.; Mehrtash, A.; Allison, T.; Arnaout, O.; Abbosh, C.; Dunn, I.F.; et al. Artificial intelligence in cancer imaging: Clinical challenges and applications. CA Cancer J. Clin. 2019, 69, 127–157. [Google Scholar] [CrossRef]
- Shaikh, F.; Franc, B.; Allen, E.; Sala, E.; Awan, O.; Hendrata, K.; Halabi, S.; Mohiuddin, S.; Malik, S.; Hadley, D.; et al. Translational Radiomics: Defining the Strategy Pipeline and Considerations for Application-Part 2: From Clinical Implementation to Enterprise. J. Am. Coll. Radiol. 2018, 15 3 Pt B, 543–549. [Google Scholar] [CrossRef]
- Azuaje, F. Artificial Intelligence for Precision Oncology: Beyond patient stratification. Npj Prec. Oncol. 2019, 3, 6. [Google Scholar] [CrossRef]
- Kingma, D.P.; Welling, M. An Introduction to Variational Autoencoders. arXiv 2019, arXiv:1906.02691. [Google Scholar] [CrossRef]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial nets. In Proceedings of the 27th International Conference on Neural Information Processing Systems - 2 (NIPS’14, Montreal, CA), Montreal, QC, Canada, 8–13 December 2014; Ghahramani, Z., Welling, M., Cortes, C., Lawrence, N.D., Weinberger, K.Q., Eds.; MIT Press: Cambridge, MA, USA, 2014; Volume 2, pp. 2672–2680. [Google Scholar]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Arindra, A.; Setio, A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B. A survey of deep learning in medical image analysis. Med Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed]
- Dominietto, M.; Tsinoremas, N.; Capobianco, E. Integrative analysis of cancer imaging readouts by networks. Mol. Oncol. 2015, 9, 1–16. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Capobianco, E.; Dominietto, M. From Medical Imaging to Radiomics: Role of Data Science for Advancing Precision Health. J. Pers. Med. 2020, 10, 15. https://doi.org/10.3390/jpm10010015
Capobianco E, Dominietto M. From Medical Imaging to Radiomics: Role of Data Science for Advancing Precision Health. Journal of Personalized Medicine. 2020; 10(1):15. https://doi.org/10.3390/jpm10010015
Chicago/Turabian StyleCapobianco, Enrico, and Marco Dominietto. 2020. "From Medical Imaging to Radiomics: Role of Data Science for Advancing Precision Health" Journal of Personalized Medicine 10, no. 1: 15. https://doi.org/10.3390/jpm10010015
APA StyleCapobianco, E., & Dominietto, M. (2020). From Medical Imaging to Radiomics: Role of Data Science for Advancing Precision Health. Journal of Personalized Medicine, 10(1), 15. https://doi.org/10.3390/jpm10010015



